Impact Factor
ISSN: 1837-9664
J Cancer 2014; 5(6):425-432. doi:10.7150/jca.8594 This issue Cite
Research Paper
ERCC1, XRCC1 and GSTP1 Single Nucleotide Polymorphisms and Survival of Patients with Colon Cancer Receiving Oxaliplatin-Based Adjuvant Chemotherapy
1. INSERM, UMR_S 938, Saint-Antoine Research Centre, F-75012, Paris, France;
2. UPMC Univ Paris 06, UMR_S 938, Saint-Antoine Research Centre, F-75012, Paris, France;
3. Department of Gastroenterology and Digestive Oncology, European Georges Pompidou Hospital, AP-HP, Paris, France;
4. Paris Sorbonne Cité, University of Paris Descartes, Paris, France;
5. Cancer Research Personalized Medicine (CARPEM), European Georges Pompidou Hospital, AP-HP, Paris, France;
6. Department of Biostatistics and Epidemiology, EA4184, Georges François Leclerc Center, Dijon, France;
7. Department of Pathology, Ambroise Paré Hospital, AP-HP, Boulogne, France;
8. EA4340, University of Versailles, Saint-Quentin-en-Yvelines, Versailles, France;
9. Department of Biology, European Georges Pompidou Hospital, AP-HP, Paris, France;
10. Department of Pathology, Saint-Antoine Hospital, AP-HP, Paris, France;
11. Department of Pathology, European Georges Pompidou Hospital, AP-HP, Paris, France;
12. Department of Pathology, Mutualiste Montsouris Institute, Paris, France;
13. Department of Medical Oncology, Mutualiste Montsouris Institute, Paris, France;
14. Department of Medical Oncology, Saint-Antoine Hospital, AP-HP, Paris, France.
Received 2014-1-15; Accepted 2014-3-26; Published 2014-5-2
Abstract
Background: While single nucleotide polymorphisms (SNP) in genes involved in DNA repair or drug metabolism have been shown to influence survival of metastatic colon cancer patients treated with FOLFOX, data on adjuvant setting are scarce.
Methods: This study evaluated the correlation between disease-free survival (DFS) of 210 unselected stage III colon cancer patients receiving FOLFOX chemotherapy, and ERCC1-118 (rs11615, c.354T>C), XRCC1-399 (rs25487, c.1196G>A) and GSTP1-105 (rs1695, c.313A>G) polymorphisms. SNP were determined on tumor DNA using a PCR-based RFLP technique.
Results: In univariate analysis, a trend towards longer DFS was observed for ERCC1 (C/T + T/T) versus (C/C) (HR=2.29; p=0.06), and XRCC1 (A/A) versus (G/G + G/A) (HR=1.61; p=0.16), but not for GSTP1 genotypes; a statistically significant p value was obtained when combining ERCC1 and XRCC1 favorable genotypes (0 versus ≥ 1 favorable genotypes, HR=2.42; p=0.02). After adjustment on tumor stage, lymph node ratio and differentiation grade, multivariate analysis showed that combining ERCC1 and XRCC1 genotypes gave a p value slightly above the threshold for statistical significance (HR=2.03; p=0.06), which was lower than for tumor stage, lymph node ratio or differentiation grade.
Conclusion: The association of ERCC1 and XRCC1 polymorphisms may influence the prognosis of stage III colon cancer patients treated with FOLFOX adjuvant chemotherapy. Yet, these findings need to be confirmed in independent prospective studies.
Keywords: colon cancer, adjuvant FOLFOX, polymorphism, ERCC1, XRCC1, GSTP1.
Introduction
Oxaliplatin is a platinum-based chemotherapeutic agent that carries 1,2-diamino-cyclohexane ring. This chemotherapeutic agent has shown in vitro and in vivo antitumor activities in colorectal cancer (CRC) (1). The administration of oxaliplatin in combination with 5-fluorouracil (5FU) (FOLFOX regimen) significantly improved progression-free survival and response rate for patients with metastatic CRC (2). More recently, the addition of oxaliplatin to 5FU was demonstrated to improve the adjuvant treatment of stage III colon cancer by reducing the risk of recurrence and increasing overall survival (3, 4). However, oxaliplatin failed to eradicate micrometastatic disease in approximately one third of stage III colon cancer patients. Oxaliplatin exerts its action by forming DNA-platinum mono-adducts, primarily with guanines, which inhibits DNA replication and transcription and induces apoptosis (1). Unlike cisplatin, oxaliplatin-induced adducts are apparently not recognized by the mismatch repair (MMR) system, but are predominantly repaired by the nucleotide excision repair (NER) and base excision repair (BER) pathways (5, 6). An enhanced DNA repair efficiency or a decreased accumulation of the cytotoxic agent may contribute to resistance to platinum drugs.
Several studies reported that single nucleotide polymorphisms (SNP) in genes involved in DNA repair, as Excision Repair Cross-Complementing group 1 (ERCC1) (7) and X-Ray Repair Cross-Complementing group 1 (XRCC1) (8, 9), or involved in drug metabolism, as glutathione S-transferases P1 (GSTP1) (10-12), may predict the clinical outcome for patients receiving oxaliplatin-based chemotherapy for metastatic CRC treatment. ERCC1 forms a heterodimeric complex with Xeroderma Pigmentosum Group F, that has an important role in the incision process of NER (13). XRCC1 interacts with many proteins involved in single-strand break repair and in BER, participating in some of the rate-limiting reactions (14). Finally, glutathione S-transferases are implicated in drug detoxification through the conjugation of glutathione to electrophilic xenobiotics. The GSTP1 isoenzyme that participates in the detoxification of platinum drugs is highly expressed in human CRC tissues and could possibly influence resistance to platinum-based chemotherapy (15). To our knowledge, the impact of polymorphisms in these genes on the survival of patients treated with oxaliplatin-based adjuvant chemotherapy has been scarcely explored yet. Thus, in the current study, we have examined the influence of ERCC1 (Asn118Asn), XRCC1 (Arg399Gln) and GSTP1 (Ile105Val) polymorphisms, separately or in combination, on disease-free survival (DFS) of patients with stage III colon cancer receiving FOLFOX adjuvant chemotherapy.
