Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(9):720-727. doi:10.7150/jca.9864 This issue Cite
Review
Utilization of Translational Bioinformatics to Identify Novel Biomarkers of Bortezomib Resistance in Multiple Myeloma
1. Gillette Children's Specialty Healthcare, St. Paul, MN;
2. Department of Genomic Sciences, University of Washington, Seattle, WA;
3. Department of Medicine, University of Minnesota, Minneapolis, MN;
4. Division of Hematology, Oncology, and Transplantation, University of Minnesota, Minneapolis, MN;
5. Department of Genetics, Cell Biology, University of Minnesota, Minneapolis, MN;
6. Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN.
Received 2014-6-10; Accepted 2014-8-20; Published 2014-9-21
Abstract
Multiple myeloma (MM) is an incurable malignant neoplasm hallmarked by a clonal expansion of plasma cells, the presence of a monoclonal protein in the serum and/or urine (M-spike), lytic bone lesions, and end organ damage. Clinical outcomes for patients with MM have improved greatly over the last decade as a result of the re-purposing of compounds such as thalidomide derivatives, as well as the development of novel chemotherapeutic agents including first and second generation proteasome inhibitors, bortezomib (Bz) and carfilzomib. Unfortunately, despite these improvements, the majority of patients relapse following treatment. While Bz, one of the most commonly used proteasome inhibitors, has been successfully incorporated into clinical practice, some MM patients have de novo resistance to Bz, and the majority of the remainder subsequently develop drug resistance following treatment. A significant gap in clinical care is the lack of a reliable clinical test that would predict which MM patients have or will subsequently develop Bz resistance. Thus, as Bz resistance remains a significant challenge, research efforts are needed to identify novel biomarkers of early Bz resistance, particularly when an early therapeutic intervention can be initiated. Recent advances in MM research indicate that genomic data can be extracted to identify novel biomarkers that can be utilized to select more effective, personalized treatment protocols for individual patients. Computationally integrating large patient databases with data from whole transcriptome profiling and laboratory-based models can potentially revolutionize our understanding of MM disease mechanisms. This systems-wide approach can provide rational therapeutic targets and novel biomarkers of risk and treatment response. In this review, we discuss the use of high-content datasets (predominantly gene expression profiling) to identify novel biomarkers of treatment response and resistance to Bz in MM.
Keywords: Multiple myeloma, biomarkers
Introduction
Translational bioinformatics and biomarkers
Personalized medicine is often discussed in the context of evaluating genetic variation as a way to better diagnose, evaluate, or effectively treat an individual patient (1). The analysis of biomarkers can provide an indication of biological action and is essential to understanding complex disease states. In many areas of medicine, biomarkers aid in early diagnosis, prognosis, and treatment selection. Current standards of care are influenced by past research, experience, and existing hospital protocols and practices. In many cases, establishment of new standards of care may be improved by the utilization of genetic data.
Utilizing translational informatics and applying genetic data to patient care has a variety of benefits such as improved treatment efficacy, increased patient safety, and reduced cost. This process may also have the potential to accelerate drug development by reducing the occurrence of unexpected safety concerns or difficulty determining efficacy in clinical trials (2). Biomarkers derived from genomic and other data can be utilized to select more effective treatment protocols (3). These interventions could benefit patients with increases in the quality of life, overall survival, and progression-free survival.
An example of how a personalized genomics approach may surpass the traditional clinical practices can be applied to the treatment of multiple myeloma (MM). This review will illustrate the above by focusing on the application of translational bioinformatics and genomic biomarker utilization to improve treatment strategies in MM. More specifically, this review will address how gene expression profiling (GEP) can be applied to address the clinical challenge of bortezomib (Bz) resistance that frequently develops when patients undergo treatment for MM.
Biomarker Development
Research and data analysis for biomarker development can be divided into 4 general stages (Figure 1). In the discovery and qualification phases, biomarkers are identified from cell lines and animal models, and they are associated with the condition of interest. The verification and validation stages involve utilizing cell lines as well as primary human data to assess the specificity of candidate biomarkers, as well as establish overall sensitivity and specificity. As research progresses from discovery to validation of biomarkers, the number of samples to be analyzed will increase as the number of analytes of interest are narrowed.
Biomarker Development Process Overview.
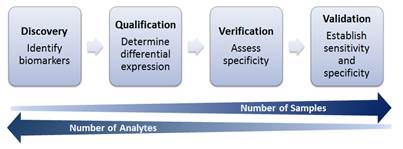
GEP analysis yields a large amount of data and lends to the utility of translational informatics methods to extract information and provide insight. Current research makes use of these data to determine related genes that may become potential targets for future research. Murine and human cell lines are utilized to identify possible biomarkers of interest through large GEP screens, and in vivo studies are conducted using mouse models. Candidate markers are determined and can be validated further through primary human data, such as retrospective and prospective clinical studies as part of biomarker discovery and validation. The addition of next-generation sequencing data and miRNA profiling information may allow for more comprehensive predictive models when combined with GEP (4).
