Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(12):3775-3782. doi:10.7150/jca.117916 This issue Cite
Research Paper
The impact of long non-coding RNA HOTTIP genetic variants on oral cancer progression and clinicopathological characteristics
1. Department of Dentistry, Changhua Christian Hospital, Changhua, Taiwan.
2. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan.
3. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan.
4. Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan.
5. Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan.
6. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan.
7. Program for Cancer Biology and Drug Discovery, China Medical University, Taichung, Taiwan.
8. Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung, Taiwan.
9. Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan.
10. School of Medicine, Chung Shan Medical University, Taichung, Taiwan.
11. Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung, Taiwan.
Received 2025-5-20; Accepted 2025-7-31; Published 2025-8-11
Abstract
Oral cancer is the sixth leading cause of cancer-related mortality worldwide. Recent studies suggest that long non-coding RNAs (lncRNAs) HOXA transcript at the distal Tip (HOTTIP) may influence oral cancer cell growth and invasion, but comprehensive genetic association studies evaluating the impact of HOTTIP single-nucleotide polymorphisms (SNPs) on oral cancer susceptibility, and clinicopathological features are lacking. In this study, we investigated the associations between SNPs in the HOTTIP gene and both oral cancer susceptibility and clinicopathological characteristics. A total of 1,192 controls and 1,205 oral cancer patients were genotyped for four HOTTIP SNPs—rs3807598, rs17501292, rs2067087, and rs1859168—using real-time polymerase chain reaction (PCR). Our results showed that among oral cancer patients aged 60 years or older, those carrying the HOTTIP rs3807598 "GC+CC" genotype had a significantly reduced risk of developing advanced clinical stage and lymph node metastasis. Additionally, carriers of the rs2067087 "CG+GG" polymorphic variants were associated with a lower risk of developing advanced clinical stages. In conclusion, our findings suggest that the HOTTIP rs3807598 and rs2067087 polymorphisms may serve as pivotal predictor for assessing oral cancer progression.
Keywords: oral cancer, HOTTIP, polymorphism
Introduction
Oral cancer is one of the most common cancers worldwide especially among males [1, 2]. In Taiwan, oral cavity and oropharynx cancers together rank sixth place in cancer incidence and the fifth place in males [3]. Tobacco smoking and smokeless tobacco use (direct or indirect exposure to tobacco products), heavy alcohol consumption, and betel quid chewing were suggested to be primary carcinogenic risk factors to oral cancer [4-6].
Long non-coding RNAs (lncRNAs) HOXA transcript at the distal Tip (HOTTIP) is located on human chromosome 7q15.2, and it is transcribed from the antisense strand at the 5' end of the HOXA gene cluster [7-9]. HOTTIP is mainly binds to the WDR5/MLL complex, driving histone H3 lysine 4 trimethylation and the transcriptional activation of the terminal gene HOXA to upregulate the expression of development-related genes [7, 10].
HOTTIP has been shown to promote cancer cell proliferation, invasion, epithelial-mesenchymal transition (EMT), and metastasis in several malignancies such as ovarian cancer, breast cancer, hepatocellular carcinoma and oral cancer [11-20]. For example, Li et al. reported that HOTTIP promotes oral cancer cell proliferation and migration by modulating microRNA-206 [16]. Moreover, Xiong et al. suggest that HOTTIP may act as an oncogene, contributing to oral cancer progression by miR-124-3p/HMGA2 axis through Wnt/β-catenin pathway [21]. Additionally, the HOTTIP single-nucleotide polymorphisms (SNPs) were indicated to be associated with cancer development and prognosis in various cancers such as breast cancer (BC) [22], colorectal cancer (CRC) [23, 24], gastric cancer (GC) [25, 26], gastrointestinal (GI) cancers [27], hepatocellular carcinoma (HCC) [28], lung cancer [29], neuroblastoma [30], and pancreatic cancer [31]. However, the associations of HOTTIP polymorphisms to oral cancer progression and clinicopathologic characteristics remained unclear. In this study, we examined four SNPs of HOTTIP rs3807598, rs17501292, rs2067087, and rs1859168, and try to elucidate their correlations to oral cancer susceptibility and clinicopathologic characteristics with environmental risk factors.