Methods
Study population
We retrospectively collected data of all consecutive patients treated with FOLFOX adjuvant chemotherapy after surgical resection of a stage III colon cancer between 06/2003 and 12/2007 in 3 University centers (Ambroise Paré, Georges Pompidou, and Saint-Antoine hospitals) and 1 private hospital (Mutualiste Montsouris Institute). Patients were eligible for this retrospective multicenter study if they met the following criteria: older than 18 years, histologically proven colon cancer, at least 1 pathologically involved lymph node, R0 resection, available tumor specimen in the pathology departments, adjuvant FOLFOX chemotherapy starting within 8 weeks after surgery and no history of sensitive peripheral neuropathy. Fourteen stage III colon cancer patients enrolled in the MOSAIC trial with available tumor and treated in these centers with FOLFOX between 11/1998 and 10/2000 were also included. Patients with rectal cancer or treated with abdominopelvic radiotherapy have been excluded. This retrospective pharmacogenetic study has been approved by the ethics committee of the Pitié-Salpétrière Hospital (Paris, France).
Treatment and follow-up
The chemotherapy cycle consisted of a 2-hour infusion of 85 mg of oxaliplatin per square meter on day 1, in addition to the standard LV5FU2 regimen (FOLFOX4) or the simplified LV5FU2 regimen (modified FOLFOX6) (3, 16). Patients were scheduled to receive one cycle of chemotherapy every two weeks for 6 months. After treatment, a control visit was performed every 3 to 6 months for 3 years, then every 6 months for 2 years, then annually, to assess potential disease recurrence. Physical examination, serum carcinoembryonic antigen assay, and abdominal plus thoracic imaging were performed at each visit.
DNA extraction and genotyping
Genomic DNA was extracted from 20-µm-thick tissue sections of frozen tumors (111 patients) or from 7-µm tissue sections of paraffin-embedded tumors when frozen samples were not available (99 patients). Areas of tumor tissues were previously delineated by microscopic examination of a reference slide stained with haematoxylin and eosin. Extraction was done using the NucleoSpin Tissue Kit (Macherey-Nagel) for frozen samples and NucleoSpin DNA FFPE Kit (Macherey-Nagel) or KAPA Express DNA Extraction Kit (Kapa Biosystems) for paraffin-embedded tissues, according the manufacturers' instructions.
SNP genotyping analyses were done on amplified tumor DNA followed by digestion with the corresponding restriction enzyme: BsrD1 (New England Biolabs) for ERCC1 (rs11615, c.354T>C, p.Asn118Asn) (17), Hpall (Fermentas) for XRCC1 (rs25487, c.1196G>A, p.Arg399Gln) and Alw26I (Fermentas) for GSTP1 (rs1695, c.313A>G, p.Ile105Val) polymorphisms. For ERCC1-118, a 239 bp fragment was amplified using 5'-GTGCGAGGAGGCAGGAGGTGTGGG-3' and 5'-GAGCTCACCTGAGGAACAGG-3', as primers. BsrD1 cleaves the ERCC1 T allele into two fragments of 155 and 84 bp. For XRCC1-399, a 242 bp was amplified using 5'-CCCCAAGTACAGCCAGGTCC-3' and 5'-CCGCTCCTCTCAGTAGTCTG-3' as primers. The XRCC1 G allele is cleaved by HpaII into two fragments of 149 and 93 bp. For GSTP1-105, a 176 bp was amplified using 5'-ACCCCAGGGCTCTATGGGAAG-3' and 5'-TGAGGGCACAAGAAGCCCCT-3' as primers. Alw26I cleaves the G allele into two fragments of 89 and 87 bp. After digestion, the PCR fragments were electrophoresed in a 2% agarose gel in the presence of GelRed™ (1/10000, Biotium Inc) and visualized using a G: BOX gel imaging system (Syngene, Ozyme).
Statistical analysis
Differences in distributions between the clinico-pathological variables listed in Table 1 were assessed with the Chi2 or the Fisher's exact test, as appropriate. The cut-off value of lymph node ratio (LNR) between metastatic and examined lymph nodes corresponded to the median. The number of FOLFOX cycles according to the 3 possible genotypes of each gene was compared using a non-parametric Kruskal-Wallis test.