Multiple Myeloma
MM, a fatal malignancy of plasma cells in the bone marrow, is diagnosed in greater than 22,000 adults in the U.S. annually (5) with a median age at diagnosis of approximately 70 years (6). The incidence rate of MM among African Americans is approximately twice as high in comparison to European Americans (7, 8). MM is hallmarked by a proliferation of clonal, malignant plasma cells that expand predominantly in the bone marrow. The malignant plasma cells accumulate in the marrow and other sites and aberrantly secrete monoclonal immunoglobulins, commonly referred to as an “M spike.” Detection, quantification, and identification of the monoclonal protein are performed by protein electrophoresis and immunofixation. Serum M-spike concentrations are used to monitor disease burden.
Accumulation of malignant plasma cells in the bone marrow results in bone marrow dysfunction leading to anemia and other cytopenias. Proliferation of these myeloma cells in the bone marrow can stimulate osteoclasts to resorb bone and form lytic lesions leading to hypercalcemia. Furthermore, the secreted monoclonal proteins can deposit and damage other tissues, such as the kidneys and can frequently lead to renal failure. Moreover, the excess protein production can lead to hyperviscosity syndrome, manifested by bleeding, blurry vision, heart failure, and neurologic symptoms.
The diagnosis of MM requires documentation of malignant plasma cells in addition to evidence of end organ damage by the malignant cell. These criteria are defined by hyperCalcemia, Renal failure, Anemia, and Bone lesions; these are sometimes described utilizing the mnemonic CRAB (9). The presence of an M-spike in the absence of end organ damage or plasmacytoma is insufficient for the diagnosis of MM and is defined as smoldering myeloma (SMM) or monoclonal gammopathy of undetermined significance (MGUS) depending on the percentage of bone marrow plasma cells.
Conventional and molecular cytogenetic and genomic profiling have been instrumental in advancing the understanding of MM pathogenesis (10) and providing a method of risk stratification. Hyperdiploidy, characterized by gains of odd-numbered chromosomes (3, 5, 7, 9, 11, 15, 19 and 21), is generally associated with a more favorable prognosis (11, 12). Non-hyperdiploid karyotypes are frequently characterized by translocations involving the immunoglobulin heavy chain locus at 14q32. Common partners include 11q13 (CCND1), 6p21 (CCND3), 16q23 (MAF), 20q12 (MAFB and 4p16 (FGFR3, MMSET), and with the exception of t(11;14)(q13;q32), these karyotypes are frequently associated with a poor prognosis (12). Common secondary aberrations include monosomy 13, deletion of 17p13 (TP53) and gains/losses of chromosome 1 (12).
Treatment of MM is complex and may include a variety of chemotherapeutic agents with multiple mechanisms of action. While a majority of patients initially respond to therapy, most patients eventually relapse or progress with treatment-refractory disease. Thus, translational bioinformatics can be potentially utilized (including gene expression profiling, oligonucleotide and SNP microarray and next generation sequencing) to look at mechanisms of drug resistance and create new diagnostic testing to improve outcomes and treatment strategies in MM.
Multiple Myeloma Treatment Overview
Within the past decade there has been an emergence of novel agents in the treatment of MM. Patients with SMM or MGUS are generally not treated, as early treatment has not been determined to increase survival or prevent onset of symptomatic MM (13). Because these conditions can progress to MM, patients with SMM or MGUS are monitored closely. MM patients are initially treated with a cocktail of agents aimed at inducing remission or optimal disease control. With the inclusion of modern agents in MM treatment, there are numerous protocols that are appropriate for initial therapy. All medically fit patients are considered for autologous stem cell transplant after medical therapy.
Selection of an initial treatment protocol relies on patient factors such as age, co-morbidities, and Eastern Cooperative Oncology Group (ECOG) performance status, as well as disease factors such as tumor burden and molecular features. Tumor burden is assessed by factors such as lytic bone disease, anemia, hypercalcemia, and serum concentration of the monoclonal protein. Karyotyping and fluorescence in situ hybridization (FISH) for a panel of MM-associated abnormalities is performed to identify prognostically-relevant chromosomal abnormalities, and patients with high risk molecular features are treated with Bz-based protocols. These protocols often incorporate a steroid and may also include a traditional cytotoxic agent (such as cyclophosphamide). Patients with very high-risk molecular features may be treated with a combination of Bz, lenalidomide, and steroid. Patients who are not candidates for stem cell transplant can be treated with stem cell-toxic agents such as melphalan.
After medical therapy, autologous hematopoietic stem cell transplant (auto-HCT) is the standard of care for all patients who meet the age criteria and have adequate organ function, performance status, and minimal co-morbidities. Allogeneic stem cell transplant is currently reserved for investigational use, as this modality has not been associated with improved survival in previous clinical trials of MM patients (14). This approach is not curative, but has been demonstrated to increase overall survival in MM (15).
Current treatment regimens for MM utilize immunomodulatory drugs (IMiDs) which are structural and functional analogs of thalidomide. The mechanism of action of thalidomide in the treatment of MM is based on its ability to enhance the immune system causing apoptosis of neoplastic cells by down-regulating anti-apoptotic pathways (16). Other second-generation IMiDs such as lenalidomide (most commonly used IMiD) and pomalidomide are derivatives of thalidomide that have more favorable toxicity and efficacy profiles and exert different effects on the immune system.