Materials and Methods
Study subjects
A total of 1205 male oral cancer patients and 1192 cancer-free controls were participated in our study. All the participants were recruited at Chung Shan Medical University Hospital, Taichung, Taiwan. For the demographic data, the age and gender were reported by each participant. The control group who enrolled in our study was those individuals who without self-reported diseases such as history of cancer of any sites. The informed written consent was provided to each patient who enrolled in this study. This project was approved by the institutional review board of Chung Shan Medical University Hospital (CSMUH No: CS1-21151).
Sample preparation and DNA extraction
For genomic DNA extraction, we collected the peripheral blood specimens from normal controls and oral cancer patients who participated in our study [32]. Each peripheral whole blood samples were preserved with EDTA containing tubes. The samples of peripheral whole blood were centrifuged under the settings of 3000 rpm, 10 minutes. The centrifuged buffy coats from the whole blood specimens were collected and further used to extract DNA. To acquire the DNA, the genomic DNA extraction assay was performed under the manufacturer's manual of QIAamp DNA blood mini kits. The DNA elution was completed with the Tris-EDTA (TE) buffer. Extracted DNA was applied for DNA template in further real-time polymerase chain reactions (PCRs).
Selection of HOTTIP SNPs
In our study, we selected four HOTTIP SNPs rs3807598, rs17501292, rs2067087, and rs1859168 based on the International HapMap Project database [33]. The SNP rs3807598 was included because previous studies have suggested that the HOTTIP rs3807598 polymorphism is associated with an increased risk of colorectal cancer (CRC) [24] and GC risk [26]. The rs17501292 variant was selected due to evidence indicating that it may improve overall survival (OS) in CRC patients, particularly in the ulcerative/invasive-type tumor subgroup [24]. The rs2067087 polymorphism was chosen because it has been reported to be associated with an elevated risk of CRC [24], increased susceptibility to HCC [28], and a potential link to GC risk [26]. Finally, rs1859168 was selected because the C > A polymorphism has been associated with reduced neuroblastoma susceptibility in Chinese children [30], while the CC genotype and C allele have been linked to increased BC risk [22]. Moreover, expression quantitative trait locus (eQTL) analysis revealed that the rs1859168 CC genotype was related to high expression of the HOTTIP gene of neuroblastoma in Chinese children [30]. Wang et al. reported that the rs2067087 and rs3807598 SNPs of HOTTIP are associated with gastric cancer risk, possibly by affecting the expression of mature HOTTIP [26].
HOTTIP SNPs genotyping
Assessment of allelic discrimination for the HOTTIP rs3807598 (assay IDs: C__30343054_10), rs17501292 (assay IDs: C__27835598_10), rs2067087 (assay IDs: C__15951120_10), and rs1859168 (assay IDs: C__11173652_30) SNP was performed with an ABI StepOne Software v2.3 Real-Time PCR System. The allelic discrimination for the four selected loci was evaluated using the TaqMan® SNP Genotyping Assay on an ABI StepOne™ Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) [34, 35]. Each 20 µL PCR reaction contained 10 µL of TaqMan® Genotyping Master Mix, 1 µL of 20× TaqMan® SNP Genotyping Assay Mix (containing allele-specific VIC® and FAM™ labeled probes and locus-specific primers), 10 ng of genomic DNA template, and nuclease-free water to adjust the final volume. The thermal cycling conditions were as follows: an initial denaturation step at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute. The analysis and calculation of the collected data of genotyping was processed with the SDS 7000 series software (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
The comparison of the age (years), betel quid chewing, cigarette smoking, alcohol drinking, tumor stage, tumor T status, lymph node status, metastasis, and cell differentiation between the oral cancer patients and the controls was evaluated with the student's t test or Chi-squared test. The association between the genotypic frequencies of HOTTIP and the clinicopathological features of oral cancer patients was assessed using multiple logistic regression models to calculate odds ratios (ORs) with 95% confidence intervals (CIs). A p < 0.05 was suggested to present statistically significant.