Clinicopathological characteristics of patients with stage III colon cancer according to ERCC1-118, XRCC1-399 and GSTP1-105 polymorphisms.
| Characteristics | All patients | ERCC1-118 (rs11615) | XRCC1-399 (rs25487) | GSTP1-105 (rs1695) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (n = 210) | (n = 202 ) | (n = 207 ) | (n = 209) | ||||||||||
| No. of patients (%) | No. of patients (%) | p | No. of patients (%) | p | No. of patients (%) | p | |||||||
| C/C | C/T | T/T | G/G | G/A | A/A | A/A | A/G | G/G | |||||
| 49 | 88 | 65 | 94 | 80 | 33 | 97 | 92 | 20 | |||||
| Age | |||||||||||||
| Median | 65.9 | 64.3 | 64.4 | 66.3 | 65.9 | 64.9 | 64.5 | 66.8 | 63.4 | 61.7 | |||
| Range | 30.5 - 82.8 | 30.5 - 82.8 | 31.1 - 81.0 | 37.8 - 82.8 | 30.8 - 82.1 | 37.8 - 82.8 | 30.5 - 77.4 | 30.8 - 81.0 | 30.5 - 82.8 | 40.9 - 81.0 | |||
| < 65 yr | 101 (48.1) | 26 (53.1) | 44 (50.0) | 29 (44.6) | 0.65 | 44 (46.8) | 40 (50.0) | 16 (48.5) | 0.92 | 39 (40.2) | 50 (54.3) | 12 (60.0) | 0.08 |
| Sex | |||||||||||||
| Female | 99 (47.1) | 23 (46.9) | 40 (45.4) | 31 (47.7) | 0.96 | 39 (41.5) | 39 (48.8) | 19 (57.6) | 0.26 | 42 (43.3) | 48 (52.2) | 9 (45.0) | 0.46 |
| Male | 111 (52.9) | 26 (53.1) | 48 (54.6) | 34 (52.3) | 55 (58.5) | 41 (51.2) | 14 (42.4) | 55 (56.7) | 44 (47.8) | 11 (55.0) | |||
| Tumor location | |||||||||||||
| Proximal | 67 (31.9) | 16 (32.7) | 28 (31.8) | 22 (33.8) | 0.97 | 31 (33.0) | 27 (33.8) | 9 (27.3) | 0.79 | 27 (27.8) | 34 (37.0) | 6 (30.0) | 0.40 |
| Distal | 143 (68.1) | 33 (67.3) | 60 (68.2) | 43 (66.2) | 63 (67.0) | 53 (66.2) | 24 (72.7) | 70 (72.2) | 58 (63.0) | 14 (70.0) | |||
| Differentiation grade | |||||||||||||
| Well / moderate | 182 (86.7) | 45 (91.8) | 76 (86.4) | 55 (84.6) | 0.50 | 82 (87.2) | 69 (86.2) | 29 (87.9) | 1.00 | 82 (84.5) | 81 (88.0) | 18 (90.0) | 0.76 |
| Poor | 28 (13.3) | 4 (8.2) | 12 (13.6) | 10 (15.4) | 12 (12.8) | 11 (13.8) | 4 (12.1) | 15 (15.5) | 11 (12.0) | 2 (10.0) | |||
| Bowel perforation / obstruction | 16 (7.6) | 6 (12.2) | 6 (6.8) | 2 (3.1) | 0.16 | 9 (9.6) | 5 (6.3) | 2 (6.1) | 0.73 | 8 (8.2) | 7 (7.6) | 0 (0.0) | 0.58 |
| Stage | |||||||||||||
| III A (T1-T2, N1) | 23 (11.0) | 9 (18.4) | 6 (6.8) | 7 (10.8) | 0.07 | 14 (14.9) | 5 (6.3) | 4 (12.1) | 0.37 | 9 (9.3) | 11 (12.0) | 3 (15.0) | 0.90 |
| III B (T3-T4, N1) | 108 (51.4) | 29 (59.2) | 43 (48.9) | 33 (50.8) | 46 (48.9) | 46 (57.5) | 15 (45.5) | 51 (52.6) | 47 (51.0) | 9 (45.0) | |||
| III C (Tx, N2) | 79 (37.6) | 11 (22.4) | 39 (44.3) | 25 (38.4) | 34 (36.2) | 29 (36.2) | 14 (42.4) | 37 (38.1) | 34 (37.0) | 8 (40.0) | |||
| Lymph node ratio | |||||||||||||
| < 0.100 | 102 (48.6) | 30 (61.2) | 43 (48.9) | 29 (44.6) | 0.20 | 44 (46.8) | 43 (53.8) | 15 (45.5) | 0.59 | 50 (51.5) | 41 (44.6) | 11 (55.0) | 0.53 |
| ≥ 0.100 | 108 (51.4) | 19 (38.8) | 45 (51.1) | 36 (55.4) | 50 (53.2) | 37 (46.2) | 18 (54.5) | 47 (48.5) | 51 (55.4) | 9 (45.0) | |||
| MMR status | |||||||||||||
| dMMR | 17 (8.1) | 6 (12.2) | 3 (3.4) | 7 (10.8) | 0.09 | 9 (9.6) | 7 (8.8) | 1 (3.0) | 0.59 | 5 (5.2) | 11 (12.0) | 1 (5.0) | 0.21 |
| pMMR | 193 (91.9) | 43 (87.8) | 85 (96.6) | 58 (89.2) | 85 (90.4) | 73 (91.2) | 32 (97.0) | 92 (94.8) | 81 (88.0) | 19 (95.0) | |||
| No. of FOLFOX cycles | |||||||||||||
| Median | 11 | 11 | 10 | 11 | 0.90 | 12 | 10 | 11 | 0.09 | 11 | 10 | 12 | 0.40 |
Abbreviations: ERCC1, Excision repair cross-complementing group 1; XRCC1, X-ray repair cross-complementing group 1; GSTP1, glutathione-S-transferase P1; dMMR, defective DNA Mismatch Repair; pMMR, proficient DNA Mismatch Repair. The cut-off value of LNR between metastatic and examined lymph nodes correponded to the median. The p values have been determined using the Chi² test or Fisher's exact test, as appropriate. The number of FOLFOX cycles according to the 3 possible genotypes of each gene was compared using a non-parametric Kruskal-Wallis test.