Proteasome inhibitors were introduced for the treatment of MM in the last decade and represent one of the newest classes of agents. Proteasome inhibitors block the ability of the proteasomal complex to degrade misfolded, damaged, or polyubiquitinated proteins. As MM cells are highly dependent on proteasome-homeostatic pathways (16), inhibition of the proteasome complex initiates a terminal unfolded protein response in MM cells (17).
Bz is the first proteasome inhibitor to be widely used in the treatment of MM. Bz inhibits the PSMB5 subunit of the proteasome and is commonly used in combination with melphalan, dexamethasone, and thalidomide derivatives (18).
The development and utilization of Bz in combination with other medications has increased survival time and improved patient outcomes. However, despite the high response rate to Bz, most patients eventually develop resistance or intolerance to Bz (23, 24) and nearly 20-30% of patients have innate or de novo resistance to Bz (primary refractory disease) (24, 25). While there remains no established standard treatment for patients who are Bz resistant (26), treatment of relapsed/refractory MM has included the use of alkylating agents, anthracyclines and corticosteroids. Other modalities now include transplant and the use of immunomodulatory drugs and other second-generation proteasome inhibitors such as carfilzomib which has some activity in 15% of Bz-refractory patients (27). For patients who were initially sensitive to Bz, response rates when re-treated with Bz may be as low as 23% (28). Furthermore, once patients develop resistance to Bz, their prognosis is often poor. Therefore, additional research is needed dedicated to discovering biomarker and genetic profiling techniques that can improve clinical decision-making in patients with refractory disease. Recent studies have attempted to distinguish patients who will respond to Bz or those who will develop resistance following initial treatment. Additional studies are aimed to identify novel chemotherapeutic options to overcome resistance.
Clinical Implications
Evaluation of resistance
Currently, there are no available testing options to measure emerging Bz resistance. However, recent studies identifying novel biomarkers of Bz resistance may indicate a potential mechanism of resistance and even suggest novel therapeutic strategies for overcoming acquired and de novo resistance (23).
Translational bioinformatics methods, such as mining GEP databases, can be used to derive novel markers of tumor classification and relate gene expression profiles to clinical outcome and chemotherapeutic response (29). Bioinformatics analysis of large databases of patient data, when coupled with advances in genetic analysis of animal and human model systems, promises to accelerate our understanding of MM disease mechanisms. Such a comprehensive understanding can promote the development of therapeutic approaches and biomarkers of risk and treatment response. Gene expression signatures can be used to predict differences in patient outcomes (30, 31), and these methods have the potential for therapeutic stratification of MM. Indeed, GEP has already been shown to provide prognostic/predictive information to patients and clinicians at the time of diagnosis (32).
Methods
References selected for inclusion in the evaluation of recent research were clinical research studies, including data mining analyses, conducted between 2008 and 2014. Studies were located by searching the following terms: profiling bortezomib resistance myeloma, bortezomib resistance myeloma, bortezomib myeloma, myeloma biomarkers clinical trial, genomics myeloma, and myeloma treatment. The following databases were utilized: PubMed, Google Scholar, Web of Science, and PLOS ONE. Studies were selected for inclusion based on methodology including model type, data evaluation method, and significance of analysis related to genomic profiling of Bz refractory MM. Ongoing clinical studies were determined from http://www.clinicaltrials.gov utilizing the search terms: bortezomib and refractory myeloma. Trials were classified as open, of known status, and were conducted in the US.
Biomarker Application to Identify Bortezomib Resistance and Develop Treatment Strategies
Recent Research
Ri, et al (33), evaluated acquired Bz resistance through the utilization of two Bz resistant human cell lines and identified mutations of the PSMB5 gene (33) suggesting that some MM cells may acquire Bz resistance by the suppression of apoptotic signals through the inhibition of unfolded protein accumulation and stress. While these mutations were seen in cell lines, additional studies sequencing primary human MM samples have not identified PSMB5 mutations (34) indicating that mutation of the Bz target molecule is not a frequent mechanism of Bz resistance.
Leung-Hagesteijn, et al (35) recently studied the role of the XBP1(S) protein in human MM and its role in Bz resistance. Looking specifically at primary human MM samples, they found that while the majority of the plasma cells express XBP1(S), there are minor subpopulations of XBP1(S)-negative pre-plasmablasts and CD20-positive B cell progenitors present in the marrow of MM patients. Moreover, the XBP1(S) subpopulations were persistent in Bz resistant patients (35). The researchers were able to show that XBP1 is not required for MM tumor cell survival (in contrast to other prior reports), but is only necessary for secretory maturation; XBP1's absence therefore likely plays a role in proteasome inhibitor resistance by decreasing the endoplasmic reticulum burden and affecting the unfolded protein response (35).
To identify genes associated with poor prognosis in MM, gene regulatory networks of the combined GEP from publically available whole-genome transcriptional profiling data from seven separate MM datasets were fully interrogated (10). Critical network analysis demonstrated that CCND1 and CCND2 were the most critical genes associated with poor prognosis and this information was used to build restricted gene models that can predict prognosis.
Zhu, et al (31), conducted a study utilizing RNAi screening approaches to identify modifiers of Bz sensitivity. Three genes were identified which when silenced, were not cytotoxic, but promoted inhibitory effects of Bz. Small molecular inhibitors of CDK5 were found to synergize with Bz and induce cytotoxicity of myeloma cells (primary and cell lines). CDK5 regulation of proteasome subunit PSMB5 was determined to be a potential route to Bz sensitization (31).
