3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(6):1082-1088. doi:10.7150/jca.17457 This issue Cite
Research Paper
IRES-Mediated Protein Translation Overcomes Suppression by the p14ARF Tumor Suppressor Protein
1. College of Pharmaceutical Sciences, Jilin University, Changchun, China;
2. Torrey Pines Institute for Molecular Studies, San Diego CA, USA;
3. College of Basic Medical Sciences, Jilin University, Changchun, China;
4. College of Life Science and Bioengineering, Beijing University of Technology, Beijing, China;
5. China-Japan Union Hospital, Jilin University, Changchun, China;
6. MRC Cancer Unit, University of Cambridge, Cambridge, United Kingdom.
Received 2016-9-4; Accepted 2017-3-6; Published 2017-4-9
Abstract
Internal ribosome entry sites (IRES elements) have attracted interest in cancer gene therapy because they can be used in the design of gene transfer vectors that provide bicistronic co-expression of two transgene products under the control of a single promoter. Unlike cellular translation of most mRNAs, a process that requires a post-translational 5' modification of the mRNA known as the cap structure, IRES-mediated translation is independent of the cap structure. The cellular conditions that may intervene to modulate IRES-mediated, cap-independent versus cap-dependent translation, however, remain poorly understood, although they could be critical to the choice of gene transfer vectors. Here we have compared the effects of the p14ARF (Alternate Reading Frame) tumor suppressor, a translational suppressor frequently overexpressed in cancer, on cap-dependent translation versus cap-independent translation from the EMCV viral IRES often used in bicistronic gene transfer vectors. We find that ectopic overexpression of p14ARF suppresses endogenous and ectopic cap-dependent protein translation, consistent with other studies. However, p14ARF has little or no effect on transgene translation initiated within an IRES element. This suggests that transgenes placed downstream of an IRES element will retain efficient translation of their gene products in the presence of high levels of ectopic or endogenous p14ARF, a finding that could be particularly relevant to therapeutic gene therapy strategies for cancer.
Keywords: p14ARF, Internal ribosome entry site (IRES), cap-dependent, protein translation.
Introduction
Initiation of protein translation from an internal ribosome entry site (IRES) is an alternative type of protein translation in which ribosomes are recruited directly to an mRNA by virtue of its secondary structure, independently of the 5' modification of the mRNA known as the cap structure, a modification required for the recruitment of the translation initiation factor protein (eIFs) to most cellular RNAs (see review [1-3]). IRES often eliminates the involvement of eIFs, activated by binding to specific sets of proteins, referred to as IRES trans-acting factors (ITAFs). IRES-mediated translation was first described in poliovirus RNA and encephalomyocarditis virus (EMCV) RNA [4, 5] ,and is now thought to contribute to the translation of certain cellular RNAs as well, particularly under abnormal cellular states triggered by hypoxia or cellular stress [6]. So far, multiple IRES were identified with different mechanisms in translational regulation. They may or may not use eIFs and ITAFs to recruit the ribosome to the message.
IRES-mediated translation has become increasingly attractive as a tool in biotechnology because it provides a means to achieve bicistronic co-expression of transduced genes under the control of a single transcriptional promoter. When an IRES segment is located between two genes in a eukaryotic mRNA molecule, it can drive translation of the downstream protein coding region independently of the 5'-cap structure bound to the message. Nevertheless, the regulation of IRES-mediated translation has not been fully explored and remains poorly understood.
In this study we asked how IRES-mediated translation is affected by the 14ARF (alternate reading frame) tumor suppressor, a translational repressor [7] induced in cells with activated oncogenes [8, 9]. p14ARF could inhibit the polysome formation and protein synthesis by interacting with the nucleolar 60 S preribosomal particle [6]. p14ARF expression is undetectable in most normal cells, but evidence suggests that its levels increase at an early stage of tumorigenesis [10] and elevated p14ARF levels have been observed in certain cancers [11-13]. This increased expression could therefore affect the outcome of some therapeutic gene transfer strategies.