Unknown data: ERCC1 polymorphism for eight patients; XRCC1 polymorphism for three patients; GSTP1 polymorphism for one patient.
The primary endpoint was DFS, defined as the time between the date of surgery and the first event (local or distant disease recurrence or death from any cause, whichever occurred first). Patients who were alive and relapse-free at the last contact were censored at the last follow-up date.
Univariate and multivariate Cox proportional hazard regression models were used to estimate the hazard ratio (HR) and 95% confidence interval (CI). The following variables were examined in univariate analyses for their relation with DFS: age, sex, tumor location, differentiation grade, bowel perforation or obstruction, tumor stage, LNR, MMR status and polymorphisms in ERCC1-118, XRCC1-399 and GSTP1-105, alone or in combination for ERCC1 and XRCC1. Yet, the ERCC1 and XRCC1 combining variable was included in the multivariate model in order to take into account the significant interaction (p < 0.05) found between ERCC1-118 and XRCC1-399 polymorphisms. MMR status was defined by immunohistochemistry as previously described (18). All variables potentially associated with DFS and having a univariate p value ≤ 0.10 were included in multivariate analyses. The multivariate Cox model was constructed according to the "one variable for 10 events" rule. The discriminatory capacity was tested using the Harrell's concordance index. Harrell's C statistic was used to estimate the proportion of correct predictions. Harrell's C index ranges from 0.5 (no discrimination) to 1 (perfect discrimination). P values < 0.05 were considered as statistically significant. All analyses were performed using Stata V11 software (StataCorp LP, College Station, TX). The cutoff date for this analysis was December 2010.
Results
Patient characteristics
A total of 210 patients were included in this study. The median number of FOLFOX cycles received was 10 for the whole population and there was no statistically significant difference in the median number of cycles received by patients according to the genotypes of any of the 3 genes analyzed (Table 1). The median follow-up was 47.3 months (95% CI, 42.8 - 48.7). At the end of follow-up, 48 patients (22.8%) had relapsed or died in the whole study population. The 3-year DFS rate was 78.3% (95% CI, 71.8 - 83.4). The clinicopathological characteristics of patients according to ERCC1-118, XRCC1-399 and GSTP1-105 polymorphisms are summarized in Table 1. The ERCC1-118, XRCC1-399 and GSPTP1-105 genotypes were successfully determined for 202, 207 and 209 patients, respectively. There was no significant correlation between clinicopathological characteristics and ERCC1-118, XRCC1-399 or GSTP1-105 polymorphisms (Table 1).
Association between the ERCC1, XRCC1 and GSTP1 polymorphisms and DFS
ERCC1-118. Among the 202 patients whose ERCC1 genotype was defined, 45 (22.3%) had relapsed or died (6 with the C/C genotype, 24 with C/T, and 15 with T/T). The 3-year DFS rate was 88.9% (95% CI, 75.3 - 95.2), 73.8% (95% CI, 63.0 - 81.9), and 78.8% (95% CI, 66.3 - 87.1) in patients with homozygous C/C, heterozygous C/T and homozygous T/T genotypes, respectively (log-rank test, p=0.12) (Fig.1A).
XRCC1-399. Among the 207 patients whose XRCC1 genotype was defined, 48 (23.2%) had relapsed or died (22 with the G/G genotype, 15 with G/A, and 11 with A/A). The 3-year DFS rate was 78.9% (95% CI, 68.9 - 86.0), 82.2% (95% CI, 71.3 - 89.3), and 66.0% (95% CI, 47.0 - 79.6) in patients with G/G, G/A and A/A genotypes, respectively (log-rank test, p=0.31) (Fig. 1B).
DFS of stage III colon cancer patients receiving FOLFOX according to ERCC1, XRCC1 and GSTP1 polymorphisms. Disease-free survival curves of stage III colon cancer patients receiving FOLFOX adjuvant chemotherapy according to ERCC1-118 (rs11615, c.354T>C) (A), XRCC1-399 (rs25487, c.1196G>A) (B) and GSTP1-105 (rs1695, c.313A>G) (C) polymorphisms. Survival curves were estimated with the Kaplan-Meier method and compared using the log-rank test.
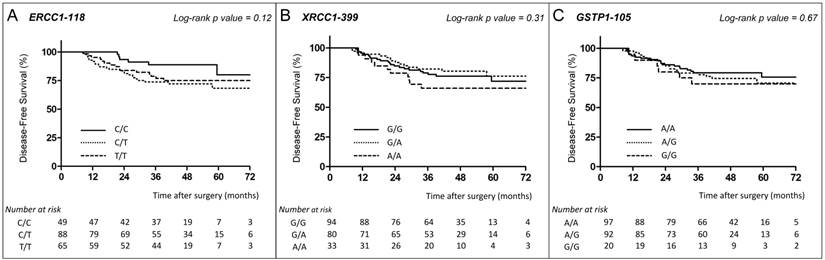
GSTP1-105. For the 209 patients whose GSTP1 genotype was defined, 48 (23%) had relapsed or died (20 with the A/A genotype, 22 with A/G, and 6 with G/G). The 3-year DFS rate was 79.4% (95% CI, 69.5 - 86.3), 79.0% (95% CI, 68.8 - 86.3), and 70.0% (95% CI, 45.1 - 85.3) in patients with A/A, A/G and G/G genotypes, respectively (log-rank test, p=0.67) (Fig. 1C).