In a recent study by Stessman, et al (30), investigators used GEP to identify genes associated with Bz resistance. This study utilized murine and human model systems to generate Bz sensitive and resistant cell lines and these cell lines were then compared using GEP to generate a gene pattern that would create a signature of sensitivity and resistance. This gene signature was used in conjunction with the connectivity map database (CMAP), a genomic database linking gene patterns associated with disease and drug treatment, to genetically predict which drugs Bz resistant cells could be subsequently sensitive to (30).
In a second study by Stessman, et al, (23) a murine MM model system was used to discover immunophenotypic signatures associated with Bz sensitivity and resistance (both acquired and innate/de novo). Specifically, in this model system, Bz resistant cell lines had genotypic and immunophenotypic features characteristic of less differentiated plasma cells including loss of surface markers CD93 and CD69; these may indicate a mechanism for Bz resistance by loss of plasma cell maturation. By querying a database of GEP data from human subjects enrolled in a phase 3 clinical trial, it was determined that CD93 expression in primary samples is associated with BLIMP-1 expression in human MM and predicts improved survival. To support the hypothesis that dedifferentiation was associated with resistance, induction of differentiation with the use of lipopolysaccharide (LPS) not only induced expression of PC maturation markers but also restored Bz sensitivity suggesting the possibility that reversing resistance by promoting plasma cell maturation prior to Bz treatment in refractory MM could be an effective treatment approach (23). Investigation of other similar approaches such as the use of CpG-oligodeoxynucleotides could be investigated as a mechanism for restoring Bz sensitivity; similar strategies have been employed for the treatment of chronic lymphocytic leukemia.
The third study by Stessman, et al (36) demonstrated that decreased expression of CXCR4, a cell surface marker with increased expression in plasma cells, was associated with Bz resistance. GEP data from Bz treated human MM patients identified decreased CXCR4 gene expression conferred a worse prognosis suggesting the possibility that CXCR4 can be used as a diagnostic biomarker to predict clinical outcomes in MM patients treated with Bz (36).
Fernandez de Larrea, et al (37), explored the role of DNA methylation of 30 genes in the bone marrow of 75 patients with relapsed MM treated with Bz correlating methylation patterns with overall survival. Lower levels of NFKB1 methylation and a higher methylated global genome was associated with longer overall survival in patients that had received Bz treatment. These results represent the potential clinical use of methylation status in the prognosis of relapsed MM in patients treated with Bz (37). Additionally, Kaiser et al (38), analyzed combined DNA methylation and GEP data to identify epigenetically repressed tumor suppressor genes related to MM prognosis. Hypermethylation on critical genes may impact response to therapy and this assessment of DNA methylation could provide a clinically useful tool for risk stratification and personalized treatment selection in myeloma.
Genetic heterogeneity in MM may also be implicated in Bz resistance and MM prognosis. The importance of heterogeneity analysis has been demonstrated as related to individualized treatment decisions (39), as well as prognostic stratification and assessment of response to treatment (7). The clonal heterogeneity in primary human MM samples was recently further characterized by whole-exome sequencing and single-cell genetic analysis (40). Using this approach, the researchers were able to show that at presentation, MM patients harbor two to six different major clones, which are related by linear and branching phylogenies (40).
Strategies for the Treatment of Bortezomib Resistant MM
The development of clinically relevant biomarkers for Bz resistant MM has been a challenge due to the variation and complexity of chromosomal abnormalities in this condition (there is no pathognomonic genetic “hit”). Novel biomarkers are being explored for their potential to impact treatment decisions utilizing a data-driven and personalized approach to care. They may lead to a change in current treatment protocols and result in a more personalized and effective approach to patient care. Current studies within the translational pipeline include determining whether CXCR4 and other biomarkers can identify Bz sensitivity and resistance in MM patients.
Summary of selected research evaluating potential biomarkers associated with bortezomib resistant multiple myeloma.