Because of the differences in the mechanisms by which CAP-dependent and IRES-mediated, CAP-independent translation are initiated, it seemed possible that p14ARF could have differential effects on these two modes of translation that could be relevant to use of IRES-containing gene transfer vectors. We report here our finding that messages translated from IRES elements display little or no suppression by p14ARF, suggesting that IRES-mediated translation may be particularly useful for gene therapy applications in certain cancers where p14ARF expression is high.
When multiple tumor suppressor genes are used in cancer gene therapy simultaneously, they sometimes interfere with each other. Our study proposed a model that when two tumor suppressor genes are constructed in one expression cassette containing IRES element, the IRES could protect the downstream gene from the inhibition induced by upstream gene, and this will make a common sense for other genes. Therefore, if we need to employ multiple tumor suppressor genes in gene therapy and to avoid the mutual interference, it is better to put genes in one cassette and introduce IRES between the genes, instead of applying two genes separately.
Materials and methods
Cell culture
All cultures were maintained at 37oC in 10% CO2 in Dulbecco's Modified Eagles Medium supplemented with nonessential amino acids, pyruvate, L-glutamine, gentamicin, and 10% FBS. DLD-1 human colon cancer cells were obtained from the Americian Type Culture Collection. OE33 human esophageal cancer cells originated from the European Collections of Cell cultures and were provided to us by Dr. Rebecca Fitzgerald, Cambridge University, UK. To obtain OE33-GFP and OE33-IRES-GFP, we stably modified OE33 cells with plasmid constructs pLXSN-GFP (for OE33-GFP) and pLXSN-IRES-GFP (for OE33-IRES-GFP), in which GFP message is translated in a cap-dependent or cap-independent manner, respectively. The GFP and IRES (ECMV) sequences were derived from pIRES-GFP (Clontech, Mountainview, CA). pLXSN was obtained from Clontech, Mounainview, CA. GFP-expressing clones were selected in G418 followed by cell sorting. Mouse Embryo Fibroblasts, nullizygous for p53 and mdm2, were generously provided by Dr. Guillermina Lozano, Department of Molecular Genetics, the University of Texas M.D. Anderson Cancer Center, Houston, TX.
Vectors and treatment conditions
The replication-defective adenoviral vectors, AdLuc, encoding firefly luciferase, and Adp53, encoding the human p53 tumor suppressor gene, were purchased from Vector Biolabs, Philadelphia, PA. The replication-defective adenoviral vectors, Adp14, encoding the human p14ARF (Alternate Reading Frame) tumor suppressor gene, and Adp14/p53, encoding both human p14ARF and p53 in a bicistronic configuration, have been described [14]. Cells were treated with adenoviral vectors for 4 hours with 50-100 plaque-forming units (pfu) per cell in culture medium containing 2% serum, as previously described [14]. Human mdm2 siRNA was purchased from Dharmacon (Chicago, Il). For siRNA treatments, 4 x106 adenoviral vector-treated cells were subjected to electroporation (200V) in the presence of 150 pmol siRNA.
Western analysis
Western analyses were carried out using 40 µg cell lysates as previously described [14, 15] using mouse monoclonal anti-human mdm2 SMP14 (Santa Cruz Biotechnology, Santa Cruz, CA).
Metabolic labeling and fluorography
GFP-expressing OE33-GFP and OE33-IRES-GFP cells were metabolically labeled for 1 hour with [35S]-L-methionine (1175 Ci/mmol, MP Biomedicals, Solon, OH), followed by immunoprecipitation of GFP protein, SDS-PAGE and fluorography as previously described [16, 17]. Mouse monoclonal anti-GFP antibody was obtained from Santa Cruz Biotechnology, Santa Cruz, CA).