Univariate analyses between covariates of interest and disease-free survival.
| n | Events | HR | IC 95% | p | Harrell's | |
|---|---|---|---|---|---|---|
| Age | 0.51 | |||||
| < 65 years | 101 | 22 | 1R | |||
| ≥ 65 years | 109 | 26 | 1.17 | [0.67 - 2.07] | 0.58 | |
| Sex | 0.53 | |||||
| Female | 99 | 21 | 1R | |||
| Male | 111 | 27 | 1.25 | [0.7 - 2.21] | 0.45 | |
| Tumor location | 0.52 | |||||
| Distal | 143 | 32 | 1R | |||
| Proximal | 67 | 16 | 1.13 | [0.62 - 2.05] | 0.70 | |
| Differentiation grade | 0.55 | |||||
| Well / Moderate | 182 | 39 | 1R | |||
| Poor | 28 | 9 | 1.87 | [0.91 - 3.87] | 0.09 | |
| Bowel perforation / obstruction | 0.52 | |||||
| Absent | 194 | 44 | 1R | |||
| Present | 16 | 4 | 1.47 | [0.53 - 4.1] | 0.46 | |
| Stage | 0.58 | |||||
| III A / III B (Tx, N1) | 131 | 23 | 1R | |||
| III C (Tx, N2) | 79 | 25 | 2.03 | [1.15 - 3.57] | 0.01 | |
| Lymph node ratio | 0.60 | |||||
| < 0.100 | 102 | 16 | 1R | |||
| ≥ 0.100 | 108 | 32 | 2.21 | [1.21 - 4.03] | 0.01 | |
| MMR status | 0.53 | |||||
| dMMR | 17 | 1 | 1R | |||
| pMMR | 193 | 47 | 4.11 | [0.57 -29.76] | 0.16 | |
| ERCC1-118 (rs11615) | 0.59 | |||||
| C/C | 49 | 6 | 1R | |||
| C/T | 88 | 24 | 2.48 | [1.02 - 6.08] | 0.14 | |
| T/T | 65 | 15 | 2.04 | [0.79 - 5.25] | ||
| XRCC1-399 (rs25487) | 0.56 | |||||
| G/G | 94 | 22 | 1R | |||
| G/A | 80 | 15 | 0.81 | [0.42 - 1.57] | 0.33 | |
| A/A | 33 | 11 | 1.47 | [0.71 - 3.04] | ||
| GSTP1-105 (rs1695) | 0.53 | |||||
| A/A | 97 | 20 | 1R | |||
| A/G | 92 | 22 | 1.21 | [0.66 - 2.22] | 0.67 | |
| G/G | 20 | 6 | 1.46 | [0.59 - 3.64] |
Abbreviations: R, reference; ERCC1, Excision repair cross-complementing group 1; XRCC1, X-ray repair cross-complementing group 1; GSTP1, glutathione-S-transferase P1; dMMR, defective DNA Mismatch Repair; pMMR, proficient DNA Mismatch Repair. The cut-off value of LNR between metastatic and examined lymph nodes correponded to the median.
Univariate and multivariate analyses of DFS
In univariate analysis, tumor stage (stage IIIC vs IIIA/B: HR=2.03; p=0.01), LNR (≥ 0.100 vs< 0.100: HR=2.21; p=0.01) and differentiation grade (poor vs well/moderate: HR=1.87; p=0.09) were potentially associated with improved DFS (threshold, 10 %) (Table 2). The genotype distribution of ERCC1-118, XRCC1-399 and GSTP1-105 polymorphisms did not show any significant correlation with DFS (Table 2). When grouping the heterozygous subjects with the homozygous genotype whose behavior was closest, we observed a trend toward improvement in DFS for ERCC1-118 (C/T + T/T vs C/C: HR=2.29; p=0.06), and for XRCC1-399 (A/A vs G/G + G/A: HR=1.61; p=0.16) (Table 3). With regard to GSTP1-105 polymorphism, no tendency could be ascribed for the heterozygous group; yet, pooling the A/G patients with the G/G or with the A/A group did not unravel any influence of GSTP1-105 polymorphism on DFS (data not shown).
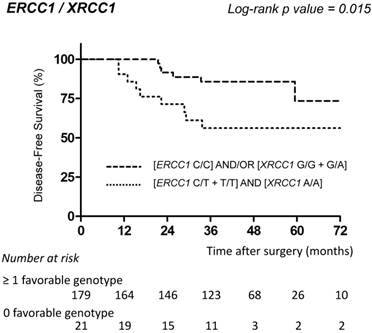
ERCC1-118 [C/C] and XRCC1-339 [G/G + G/A] were identified as favorable genotypes, whereas ERCC1-118 [C/T + T/T] and XRCC1-339 [A/A] represented unfavorable genotypes. In univariate analysis, patients with at least one favorable genotype had a significantly longer DFS compared to patients with no favorable genotype (0 vs ≥ 1 favorable genotype: HR=2.42; p=0.02) (Table 3; Fig. 2).
Multivariate analyses included all variables associated with DFS and having a p value ≤ 0.10 in the univariate analysis, i.e. tumor stage, LNR, differentiation grade, and the combination of ERCC1-118 and XRCC1-339 polymorphisms because of the significant interaction between ERCC1-118 and XRCC1-399 (p < 0.05). Multivariate analysis showed that none of these four variables was significantly associated with DFS (Table 4). Nevertheless, the combination of ERCC1-118 and XRCC1-399 polymorphisms was the variable associated with the lowest p value slightly above the threshold for statistical significance (0 vs ≥ 1 favorable genotype: HR=2.03; p=0.06) (Table 4).
DFS of stage III colon cancer patients receiving FOLFOX according to combined ERCC1/XRCC1 polymorphisms. Disease-free survival curves of stage III colon cancer patients receiving FOLFOX adjuvant chemotherapy according to the combination of ERCC1-118 (rs11615, c.354T>C) and XRCC1-399 (rs25487, c.1196G>A) polymorphisms. Survival curves were estimated with the Kaplan-Meier method and compared using the log-rank test.