| Author | Title | Result |
|---|---|---|
| Stessman et al, 2013 | Bortezomib resistance can be reversed by induced expression of plasma cell maturation markers in a mouse in vitro model of multiple myeloma | Certain genes were identified as biomarkers and may indicate a mechanism for Bz resistance through the loss of PC maturation. Induced PC maturation in both acquired and innate resistant cells restored Bz sensitivity |
| Stessman et al, 2013 | Profiling bortezomib resistance identifies secondary therapies in a mouse myeloma model | Identified 23 gene signature that distinguished between BzS and BzR mouse cell lines and significantly predicted differences in patient outcomes in a clinical trial utilizing Bz |
| Stessman et al, 2013 | Reduced CXCR4 expression is associated with extramedullary disease in a mouse model of myeloma and predicts poor survival in multiple myeloma patients treated with bortezomib | Determined that low CXCR4 is associated with Bz resistance and poor outcomes. Supports the use of CXCR4 as a diagnostic biomarker that predicts clinical outcome in patients treated with Bz |
| Ri et al, 2010 | Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress | Preventing the accumulation of misfolded proteins and avoidance of ER stress has an important role in Bz resistance by suppressing apoptosis-inducing signals in MM cells |
| Agnelli et al, 2011 | The reconstruction of transcriptional networks reveals critical genes with implications for clinical outcome of multiple myeloma | Gene signatures demonstrated to predict survival were determined utilizing data from 7 MM datasets |
| Zhu et al, 2011 | RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5 | Identified 37 genes which when silenced are not directly cytotoxic but do synergistically promote inhibitory effects of Bz. Combinations of Bz with other proteasome inhibitor drugs or combinations with inhibitors of CDK5 make sense to explore for response prediction |
| Fernandez de Larrea et al, 2013 | Impact of global and gene-specific DNA methylation pattern in relapsed multiple myeloma patients treated with bortezomib | Combination of highly methylated global genome with low NFkB1 methylation status defined a specific subset of patients with better prognosis |
| Kaiser et al, 2013 | Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma | Assessment of DNA methylation of certain genes could provide a clinically useful tool for risk determination and individualized treatment selection in MM |
| Lohr et al, 2014 | Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy | Heterogeneity analysis displays clinical utility for treatment decisions |
| Bolli et al, 2013 | Heterogeneity of genomic evolution and mutational profiles in multiple myeloma | The heterogeneity of the MM genome may impact prognosis stratification treatment approach and assessment of treatment response |
There is no standardized treatment approach for Bz refractory MM. While several approaches have been studied, attempts to identify molecular targets that can provide insight into the mechanism behind resistance is critically needed. For example, one potential strategy suggests that promoting malignant plasma cell maturation prior to Bz treatment could reverse resistance (23); however, while this approach has been successful in experimental systems, efficacy in clinical trials has not yet been studied.
Another potential option to overcome resistance Bz is the addition of another chemotherapeutic agent in conjunction to Bz treatment. For example, signaling through the IGF-1/IGF-1R axis contributes to acquired resistance and suggested that combining Bz with IGF-1R inhibitors could be a strategy to prevent or overcome Bz resistance (41).
Treatment with panobinostat is an option currently under investigation to overcome Bz resistance. PANORAMA 2 is a phase II clinical trial of panobinostat in combination with Bz and dexamethasone to treat relapsed and Bz-refractory MM (42). Panobinostat is a pan-deacetylase inhibitor that works with Bz to inhibit the aggresome and proteasome pathways (43). A 35.4% response rate was observed in this study with a clinical benefit rate of 52.7%. This combination is also being studied in a phase 3 trial, PANORAMA 2 (42). This was further supported by preclinical research models that predicted that panobinostat could be helpful in some Bz resistant MM cell lines (30).
The strategies discussed above suggest that the utilization of data-driven technologies has the potential to improve treatment selection and efficacy in MM. These techniques require bioinformatics methods to extract clinically relevant information from vast amounts of genomic data, as well as to determine which data are helpful to collect prospectively and utilize in future biomarker research.
Clinical Trials
Currently, there are 37 open clinical trials investigating treatment resistant MM in the United States (Table 2). The majority of studies focus on the combination of Bz and other chemotherapeutic agents as a way to treat patients with relapsed and refractory MM.
Conclusion and Future Directions
MM is generally considered an invariably fatal disease, and the incidence of drug resistance to Bz in MM is a significant problem that can be improved by defining and utilizing more specific and clinically useful biomarkers. Current strategies for novel genotypic and/or immunophenotypic biomarker development are in the early stages of development and validation, but there are several promising markers to predict Bz resistance. Ideally, early detection of Bz resistance could lead to an effective therapeutic intervention. Among the clinical trials currently underway, many focus on the development of additional medications that would work synergistically with Bz to achieve a treatment effect.
Summary of Current Bortezomib Clinical Studies in Relapsed/Refractory MM.