Polysome fractionation and analysis
Post-mitochondrial cytoplasmic fractions were sedimented on 17 ml 10-50% sucrose gradients following published procedures [18]. 1.5 ml fractions were collected and RNA was extracted and analyzed by semi-quantitative RT-PCR as previously described [14, 15]. p53 primers have been previously described [19]. The RT-PCR products were resolved on a 1.0% agarose gel stained with ethidium bromide, and band intensities were quantitated digitally and represented as a percent of total intensity. Results of each polysome analysis were confirmed by at least one independent repeat experiment.
Results and Discussion
The total level of a given protein subjects to two events, synthesis and degradation. It is known that p14ARF has a specific effect on p53 protein stability by binding with mdm2, the major inhibitor of p53 [20], therefore, our study focused on detection of the rates of p53 synthesis rather than total accumulation. Rates of protein are related to the number of ribosomes per message, with rapidly synthesized proteins being derived from messages that are heavily occupied with ribosomes and that sediment more rapidly. Alternatively, rates of protein synthesis rates are related to the incorporation of [35S]-methionine during a pulse.
We first investigated how overexpressed ectopic p14ARF affects the translation rate of an endogenous gene by examining the distribution of endogenous p53 message in polysomes from p14ARF-transduced DLD-1 human colon cancer cells, following sedimentation of post-mitochondrial cytoplasmic fractions on sucrose density gradients. A typical sedimentation profile of polysomes from DLD-1 cells, monitored by absorbance at 260 nm, is shown in Figure 1a, where those with heavier ribosome occupation appear in the rapidly sedimenting polysome region towards the right of the profile, and 60S ribosomal subunit of 80S initiation complex located in the left. The staining profile of ribosomal RNA is shown underneath it. Figure 1b shows the distribution of endogenous p53 message in polysomes of DLD-1 cells 24 hours post-treatment with 50 plaque-forming units (pfu) per cell of either a control replication-defective adenoviral vector encoding firefly luciferase (AdLuc) or a replication-defective adenoviral vector encoding p14ARF (Adp14). As we have previously reported, expression of p14ARF protein in DLD-1 cells is low to undetectable [14]. Treatment with Adp14 leads to about a 20-fold increase in p14ARF message levels by 24 hours post treatment, and results in readily detectable levels of p14ARF protein on Western blots (data not shown). Suppression of cell viability by Adp14 treatment only becomes evident after 72 hours [14]. We found that in cells treated with AdLuc, endogenous p53 message was broadly distributed across the polysome profile, peaking in fraction 5, indicating efficient recruitment of message to the heavier polysome region and efficient translation. In contrast, in cells treated with Adp14, endogenous p53 message shifted to the left of the profile, peaking in fraction2, a region occupied by monosomes and small polysomes associated with slowly translated messages (Figure 1b). Figure 1c shows the correspondent polysome profiles from the cells treated with Adluc and Adp14. Thus, endogenous p53 message is less efficiently translated in cells overexpressing ectopic p14ARF, consistent with previous reports that p14ARF can inhibit translation initiation [7].
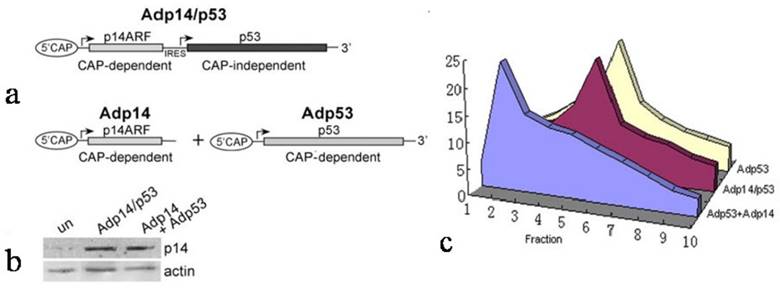
However, a different result was observed when we examined ectopic expression of p53 following treatment of DLD-1 cells with a bicistronic adenoviral vector encoding both p14ARF and p53 in a single transcript (Adp14/p53), where the p53 open reading frame is situated downstream of an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES). We found that in DLD-1 cells treated either with Adp14/p53 or with the Adp53 single gene vector (Figure 2a), the ectopic p53 message was broadly distributed across the polysome profile, both peaking in fraction 5. Relative polysome profiles for the vector treatment were shown in Figure 2b. Treatment conditions, as previously shown, were such that p53 message levels increased by some 20-fold by 24 hours post-treatment of DLD-1 cells with either Adp53 or Adp14/p53 [14]. Message levels of p14ARF also increased by some 20-fold in Adp14/p53-treated cells [14].