Results
The distribution of demographical characteristics in 1192 controls and 1205 male patients with oral cancer was listed in Table 1. In the current study, we observed that the distributions of age (years) < 60 was 774 (64.9%) in controls and 740 (61.4%) in oral cancer patients, and the age ≥ 60 in controls and oral cancer patients was 418 (35.1%) and 465 (38.6%), respectively. The distributions of environmental risk factors exposure between the controls and oral cancer patients were 198 (16.6%) and 849 (70.5%) in betel quid chewing (p < 0.001), 634 (53.2%) and 970 (80.5%) in cigarette smoking (p < 0.001), and 235 (19.7%) and 495 (41.1%) in alcohol drinking (p < 0.001), respectively.
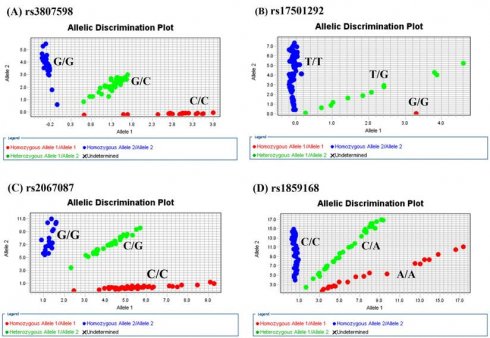
The genotype distributions of HOTTIP gene polymorphisms in 1192 controls and 1205 male patients with oral cancer were listed in Table 2. As shown in Figure 1, the most frequently occurring alleles were G/C for rs3807598, T/T for rs17501292, and C/G for rs2067087, C/A for rs1859168 (Figure 1). The adjusted odds ratios (AOR) with their 95% confidence intervals (CIs) were estimated by multiple logistic regression models. After adjustment for the effects of age, betel quid chewing, cigarette smoking, and alcohol drinking, no significant associations were found between the oral cancer patients and the controls (Table 2).
We further analyzed the odds ratio (OR) and 95% CIs of clinical statuses associated with genotypic frequencies of HOTTIP SNPs in male oral cancer patients. For HOTTIP rs3807598, no significant association was found between the HOTTIP rs3807598 polymorphisms and clinical statuses in oral cancer patients (Table 3). For HOTTIP rs2067087, in male oral cancer patients, a significant association was found between HOTTIP rs2067087 “CG+GG” genotype and metastasis [OR (95% CI): 0.115 (0.013-0.985); p = 0.017] (Table 4). Moreover, in male oral cancer patients with age ≥ 60, a significant association was found in those individuals who carried the HOTTIP rs3807598 polymorphic variant C, with a lower risk of clinical stage [OR (95% CI): 0.590 (0.393-0.886); p = 0.011], and lymph node metastasis [OR (95% CI): 0.649 (0.422-0.997); p = 0.047] (Table 5). Additionally, statistically significant association was found between the HOTTIP rs2067087 “CG+GG” polymorphic variants and clinical stage in male oral cancer patients with age ≥ 60 [OR (95% CI): 0.653 (0.444-0.962); p = 0.030] (Table 6). However, for HOTTIP 17501292 and rs1859168, no significant association was found between these SNPs in male oral cancer patients and patients with age ≥ 60 (data not shown).
The distributions of demographical characteristics in 1192 controls and 1205 male patients with oral cancer.
| Variable | Controls (N=1192) | Patients (N=1205) | p value |
|---|---|---|---|
| Age (yrs) | |||
| < 60 | 774 (64.9%) | 740 (61.4%) | p = 0.074 |
| > 60 | 418 (35.1%) | 465 (38.6%) | |
| Betel quid chewing | |||
| No | 994 (83.4%) | 356 (29.5%) | |
| Yes | 198 (16.6%) | 849 (70.5%) | p < 0.001* |
| Cigarette smoking | |||
| No | 558 (46.8%) | 235 (19.5%) | |
| Yes | 634 (53.2%) | 970 (80.5%) | p < 0.001* |
| Alcohol drinking | |||
| No | 957 (80.3%) | 710 (58.9%) | |
| Yes | 235 (19.7%) | 495 (41.1%) | p < 0.001* |
| Stage | |||
| I+II | 552 (45.8%) | ||
| III+IV | 653 (54.2%) | ||
| Tumor T status | |||
| T1+T2 | 571 (47.4%) | ||
| T3+T4 | 634 (52.6%) | ||
| Lymph node status | |||
| N0 | 813 (67.5%) | ||
| N1+N2+N3 | 392 (32.5%) | ||
| Metastasis | |||
| M0 | 1199 (99.5%) | ||
| M1 | 6 (0.5%) | ||
| Cell differentiation | |||
| Well differentiated | 193 (16.0%) | ||
| Moderately or poorly differentiated | 1012 (84.0%) |
* p value < 0.05 as statistically significant.
Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with HOTTIP genotypic frequencies.
| Variable | Controls (N=1192) (%) | Patients (N=1205) (%) | AOR (95% CI) | p value |
|---|---|---|---|---|
| rs3807598 | ||||
| GG | 302 (25.3%) | 347 (28.8%) | 1.000 (reference) | |
| GC | 617 (51.8%) | 571 (47.4%) | 0.818 (0.651-1.029) | p=0.087 |
| CC | 273 (22.9%) | 287 (23.8%) | 0.950 (0.725-1.244) | p=0.707 |
| GC+CC | 890 (74.7%) | 858 (71.2%) | 0.858 (0.692-1.064) | p=0.163 |
| rs17501292 | ||||
| TT | 1061 (89.0%) | 1065 (88.4%) | 1.000 (reference) | |
| TG | 128 (10.7%) | 136 (11.3%) | 1.117 (0.824-1.515) | p=0.477 |
| GG | 3 (0.3%) | 4 (0.3%) | 2.139 (0.410-11.150) | p=0.367 |
| TG+GG | 131 (11.0%) | 140 (11.6%) | 1.122 (0.830-1.516) | p=0.456 |
| rs2067087 | ||||
| CC | 430 (36.1%) | 442 (36.7%) | 1.000 (reference) | |
| CG | 573 (48.1%) | 511 (42.4%) | 0.825 (0.666-1.021) | p=0.077 |
| GG | 189 (15.8%) | 252 (20.9%) | 1.251 (0.951-1.648) | p=0.110 |
| CG+GG | 762 (63.9%) | 763 (63.3%) | 0.945 (0.775-1.152) | p=0.578 |
| rs1859168 | ||||
| CC | 432 (36.2%) | 480 (39.8%) | 1.000 (reference) | |
| CA | 591 (49.6%) | 533 (44.2%) | 0.795 (0.628-1.018) | p=0.056 |
| AA | 169 (14.2%) | 192 (16.0%) | 1.064 (0.796-1.422) | p=0.676 |
| CA+AA | 760 (63.8%) | 725 (60.2%) | 0.836 (0.687-1.018) | p=0.075 |
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, betel quid chewing, cigarette smoking, and alcohol drinking.
Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of HOTTIP rs3807598 in male oral cancer patients.