| Category | Study |
|---|---|
| Combination of bortezomib and another medication for relapsed or refractory myeloma | A Study to Evaluate the Use of Chloroquine in Combination With VELCADE and Cyclophosphamide in Patients With Relapsed and Refractory Multiple Myeloma |
| Study to Determine the Maximum Tolerated Dose for the Combination of Pomalidomide, Bortezomib and Low-Dose Dexamethasone in Subjects With Relapsed or Refractory Multiple Myeloma | |
| Pomalidomide, Bortezomib, and Dexamethasone in Treating Patients With Relapsed or Refractory Multiple Myeloma | |
| Vorinostat, Bortezomib and Dexamethasone in Relapsed/Refractory Multiple Myeloma | |
| Open-label Study of TH-302 and Dexamethasone With or Without Bortezomib in Subjects With Relapsed/Refractory Multiple Myeloma | |
| Study of ACY-1215 Alone and in Combination With Bortezomib and Dexamethasone in Relapsed/Refractory Multiple Myeloma | |
| Combination Plerixafor (AMD3100)and Bortezomib in Relapsed or Relapsed/Refractory Multiple Myeloma | |
| A Phase I Study Of Panobinostat/Lenalidomide/Bortezomib/Dex for Relapsed And Relapsed/Refractory Multiple Myeloma | |
| Aurora A Kinase Inhibitor MLN8237 and Bortezomib in Treating Patients With Relapsed or Refractory Multiple Myeloma | |
| Anti-CXCR4 (BMS-936564) Alone and in Combination With Lenalidomide/Dexamethasone or Bortezomib/Dexamethasone in Relapsed/Refractory Multiple Myeloma | |
| A Study Evaluating ABT-199 in Multiple Myeloma Subjects Who Are Receiving Bortezomib and Dexamethasone as Standard Therapy Study of Bortezomib and Dexamethasone With or Without Elotuzumab to Treat Relapsed or Refractory Multiple Myeloma | |
| Safety and Efficacy of Pomalidomide, Bortezomib and Low-dose Dexamethasone in Subjects With Relapsed or Refractory Multiple Myeloma | |
| Vorinostat in Combination With Bortezomib, Doxorubicin and Dexamethasone (VBDD) in Patients With Refractory or Relapsed Multiple Myeloma (MM) | |
| Bendamustine, Wkly Bortezomib, Lenalidomide and Dexamethasone for Multiple Myeloma | |
| A Trial of ASP7487 (OSI-906) in Combination With Bortezomib for the Treatment of Relapsed Multiple Myeloma | |
| A Phase I Study of Ganetespib +/- Bortezomib in Patients With Relapsed and/or Refractory Multiple Myeloma | |
| Phase I Bortezomib (VELCADE) in Combo With Pralatrexate in Relapsed/Refractory MM | |
| A Study of ARRY-520 and Bortezomib Plus Dexamethasone in Patients With Relapsed/Refractory Multiple Myeloma | |
| A Phase 3 Study to Evaluate Efficacy and Safety of Masitinib in Patients With Relapse or Refractory Multiple Myeloma | |
| Dinaciclib, Bortezomib, and Dexamethasone in Treating Patients With Relapsed Multiple Myeloma | |
| Ph 1b Study to Evaluate GSK2110183 in Combination With Bortezomib and Dexamethasone in Subjects With Multiple Myeloma | |
| Bendamustine in Combination With Bortezomib and Pegylated Liposomal Doxorubicin for Multiple Myeloma | |
| Phase 3 Study With Carfilzomib and Dexamethasone Versus Velcade and Dexamethasone for Relapsed Multiple Myeloma Patients | |
| Addition of Daratumumab to Combination of Bortezomib and Dexamethasone in Participants With Relapsed or Refractory Multiple Myeloma | |
| Study of Bortezomib and Dexamethasone With or Without Elotuzumab to Treat Relapsed or Refractory Multiple Myeloma | |
| Study of Plitidepsin (Aplidin®) in Combination With Bortezomib and Dexamethasone in Patients With Multiple Myeloma | |
| Study of Treatment for Newly Diagnosed Multiple Myeloma Patients Older Than 65 Years With Sequential Melphalan/Prednisone/Velcade (MPV) Followed by Revlimid/Low Dose Dexamethasone (Rd) Versus Alternating Velcade/Melphalan/Prednisone (MPV) With Revlimid/Low Dose Dexamethasone | |
| Evaluation of a new drug for relapsed or refractory myeloma | A Phase I/IIa Study of Human Anti-CD38 Antibody MOR03087 in Relapsed/Refractory Multiple Myeloma |
| Study of LY2127399 in Japanese Participants With Relapsed or Refractory Multiple Myeloma | |
| Proteasome Inhibitor MLN9708 in Treating Patients With Relapsed Multiple Myeloma That Is Not Refractory to Bortezomib | |
| A Phase 2 Trial of Filanesib in Relapsed/Refractory Multiple Myeloma (AfFIRM) | |
| Subcutaneous Bortezomib Dosing | Subcutaneous (SC) Bortezomib-Regimens for Patients With RR MM Failing Prior IV Bortezomib-Containing Regimens |
| Maintenance Therapy With Subcutaneous Bortezomib | |
| Transplant and Bortezomib | Autologous or Syngeneic Stem Cell Transplant Followed by Donor Stem Cell Transplant and Bortezomib in Treating Patients With Newly Diagnosed High-Risk, Relapsed, or Refractory Multiple Myeloma |
| Bortezomib, Total Marrow Irradiation, Fludarabine Phosphate, and Melphalan in Treating Patients Undergoing Donor Peripheral Blood Stem Cell Transplant For High-Risk Stage I or II Multiple Myeloma | |
| Lenalidomide After Donor Stem Cell Transplant and Bortezomib in Treating Patients With High Risk Multiple Myeloma | |
| Bortezomib, Melphalan, and Total-Body Irradiation Before Stem Cell Transplant in Treating Patients With Multiple Myeloma |
Translational informatics has the potential to improve treatment strategies for malignancies such as MM by evaluating large datasets of genetic findings that could provide insight into differing treatment effects between patients. This evaluation and utilization of clinically relevant biomarkers has the potential to change the way MM is treated by moving towards a personalized and evidence-based approach. Recent research into the development of a diagnostic procedure to determine if a patient will be responsive to treatment has demonstrated that this may soon be a possibility (23). However, there is still a need for further clinical studies comparing the use and combination of medications in refractory and relapsed MM as well as defining similarities and differences between de novo and acquired resistance. It is also likely that the integration of laboratory results into patient EHR systems could improve care and provide valuable data, especially in relation to research centered on relapsed or refractory MM (44). Overall, future research is needed with a focus on the verification and validation of the potential biomarkers that have been determined by the previously discussed variety of genomic profiling studies.