We ruled out a possible role of the mdm2 protein in maintaining p53 translation rates in Adp14/p53-treated cells. The mdm2 gene is induced by p53 [21], and mdm2 protein can in turn stimulate p53 translation [22]. Induced mdm2 could therefore potentially offset the possible negative effects of over-expressed ectopic p14ARF. However, we found that siRNA treatment to downregulate mdm2 protein levels in Adp14/p53-treated cells by 48 hours post-treatment (Figure 2c, lane 4 versus lane 3) had no effect on the 48 hour polysome distribution of ectopic p53 (Figure 2a). This indicates that the translation of p53 from the bicistronic construct is intrinsically less susceptible p14ARF-mediated translational suppression than is translation of the endogenous p53 transcript, independently of mdm2.
Effect of p14ARF on endogenous p53 translation in DLD-1 human colon cancer cells. (a) Typical O.D.260 profile of fractionated polysomes and ribosomal RNA staining profile from 2 x 107 DLD-1 cells. Arrow indicates direction of sedimentation. (b) Distribution of endogenous p53 message in fractions of polysomes, prepared from 2 x 107 DLD-1 cells 24 hours post-treatment (50 pfu/per cell, 4 hours) with AdLuc control vector or Adp14. Fractions (1.5 ml) were collected from the top of the gradient, RNA was purified, and subjected to semi-quantitative RT-PCR analysis of p53. (c) Relative polysome profiles from the cells treated with Adluc and Adp14.
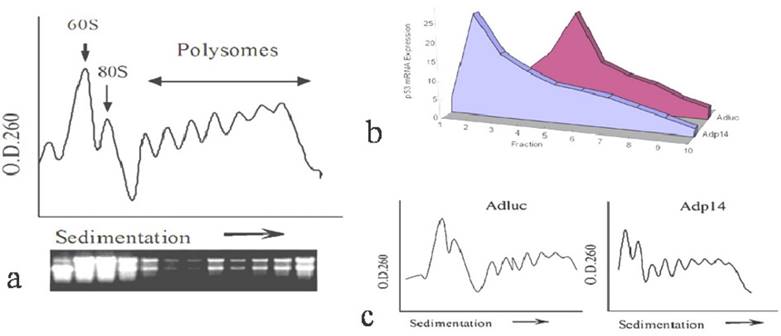
Effect of p14ARF on ectopic p53 translation in DLD-1 human colon cancer cells. Distribution of ectopic p53 message in various polysome fractions from 4 x 106 DLD-1 cells, 48 hours post-treatment (50 pfu/cell, 4 hours) with Adp53, or Adp14/p53 bicistronic vector , or with Adp14/p53 bicistronic vector followed by electroporation with human mdm2 siRNA. (b) Relative polysome profiles for all the adenovirus treatment, seperately. (c) Western analysis of human mdm2 protein expression in DLD-1 cells, 48 hours after treatment with the indicated vectors +/- mdm2 siRNA.