Univariate analyses between ERCC1-118 and XRCC1-339 polymorphisms, separately or in combination, with disease-free survival.
| n | Events | HR | IC 95% | P | Harrell's | |
|---|---|---|---|---|---|---|
| ERCC1-118 (rs11615) | 0.57 | |||||
| C/C | 49 | 6 | 1R | |||
| C/T + T/T | 153 | 39 | 2.29 | [0.97 - 5.41] | 0.06 | |
| XRCC1-399 (rs25487) | 0.54 | |||||
| G/G + G/A | 174 | 37 | 1R | |||
| A/A | 33 | 11 | 1.61 | [0.82 - 3.12] | 0.16 | |
| ERCC1-118 (rs11615) AND XRCC1-339 (rs25487) | 0.56 | |||||
| ≥ 1 genotype favorable: [ERCC1 C/C] AND/OR [XRCC1 G/G + G/A] | 179 | 36 | 1R | |||
| 0 genotype favorable: [ERCC1 C/T + T/T] AND [XRCC1 A/A] | 21 | 9 | 2.42 | [1.16 - 5.03] | 0.02 |
Abbreviations: R, reference; ERCC1, Excision repair cross-complementing group 1; XRCC1, X-ray repair cross-complementing group 1.
Prognostic factors for disease-free survival in multivariate analysis.
| n | Events | HR | IC 95% | P | |
|---|---|---|---|---|---|
| Differentiation grade | |||||
| Well / Moderate | 174 | 36 | 1R | ||
| Poor | 26 | 9 | 1.58 | [0.74 - 3.40] | 0.24 |
| Stage | |||||
| III A / III B (Tx, N1) | 127 | 23 | 1R | ||
| III C (Tx, N2) | 73 | 22 | 1.28 | [0.62 - 2.64] | 0.51 |
| Lymph node ratio | |||||
| < 0.100 | 102 | 16 | 1R | ||
| ≥ 0.100 | 98 | 29 | 1.62 | [0.74 - 3.56] | 0.23 |
| ERCC1-118 (rs11615) AND XRCC1-399 (rs25487) | |||||
| ≥ 1 genotype favorable: [ERCC1 C/C] AND/OR [XRCC1 G/G + G/A] | 179 | 36 | 1R | ||
| 0 genotype favorable: [ERCC1 C/T + T/T] AND [XRCC1 A/A] | 21 | 9 | 2.03 | [0.96 - 4.28] | 0.06 |
Abbreviations: R, reference; ERCC1, Excision repair cross-complementing group 1; XRCC1, X-ray repair cross-complementing group 1. The cut-off value of LNR between metastatic and examined lymph nodes corresponded to the median.
A multivariate Cox model was constructed according to the "one variable for 10 events" rule. The discriminatory capacity was tested using the Harrell's concordance index. Harrell's C statistic was used to estimate the proportion of correct predictions. Harrell's C index ranges from 0.5 (no discrimination) to 1 (perfect discrimination).
Ten patients were excluded from the multivariate model because they had at least one missing polymorphism data (Harrell's C=0.65).
Discussion
The present study indicates that pharmacogenetic profiling of genes involved in DNA repair may influence clinical outcome of patients receiving FOLFOX adjuvant treatment for stage III colon cancer. This study focused on ERCC1-118, XRCC1-399 and GSTP1-105 genes polymorphisms because previously published data suggested a potential association between their SNPs and clinical outcome of patient treated with oxaliplatin-based chemotherapy for advanced CRC (7-12, 17). In our study, patients with no favorable genotype among ERCC1-118 and XRCC1-399 combination polymorphism have a shorter DFS upon FOLFOX adjuvant chemotherapy. However, the prognostic effect of this combined genotype analysis did not reach statistical significance in multivariate analysis, possibly because the number of patients in our series was not large enough, notably for the group of patients having no favorable genotype. Nevertheless, regarding the association with DFS in multivariate analysis, the p value obtained when combining ERCC1 and XRCC1 genotypes was lower than for the other well-recognized prognostic factors (differentiation grade, LNR or tumor stage). A second possible explanation is that the polymorphism might be in linkage disequilibrium with another factor influencing survival to platinum-based chemotherapy within the same gene or in a gene nearby, such as ERCC2, as previously suggested (11).
Several studies reported that SNP in ERCC1 could predict the clinical outcome for patients receiving platinum-based chemotherapy for treatment of various tumors, including colorectal, gastric or lung cancers (19). Previous data suggested that T allele compared to C allele at codon 118 of ERCC1 gene, both coding for asparagine, was associated with higher ERCC1 mRNA level resulting in resistance to platinum drugs (20). DFS benefit for colon cancer patients with ERCC1-118 C/C in our study may support these findings. However, other analysis performed in ovarian cancer cells lines did not confirm these results (21), and the relationship between ERCC1-118 SNP and clinical outcome of patients receiving oxaliplatin-based chemotherapy for advanced CRC remains controversial. Regarding patients with metastatic colorectal cancer, several studies reported that ERCC1-118 C/C homozygote genotype was associated with better clinical outcome (7, 9, 11, 12, 22-25), whereas others showed no correlation between genotype and survival (26, 27). In adjuvant setting, there is to date only one published study that evaluated the impact of ERCC1-118 SNP on survival of patients with stage III colon cancer (28). This retrospective study included 98 patients with stage III colon cancer receiving oxaliplatin-based (n = 53) or Mayo regimen (leucovorin and bolus fluorouracil) (n = 45) adjuvant chemotherapy. In univariate and multivariate survival analyses, ERCC1-118 polymorphism did not influence DFS, whatever the treatment administered (28).