MM treatment options as well as clinical outcomes have improved greatly over the last decade due to the introduction of novel chemotherapeutic agents such as Bz. However, there is variation in the treatment or evaluation for resistant/refractory MM. Bioinformatics methods have been used to analyze high complexity data sets, such as GEP, and have provided important insights into the pathophysiology of Bz resistance as well as potential biomarkers to predict treatment response. Further biomarker development and validation may change the way in which refractory and relapsed MM is treated. The progression and availability of genomics and proteomics has increased our ability to identify new biomarkers for diagnosis, classification, and treatment stratification in MM.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Schiess R, Wollscheid B, Aebersold R. Targeted proteomic strategy for clinical biomarker discovery. Mol Oncol. 2009;3(1):33-44
2. Kelloff GJ, Sigman CC. New science-based endpoints to accelerate oncology drug development. Eur J Cancer. 2005;41(4):491-501
3. Lussier YA, Stadler WM, Chen JL. Advantages of genomic complexity: bioinformatics opportunities in microRNA cancer signatures. J Am Med Inform Assoc. 2012;19(2):156-60
4. Amin SB, Yip WK, Minvielle S, Broyl A, Li Y, Hanlon B. et al. Gene expression profile alone is inadequate in predicting complete response in multiple myeloma. Leukemia. 2014
5. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11-30
6. Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12(1):20-37
7. Bolli N, Avet-Loiseau H, Wedge DC, Van Loo P, Alexandrov LB, Martincorena I. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997
8. Baker A, Braggio E, Jacobus S, Jung S, Larson D, Therneau T. et al. Uncovering the biology of multiple myeloma among African Americans: a comprehensive genomics approach. Blood. 2013;121(16):3147-52
9. Talamo G, Farooq U, Zangari M, Liao J, Dolloff NG, Loughran TP Jr. et al. Beyond the CRAB symptoms: a study of presenting clinical manifestations of multiple myeloma. Clin Lymphoma Myeloma Leuk. 2010;10(6):464-8
10. Agnelli L, Forcato M, Ferrari F, Tuana G, Todoerti K, Walker BA. et al. The reconstruction of transcriptional networks reveals critical genes with implications for clinical outcome of multiple myeloma. Clin Cancer Res. 2011;17(23):7402-12
11. Landgren O, Morgan GJ. Biologic frontiers in multiple myeloma: from biomarker identification to clinical practice. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(4):804-13
12. Sawyer JR. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer genetics. 2011;204(1):3-12
13. Rajkumar SV, Gertz MA, Kyle RA, Greipp PR. Current therapy for multiple myeloma. Mayo Clin Proc. 2002;77(8):813-22
14. Freytes CO, Vesole DH, Lerademacher J, Zhong X, Gale RP, Kyle RA. et al. Second transplants for multiple myeloma relapsing after a previous autotransplant-reduced-intensity allogeneic vs autologous transplantation. Bone Marrow Transplant. 2013
15. Rajkumar SV, Gahrton G, Bergsagel PL. Approach to the treatment of multiple myeloma: a clash of philosophies. Blood. 2011;118(12):3205-11
16. Castelli R, Gualtierotti R, Orofino N, Losurdo A, Gandolfi S, Cugno M. Current and Emerging Treatment Options for Patients with Relapsed Myeloma. Clin Med Insights Oncol. 2013;7:209-19
17. Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. Proteasome inhibition as a novel therapeutic target in human cancer. J Clin Oncol. 2005;23(3):630-9
18. Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K. Latest advances and current challenges in the treatment of multiple myeloma. Nat Rev Clin Oncol. 2012;9(3):135-43
19. Siegel DS, Martin T, Wang M, Vij R, Jakubowiak AJ, Lonial S. et al. A phase 2 study of single-agent carfilzomib (PX-171-003-A1) in patients with relapsed and refractory multiple myeloma. Blood. 2012;120(14):2817-25
20. Moreau P, Pylypenko H, Grosicki S, Karamanesht I, Leleu X, Grishunina M. et al. Subcutaneous versus intravenous administration of bortezomib in patients with relapsed multiple myeloma: a randomised, phase 3, non-inferiority study. Lancet Oncol. 2011;12(5):431-40
21. Attal M, Lauwers-Cances V, Marit G, Caillot D, Moreau P, Facon T. et al. Lenalidomide maintenance after stem-cell transplantation for multiple myeloma. N Engl J Med. 2012;366(19):1782-91
22. McCarthy PL, Owzar K, Hofmeister CC, Hurd DD, Hassoun H, Richardson PG. et al. Lenalidomide after stem-cell transplantation for multiple myeloma. N Engl J Med. 2012;366(19):1770-81
23. Stessman HA, Mansoor A, Zhan F, Linden MA, Van Ness B, Baughn LB. Bortezomib resistance can be reversed by induced expression of plasma cell maturation markers in a mouse in vitro model of multiple myeloma. PLoS One. 2013;8(10):e77608