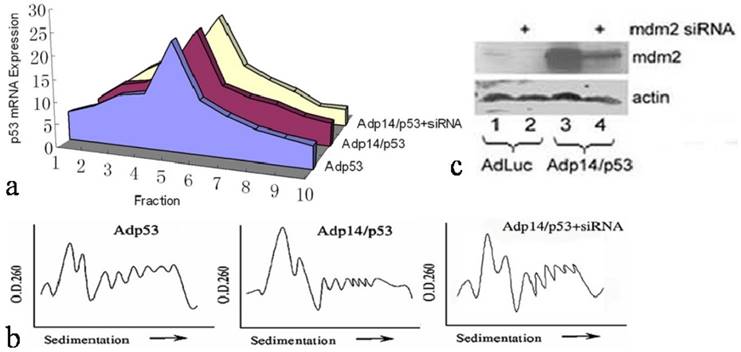
We therefore considered the possibility that the failure of p14ARF to suppress ectopic p53 translation in Adp14/p53-treated cells derived from the characteristics of the bicistronic vector itself, and in particular, the presence of the IRES in the bicistronic vector and the fact that translation from an IRES element was cap-independent, while translation of the endogenous message was cap-dependent. To test this, we used p14ARF and p53 single gene and bicistronic adenoviral vectors that would allow us to compare these two modes of translation for ectopic p53 under similar conditions (see diagram in Figure 3a). Translation of ectopic p53 message was monitored in p53/mdm2 doubly nullizygous mouse embryo fibroblasts (DN-MEFs), which lack endogenous p53 message. Polysome analyses were carried out 24 hours post-treatment with (a) Adp53 alone, where p53 translation is cap-dependent, or (b) with a combination of Adp53 plus Adp14, or (c) with the Adp14/p53 bicistronic vector, where p53 translation is cap-independent. Treatment conditions for Adp14 or Adp14/p53 were 100 pfu/cell and produced the same level of p14ARF protein (Figure 3b). We found that ectopic p53 message in Adp53-treated DN-MEFs was efficiently translated, as indicated by the broad distribution of message across the polysome profile, peaking in fraction 5 (Figure 3c). The results are consistent with the results with Adp53-treated DLD-1 cells (Figure 2a). We found that ectopic p53 message was less efficiently translated in Adp53 + Adp14-treated DN-MEFs, as indicated by the shift in the polysome distribution of ectopic p53 message toward the left of the gradient, peaking in fraction 2 (Figure 3c). Thus, cap-dependent translation of ectopic p53 is less efficient in the presence of overexpressed ectopic p14ARF. In contrast, we found that ectopic p53 message in Adp14/p53-treated DN-MEFs, was efficiently translated, despite the overexpression of p14ARF, as indicated by the polysome profile peaking in fraction 5 (Figure 3c). The profile is similar to that observed with Adp53-treated DN-MEFs and Adp14/p53-treated DLD-1 cells. Thus, IRES-mediated, cap-independent translation of ectopic p53 is efficient, even in the presence of overexpressed ectopic p14ARF.
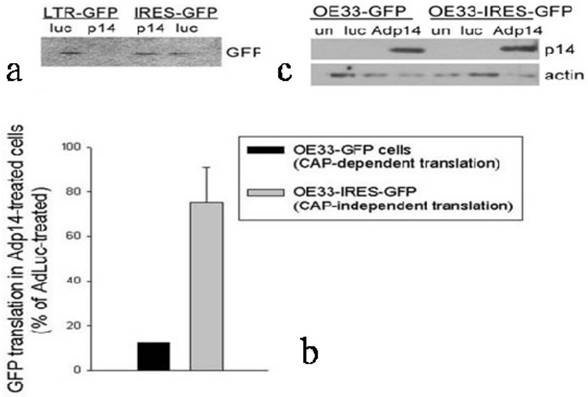
To confirm and extend these observations to another cell line and to another gene, and to use a different method to evaluate protein synthesis rates, we prepared stably-modified variants of the OE33 human esophageal adenocarcinoma cell line, in which the Green Fluorescent Protein (GFP) gene was placed either downstream of a viral LTR promoter (OE33-GFP, providing cap-dependent translation of GFP) or an IRES (OE33-IRES-GFP, providing cap-independent translation of GFP). The two modified clones were treated with Adp14 (100 pfu/cell) and 2 days later the rate of GFP protein synthesis was monitored by pulsing cells with [35S]-methionine, followed by SDS/PAGE and fluorography of immunoprecipitated GFP. A typical fluorograph of [35S]-methionine incorporation into GFP is shown in Figure 4a, where each lane represents 105 cell equivalents. A bar graph representing an average of two experiments is shown in Figure 4b. A Western blot confirming that p14ARF was induced to similar levels 24 hours post-infection of both cell lines is shown in Figure 4c. The results show that cap-dependent translation of GFP in OE33-GFP cells modified with an LTR-GFP construct is suppressed on the average by some 90% in Adp14 treated cells compared to AdLuc-treated cells (Figure 4b, black bar), while cap-independent translation of GFP in OE33-IRES-GFP cells modified with an IRES-GFP construct shows only weak to no suppression by Adp14 (Figure 4b, gray bar). The ability of IRES-mediated protein translation to escape suppression by p14ARF appears therefore to be a general phenomenon with potential relevance to any gene transfer protocol involving p14ARF.