Several authors attempted to identify combination of genomic variants occurring in several genes involved in DNA repair or metabolism drugs that may help predicting efficacy of platinum-based chemotherapy for CRC patients (11, 12, 22). Liang et al evaluated the prognostic value of ERCC1-118 and XRCC1-399 SNP, separately and in combination, in 113 patients receiving oxaliplatin-based chemotherapy for metastatic CRC (26). This retrospective study reported that the favorable genotypes of ERCC1-118 and XRCC1-399 polymorphisms, in combination but not individually, were an independent prognostic factor for disease control rate and overall survival (26). However while favorable genotypes for ERCC1-118 SNP were identical in both studies (C/C homozygous), the XRCC1-399 favorable genotype (A/A homozygous) differed from ours (G/G + G/A) (26).
The first study evaluating the value of XRCC1-399 as a pharmacogenetic marker was performed in 61 patients with advanced CRC receiving 5FU/oxaliplatin chemotherapy (8). The authors showed that 73% (8/11) of responders had a G/G genotype and 27% (3/11) were G/A, while 66% (33/50) of non-responders showed A/A or G/A genotypes (p=0.038) (8). Patients carrying at least one A allele were at a 5.2 fold increased risk to be resistant to oxaliplatin-based chemotherapy (8). More recently, another study reported similar results in 62 patients treated with oxaliplatin-based chemotherapy for metastatic CRC (9). In keeping, we observed in our study that DFS of patients with XRCC1-399 A/A genotype was shorter. However, other studies including between 61 and 166 patients failed to detect a significant prognostic impact of XRCC1-399 SNP in metastatic setting (11, 22, 24, 29). This indicates that XRCC1 genotype is probably not strongly associated with clinical outcome, though subtle differences have been reported in some studies.
GSTP1 is a member of a superfamily of metabolic enzymes involved in the detoxification of platinum compounds. Stoehlmacher et al have reported in two studies that GSTP1-105 G allele coding a Valine was associated with increased survival of patients with advanced CRC receiving 5-FU/oxaliplatin chemotherapy (10, 11). More recently, Chen et al confirmed these results in a series of Asian metastatic CRC patients, with higher tumor response and longer survival seen in the group of patients with at least one GSTP1-105 G allele (12). However, the association between GSTP1-105 polymorphism and efficacy in the context of oxaliplatin-based treatment remains controversial. Indeed, some studies did not confirm the prognostic value of GSTP1-105 SNP in metastatic (22, 25) or adjuvant (28) oxaliplatin-based treatment.
In conclusion, this study suggests that pharmacogenetic profiling of genes involved in DNA repair may have an impact on clinical outcome of patients receiving FOLFOX adjuvant treatment for stage III colon cancer. In univariate analysis, combination of ERCC1-118 and XRCC1-399 emerged as promising prognostic markers for DFS, although this combined genotype factor did not reach statistical significance in multivariate analysis. Future prospective studies with large sample sizes are required to confirm and extend our findings.
Acknowledgements
We thank Thomas Le Picard for his technical help. The authors thank all their colleagues who helped them for data collection.
Funding
Claire Goumard was a recipient of a grant of «Institut National du Cancer» (INCa). The funding source had no role in study design, data collection, data analysis, data interpretation or the writing of the report.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Raymond E, Chaney SG, Taamma A, Cvitkovic E. Oxaliplatin: a review of preclinical and clinical studies. Ann Oncol. 1998;9:1053-71
2. de Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J. et al. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J Clin Oncol. 2000;18:2938-47
3. Andre T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T. et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343-51
4. Kuebler JP, Wieand HS, O'Connell MJ, Smith RE, Colangelo LH, Yothers G. et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: results from NSABP C-07. J Clin Oncol. 2007;25:2198-204
5. Chaney SG, Campbell SL, Bassett E, Wu Y. Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit Rev Oncol Hematol. 2005;53:3-11
6. Zaanan A, Meunier K, Sangar F, Flejou JF, Praz F. Microsatellite instability in colorectal cancer: from molecular oncogenic mechanisms to clinical implications. Cell Oncol (Dordr). 2011;34:155-76
7. Yin M, Yan J, Martinez-Balibrea E, Graziano F, Lenz HJ, Kim HJ. et al. ERCC1 and ERCC2 polymorphisms predict clinical outcomes of oxaliplatin-based chemotherapies in gastric and colorectal cancer: a systemic review and meta-analysis. Clin Cancer Res. 2011;17:1632-40
8. Stoehlmacher J, Ghaderi V, Iobal S, Groshen S, Tsao-Wei D, Park D. et al. A polymorphism of the XRCC1 gene predicts for response to platinum based treatment in advanced colorectal cancer. Anticancer Res. 2001;21:3075-9
9. Lv H, Li Q, Qiu W, Xiang J, Wei H, Liang H. et al. Genetic polymorphism of XRCC1 correlated with response to oxaliplatin-based chemotherapy in advanced colorectal cancer. Pathol Oncol Res. 2012;18:1009-14