24. Kumar S, Rajkumar SV. Many facets of bortezomib resistance/susceptibility. Blood. 2008;112(6):2177-8
25. Moreau P, Richardson PG, Cavo M, Orlowski RZ, San Miguel JF, Palumbo A. et al. Proteasome inhibitors in multiple myeloma: 10 years later. Blood. 2012;120(5):947-59
26. van de Donk NW, Lokhorst HM, Dimopoulos M, Cavo M, Morgan G, Einsele H. et al. Treatment of relapsed and refractory multiple myeloma in the era of novel agents. Cancer Treat Rev. 2011;37(4):266-83
27. Berenson JR, Hilger JD, Yellin O, Dichmann R, Patel-Donnelly D, Boccia RV. et al. Replacement of bortezomib with carfilzomib for multiple myeloma patients progressing from bortezomib combination therapy. Leukemia. 2014
28. Conner TM, Doan QD, Walters IB, LeBlanc AL, Beveridge RA. An observational, retrospective analysis of retreatment with bortezomib for multiple myeloma. Clin Lymphoma Myeloma. 2008;8(3):140-5
29. Shaughnessy JD Jr, Qu P, Usmani S, Heuck CJ, Zhang Q, Zhou Y. et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118(13):3512-24
30. Stessman HA, Baughn LB, Sarver A, Xia T, Deshpande R, Mansoor A. et al. Profiling bortezomib resistance identifies secondary therapies in a mouse myeloma model. Mol Cancer Ther. 2013;12(6):1140-50
31. Zhu YX, Tiedemann R, Shi CX, Yin H, Schmidt JE, Bruins LA. et al. RNAi screen of the druggable genome identifies modulators of proteasome inhibitor sensitivity in myeloma including CDK5. Blood. 2011;117(14):3847-57
32. van Laar R FR, Brown N, Ramsey J, Riccitelli S, Heuck C, Barlogie B, Shaughnessy JD Jr. Translating a gene expression signature for multiple Myeloma prognosis into a robust high-throughput assay for clinical use. BMC Medical Genomics. 2014
33. Ri M, Iida S, Nakashima T, Miyazaki H, Mori F, Ito A. et al. Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia. 2010;24(8):1506-12
34. Lichter DI, Danaee H, Pickard MD, Tayber O, Sintchak M, Shi HL. et al. Sequence analysis of beta-subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood. 2012;120(23):4513-6
35. Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, Reece DE. et al. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer cell. 2013;24(3):289-304
36. Stessman HA, Mansoor A, Zhan F, Janz S, Linden MA, Baughn LB. et al. Reduced CXCR4 expression is associated with extramedullary disease in a mouse model of myeloma and predicts poor survival in multiple myeloma patients treated with bortezomib. Leukemia. 2013;27(10):2075-7
37. Fernandez de Larrea C, Martin-Antonio B, Cibeira MT, Navarro A, Tovar N, Diaz T. et al. Impact of global and gene-specific DNA methylation pattern in relapsed multiple myeloma patients treated with bortezomib. Leuk Res. 2013;37(6):641-6
38. Kaiser MF, Johnson DC, Wu P, Walker BA, Brioli A, Mirabella F. et al. Global methylation analysis identifies prognostically important epigenetically inactivated tumor suppressor genes in multiple myeloma. Blood. 2013;122(2):219-26
39. Lohr JG, Stojanov P, Carter SL, Cruz-Gordillo P, Lawrence MS, Auclair D. et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91-101
40. Melchor L, Brioli A, Wardell CP, Murison A, Potter NE, Kaiser MF. et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia. 2014;28(8):1705-15
41. Kuhn DJ, Berkova Z, Jones RJ, Woessner R, Bjorklund CC, Ma W. et al. Targeting the insulin-like growth factor-1 receptor to overcome bortezomib resistance in preclinical models of multiple myeloma. Blood. 2012;120(16):3260-70
42. Richardson PG, Schlossman RL, Alsina M, Weber DM, Coutre SE, Gasparetto C. et al. PANORAMA 2: panobinostat in combination with bortezomib and dexamethasone in patients with relapsed and bortezomib-refractory myeloma. Blood. 2013;122(14):2331-7
43. Wolf JL, Siegel D, Goldschmidt H, Hazell K, Bourquelot PM, Bengoudifa BR. et al. Phase II trial of the pan-deacetylase inhibitor panobinostat as a single agent in advanced relapsed/refractory multiple myeloma. Leuk Lymphoma. 2012;53(9):1820-3
44. Gundlapalli AV, Delgado JC, Jackson BR, Tricot GJ, Hill HR. Composite patient reports: a laboratory informatics perspective and pilot project for personalized medicine and translational research. Summit on Translat Bioinforma. 2009;2009:39-43
Author contact
![]() Corresponding author: Michael A. Linden, MD, PhD. University of Minnesota, Department of Laboratory Medicine and Pathology MMC 609, 420 Delaware St SE, Minneapolis, MN 55455, 612-273-5629 (tel), 612-624-6662 (fax), E-mail: linde013edu.
Corresponding author: Michael A. Linden, MD, PhD. University of Minnesota, Department of Laboratory Medicine and Pathology MMC 609, 420 Delaware St SE, Minneapolis, MN 55455, 612-273-5629 (tel), 612-624-6662 (fax), E-mail: linde013edu.

 Global reach, higher impact
Global reach, higher impact