Effect of p14ARF on cap-dependent versus cap-independent translation of p53 in p53-null, mdm2-null MEFs (DN-MEFs). (a) Diagram illustrating the two modes of p53 translation achieved with either the bicistronic adenoviral vector, Adp14/p53 (top), where p53 is translated from the IRES in a cap-independent manner, or the p53 single gene vector, Adp53 (bottom), where p53 message is expressed from the CMV promoter and translated in a cap-dependent manner. (b) Western blot of p14ARF protein levels in untreated DN-MEFs (control), and in DN-MEFs, 24 hours post-treatment with 100 pfu/cell of Adp14 or Adp14/p53. Each lane represents 40 µg cellular lysates. (c) Distribution of ectopic p53 message in various fractions of polysomes from 4 x 106 p53/mdm2 doubly nullizygous mouse embryo fibroblasts (DN-MEFs), 24 hours after treatment with Adp53 alone (100 pfu/cell), or with a combination of Adp53 plus Adp14 single gene vectors (100 pfu/cell each), or with Adp14/p53 bicistronic vector (100 pfu/cell).
Effect of p14ARF on cap-dependent versus cap-independent GFP translation in OE33 human esophageal adenocarcinoma cells. (a) GFP protein synthesized in OE33-GFP and OE33-IRES-GFP cells during a 1-hour pulse with [35S]-methionine carried out 24 hours post-infection with AdLuc or Adp14 at 100 pfu/cell, followed by immunoprecipitation with anti-GFP, SDS-PAGE and fluorography. Each lane represents 105 cell equivalents. (b) Bar graph representing the average of two [35S]-methionine uptake experiments described in part (A). Bars represent average incorporation of [35S]-methionine into GFP in Adp14-treated cells relative to AdLuc-treated cells, with standard deviations shown. (c) Western blot of p14ARF protein levels in AdLuc-treated or Adp14-treated OE33-GFP and OE33-IRES-GFP, 24 hours post-treatment with 100 pfu/cell of Adp14 or Adp14/p53. each lane represents 40 µg cellular lysates.
Multiple tumor suppressor genes and molecular pathways have been disrupted in cancer. However, the current strategy of cancer therapy usually focuses on repairing a particular gene or pathway, leading to a limited therapeutic effect. p53 and p14ARF have been reported to be loss of function in about 50% and 40% of cancer patients separately. In our study, we choose p53 and p14ARF as our selected target, and our results suggest that for gene transfer applications where p14ARF is co-expressed with another transgene, it may be necessary to place the second transgene downstream of an IRES element to insure maximum expression of its gene product. Similarly, for cancer cells with a disabled apoptotic pathways, and characterized by high level expression of endogenous p14ARF due to chronic oncogene activation, it may be necessary to employ gene transfer vectors that incorporate IRES elements in order to ensure maximum transgene expression following gene transfer. However, we didn't check other viral IRES types, whether our conclusion applied to EMCV IRES only, or other IRES elements, is still unknown.