10. Stoehlmacher J, Park DJ, Zhang W, Groshen S, Tsao-Wei DD, Yu MC. et al. Association between glutathione S-transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst. 2002;94:936-42
11. Stoehlmacher J, Park DJ, Zhang W, Yang D, Groshen S, Zahedy S. et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344-54
12. Chen YC, Tzeng CH, Chen PM, Lin JK, Lin TC, Chen WS. et al. Influence of GSTP1 I105V polymorphism on cumulative neuropathy and outcome of FOLFOX-4 treatment in Asian patients with colorectal carcinoma. Cancer Sci. 2010;101:530-5
13. McNeil EM, Melton DW. DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res. 2012;40:9990-10004
14. Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48-63
15. Ban N, Takahashi Y, Takayama T, Kura T, Katahira T, Sakamaki S. et al. Transfection of glutathione S-transferase (GST)-pi antisense complementary DNA increases the sensitivity of a colon cancer cell line to adriamycin, cisplatin, melphalan, and etoposide. Cancer Res. 1996;56:3577-82
16. de Gramont A, Louvet C, Andre T, Tournigand C, Krulik M. A review of GERCOD trials of bimonthly leucovorin plus 5-fluorouracil 48-h continuous infusion in advanced colorectal cancer: evolution of a regimen. Groupe d'Etude et de Recherche sur les Cancers de l'Ovaire et Digestifs (GERCOD). Eur J Cancer. 1998;34:619-26
17. Viguier J, Boige V, Miquel C, Pocard M, Giraudeau B, Sabourin JC. et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res. 2005;11:6212-7
18. Zaanan A, Cuilliere-Dartigues P, Guilloux A, Parc Y, Louvet C, de Gramont A. et al. Impact of p53 expression and microsatellite instability on stage III colon cancer disease-free survival in patients treated by 5-fluorouracil and leucovorin with or without oxaliplatin. Ann Oncol. 2010;21:772-80
19. Kirschner K, Melton DW. Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res. 2010;30:3223-32
20. Park DJ, Zhang W, Stoehlmacher J, Tsao-Wei D, Groshen S, Gil J. et al. ERCC1 gene polymorphism as a predictor for clinical outcome in advanced colorectal cancer patients treated with platinum-based chemotherapy. Clin Adv Hematol Oncol. 2003;1:162-6
21. Yu JJ, Lee KB, Mu C, Li Q, Abernathy TV, Bostick-Bruton F. et al. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int J Oncol. 2000;16:555-60
22. Ruzzo A, Graziano F, Loupakis F, Rulli E, Canestrari E, Santini D. et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol. 2007;25:1247-54
23. Martinez-Balibrea E, Abad A, Aranda E, Sastre J, Manzano JL, Diaz-Rubio E. et al. Pharmacogenetic approach for capecitabine or 5-fluorouracil selection to be combined with oxaliplatin as first-line chemotherapy in advanced colorectal cancer. Eur J Cancer. 2008;44:1229-37
24. Pare L, Marcuello E, Altes A, del Rio E, Sedano L, Salazar J. et al. Pharmacogenetic prediction of clinical outcome in advanced colorectal cancer patients receiving oxaliplatin/5-fluorouracil as first-line chemotherapy. Br J Cancer. 2008;99:1050-5
25. Boige V, Mendiboure J, Pignon JP, Loriot MA, Castaing M, Barrois M. et al. Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000-05. J Clin Oncol. 2010;28:2556-64
26. Liang J, Jiang T, Yao RY, Liu ZM, Lv HY, Qi WW. The combination of ERCC1 and XRCC1 gene polymorphisms better predicts clinical outcome to oxaliplatin-based chemotherapy in metastatic colorectal cancer. Cancer Chemother Pharmacol. 2010;66:493-500
27. Spindler KL, Andersen RF, Jensen LH, Ploen J, Jakobsen A. EGF61A>G polymorphism as predictive marker of clinical outcome to first-line capecitabine and oxaliplatin in metastatic colorectal cancer. Ann Oncol. 2010;21:535-9
28. Farina Sarasqueta A, van Lijnschoten G, Lemmens VE, Rutten HJ, van den Brule AJ. Pharmacogenetics of oxaliplatin as adjuvant treatment in colon carcinoma: are single nucleotide polymorphisms in GSTP1, ERCC1, and ERCC2 good predictive markers? Mol Diagn Ther. 2011;15:277-83
29. Chua W, Goldstein D, Lee CK, Dhillon H, Michael M, Mitchell P. et al. Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer. 2009;101:998-1004
Author contact
![]() Corresponding author: Françoise Praz, PhD, Centre de Recherche Saint-Antoine, INSERM-UPMC UMR_S 938, Bâtiment Kourilsky, 184 rue du Faubourg Saint-Antoine, 75571 Paris cedex 12, France. Telephone: 33 1 49 28 66 75; Fax: 33 1 43 43 10 65; E-mail : francoise.prazfr. And Aziz Zaanan, MD, PhD, Department of Gastroenterology and Digestive Oncology, Hôpital Européen Georges Pompidou, 20 rue Leblanc, 75015, Paris, France. Telephone: 33 1 56 09 50 64; Fax: 33 1 56 09 50 69; E-mail: aziz.zaananaphp.fr.
Corresponding author: Françoise Praz, PhD, Centre de Recherche Saint-Antoine, INSERM-UPMC UMR_S 938, Bâtiment Kourilsky, 184 rue du Faubourg Saint-Antoine, 75571 Paris cedex 12, France. Telephone: 33 1 49 28 66 75; Fax: 33 1 43 43 10 65; E-mail : francoise.prazfr. And Aziz Zaanan, MD, PhD, Department of Gastroenterology and Digestive Oncology, Hôpital Européen Georges Pompidou, 20 rue Leblanc, 75015, Paris, France. Telephone: 33 1 56 09 50 64; Fax: 33 1 56 09 50 69; E-mail: aziz.zaananaphp.fr.

 Global reach, higher impact
Global reach, higher impact