Further studies will be needed to determine whether selective suppression of cap-dependent translation by p14ARF involves a specific interaction of p14ARF with a translational regulatory protein, and whether cellular IRES elements are also resistant to p14ARF-mediated translational suppression. In preliminary work, we have been unable to find evidence for the binding of p14ARF to the eIF4E binding protein, an upstream regulator of the eIF4E translation initiation factor, suggesting that p14ARF does not exert its differential effects via the eIF4E initiation complex (Huang Y., et al, unpublished observations).
Conclusions
From the conclusion of our study, we can use the Adp14/p53 in which two tumor suppressor genes were expressed in one cassette including IRES for colon cancer treatment, and achieve a significant effect of tumor suppression. Also, the ability of p14ARF to selectively suppress cap-dependent translation could have broader relevance to cellular regulation. CAP-independent translation from cellular IRES elements, though normally weak [23], is emerging as an important cellular mechanism for maintaining expression of critical gene products under conditions where cap-dependent translation is suppressed, for example during the cellular stress response [24]. Thus the induction and subcellular redistribution of p14ARF in response to certain types of DNA damage [17, 25] could have a dual consequences for the cellular stress response, the first involving its well-described role in stabilizing the p53 protein, and the second involving selective suppression of cap-dependent protein translation followed by a redirection of cellular translation towards IRES-mediated translation of stress response proteins.
Abbreviations
ARF, alternate reading frame; IRES, Internal ribosome entry site; EIF, eukaryotic translation initiation factor; ITAF, IRES trans-acting factor; EMCV, encephalomyocarditis virus; Luc, luceriferase; DN, double null; MEF, mouse embryonic fibroblast
Acknowledgements
We thank Dr. Guillermina Lozano, Department of Molecular Genetics, The University of Texas M. D. Anderson Cancer Center, Houston, TX, for the generous gift of p53-null, mdm2-null mouse embryo fibroblasts. Grant support provided by NIH/NCI CA111868 (USA), and NSFC 81071662, 81428016, 81472209; Beijing Science foundation Z151100003915073 (China); Jilin Science foundation 20140414061GH.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fitzgerald KD, Semler BL. Bridging ires elements in mrnas to the eukaryotic translation apparatus. Biochim Biophys Acta. 2009;1789:518-528
2. Pain VM. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747-771
3. Kieft JS. Viral ires rna structures and ribosome interactions. Trends Biochem Sci. 2008;33:274-283
4. Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mrna molecules. Genes Dev. 2001;15:1593-1612
5. Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mrna directed by a sequence derived from poliovirus rna. Nature. 1988;334:320-325
6. Pickering BM, Willis AE. The implications of structured 5' untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39-47
7. Rizos H, McKenzie HA, Ayub AL, Woodruff S, Becker TM, Scurr LL, Stahl J, Kefford RF. Physical and functional interaction of the p14arf tumor suppressor with ribosomes. J Biol Chem. 2006;281:38080-38088
8. Palmero I, Pantoja C, Serrano M. P19arf links the tumour suppressor p53 to ras. Nature. 1998;395:125-126
9. Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the arf tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424-2433
10. Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, Roussel MF, Sherr CJ. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci U S A. 2003;100:15930-15935
11. Garcia JF, Villuendas R, Sanchez-Beato M, Sanchez-Aguilera A, Sanchez L, Prieto I, Piris MA. Nucleolar p14(arf) overexpression in reed-sternberg cells in hodgkin's lymphoma: Absence of p14(arf)/hdm2 complexes is associated with expression of alternatively spliced hdm2 transcripts. Am J Pathol. 2002;160:569-578
12. Lee YK, Park JY, Kang HJ, Cho HC. Overexpression of p16ink4a and p14arf in haematological malignancies. Clin Lab Haematol. 2003;25:233-237
13. Sanchez-Aguilera A, Sanchez-Beato M, Garcia JF, Prieto I, Pollan M, Piris MA. P14(arf) nuclear overexpression in aggressive b-cell lymphomas is a sensor of malfunction of the common tumor suppressor pathways. Blood. 2002;99:1411-1418
14. Huang Y, Tyler T, Saadatmandi N, Lee C, Borgstrom P, Gjerset RA. Enhanced tumor suppression by a p14arf/p53 bicistronic adenovirus through increased p53 protein translation and stability. Cancer Res. 2003;63:3646-3653
15. Saadatmandi N, Tyler T, Huang Y, Haghighi A, Frost G, Borgstrom P, Gjerset RA. Growth suppression by a p14(arf) exon 1beta adenovirus in human tumor cell lines of varying p53 and rb status. Cancer Gene Ther. 2002;9:830-839
16. Gjerset RA, Arya J, Volkman S, Haas M. Association of induction of a fully tumorigenic phenotype in murine radiation-induced t-lymphoma cells with loss of differentiation antigens, gain of cd44, and alterations in p53 protein levels. Mol Carcinog. 1992;5:190-198
17. Lee C, Smith BA, Bandyopadhyay K, Gjerset RA. DNA damage disrupts the p14arf-b23(nucleophosmin) interaction and triggers a transient subnuclear redistribution of p14arf. Cancer Res. 2005;65:9834-9842
18. Taha C, Liu Z, Jin J, Al-Hasani H, Sonenberg N, Klip A. Opposite translational control of glut1 and glut4 glucose transporter mrnas in response to insulin. Role of mammalian target of rapamycin, protein kinase b, and phosphatidylinositol 3-kinase in glut1 mrna translation. J Biol Chem. 1999;274:33085-33091
19. Gjerset RA, Turla ST, Sobol RE, Scalise JJ, Mercola D, Collins H, Hopkins PJ. Use of wild-type p53 to achieve complete treatment sensitization of tumor cells expressing endogenous mutant p53. Mol Carcinog. 1995;14:275-285
20. Zhang Y, Xiong Y, Yarbrough WG. Arf promotes mdm2 degradation and stabilizes p53: Arf-ink4a locus deletion impairs both the rb and p53 tumor suppression pathways. Cell. 1998;92:725-734
21. Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126-1132
22. Yin Y, Stephen CW, Luciani MG, Fahraeus R. P53 stability and activity is regulated by mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462-467
23. Owens GC, Chappell SA, Mauro VP, Edelman GM. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:1471-1476
24. Tinton SA, Buc-Calderon PM. Hypoxia increases the association of 4e-binding protein 1 with the initiation factor 4e in isolated rat hepatocytes. FEBS Lett. 1999;446:55-59
25. Khan SH, Moritsugu J, Wahl GM. Differential requirement for p19arf in the p53-dependent arrest induced by DNA damage, microtubule disruption, and ribonucleotide depletion. Proc Natl Acad Sci U S A. 2000;97:3266-3271
Author Biography
Dr. Yinghui Huang is the dean and distinguished professor in the college of life science and bioengineering at Beijing University of Technology. He got his medical degree from Hubei Medical University in China and Ph.D. degree from University of Cambridge in UK. Dr. Huang also has 15 years of research experience in USA. He has coauthored about 30 publications including cancer research, cell reports, gut, and molecular therapy. His team has received many awards in China. The current research in Dr. Huang's group focuses on cancer epigenetics and cancer gene therapy.
![]() Corresponding author: Yinghui Huang, M.D., Ph.D., College of Life Science and Bioengineering, Beijing University of Technology, Beijing, China Phone: +86-15904420535; Fax: +86-10-67396137; email: yhuangedu.cn.
Corresponding author: Yinghui Huang, M.D., Ph.D., College of Life Science and Bioengineering, Beijing University of Technology, Beijing, China Phone: +86-15904420535; Fax: +86-10-67396137; email: yhuangedu.cn.

 Global reach, higher impact
Global reach, higher impact