| Variable | GG (N=347) | GC+CC (N=858) | OR (95% CI) | p value |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I+II | 144 (41.5%) | 408 (47.6%) | 1.000 (reference) | p=0.056 |
| Stage III+IV | 203 (58.5%) | 450 (52.4%) | 0.782 (0.608-1.007) | |
| Tumor size | ||||
| ≦ T2 | 165 (47.6%) | 406 (47.3%) | 1.000 (reference) | p=0.942 |
| > T2 | 182 (52.4%) | 452 (52.7%) | 1.009 (0.786-1.296) | |
| Lymph node metastasis | ||||
| No | 221 (63.7%) | 592 (69.0%) | 1.000 (reference) | p=0.075 |
| Yes | 126 (36.3%) | 266 (31.0%) | 0.788 (0.606-1.024) | |
| Metastasis | ||||
| M0 | 343 (98.8%) | 856 (99.8%) | 1.000 (reference) | p=0.061 |
| M1 | 4 (1.2%) | 2 (0.2%) | 0.200 (0.037-1.099) | |
| Cell differentiated grade | ||||
| Well | 51 (14.7%) | 142 (16.6%) | 1.000 (reference) | p=0.427 |
| Moderate or poor | 296 (85.3%) | 716 (83.4%) | 0.869 (0.614-1.230) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
Discussion
In this study, we discovered the associations between the HOTTIP SNPs and oral cancer. Heavy alcohol consumption, betel quid chewing, and tobacco smoking are well-known risk factors responsible for oral cancer carcinogenesis, disease development, and progression [3, 36-38]. About 90% of oral cancer was oral squamous cell carcinoma (OSCC) [16, 39]. In our study, statistically significant associations of these risk factors including betel quid chewing, cigarette smoking, and alcohol drinking were found in 1205 male patients with oral cancer compared with 1192 controls, respectively. For the correlations of these risk factors to HOTTIP expression, a previous study has suggested that the lncRNAs HOTTIP was over expressed in extracellular vesicles (EVs) from smokers and NSCLC patients [40], and HOTTIP was found to be overexpressed in squamous cell carcinoma and in smokers [41]. Therefore, although the information of alcohol consumption, betel quid chewing, and their synergistic effect combined with tobacco smoking to HOTTIP expression remained unclear till date, and the lncRNAs HOTTIP was suggested to be involved in disease development and progression, proliferation, migration, and invasion of oral cancer [15, 16, 42]. It can be proposed that the tobacco smoking may play a vital role to influence the HOTTIP expression in OSCC patients.
Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of HOTTIP rs2067087 in male oral cancer patients.
| Variable | CC (N=442) | CG+GG (N=763) | OR (95% CI) | p value |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I+II | 190 (43.0%) | 362 (47.4%) | 1.000 (reference) | p=0.134 |
| Stage III+IV | 252 (57.0%) | 401 (52.6%) | 0.835 (0.767-1.037) | |
| Tumor size | ||||
| ≦ T2 | 206 (46.6%) | 365 (47.8%) | 1.000 (reference) | p=0.680 |
| > T2 | 236 (53.4%) | 398 (52.2%) | 0.952 (0.753-1.204) | |
| Lymph node metastasis | ||||
| No | 298 (67.4%) | 515 (67.5%) | 1.000 (reference) | p=0.978 |
| Yes | 144 (32.6%) | 248 (32.5%) | 0.997 (0.776-1.280) | |
| Metastasis | ||||
| M0 | 437 (98.9%) | 762 (99.9%) | 1.000 (reference) | p=0.017* |
| M1 | 5 (1.1%) | 1 (0.1%) | 0.115 (0.013-0.985) | |
| Cell differentiated grade | ||||
| Well | 77 (17.4%) | 116 (15.2%) | 1.000 (reference) | p=0.312 |
| Moderate or poor | 365 (82.6%) | 647 (84.8%) | 1.177 (0.858-1.613) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
* p value < 0.05 as statistically significant.
Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of HOTTIP rs3807598 in male oral cancer patients with age > 60.
| Variable | GG (N=136) | GC+CC (N=329) | OR (95% CI) | p value |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I+II | 53 (39.0%) | 171 (52.0%) | 1.000 (reference) | p=0.011* |
| Stage III+IV | 83 (61.0%) | 158 (48.0%) | 0.590 (0.393-0.886) | |
| Tumor size | ||||
| ≦ T2 | 66 (48.5%) | 159 (48.3%) | 1.000 (reference) | p=0.969 |
| > T2 | 70 (51.5%) | 170 (51.7%) | 1.008 (0.676-1.504) | |
| Lymph node metastasis | ||||
| No | 88 (64.7%) | 243 (73.9%) | 1.000 (reference) | p=0.047* |
| Yes | 48 (35.3%) | 86 (26.1%) | 0.649 (0.422-0.997) | |
| Metastasis | ||||
| M0 | 135 (99.3%) | 328 (99.7%) | 1.000 (reference) | p=0.518 |
| M1 | 1 (0.7%) | 1 (0.3%) | 0.412 (0.026-6.628) | |
| Cell differentiated grade | ||||
| Well | 22 (16.2%) | 54 (16.4%) | 1.000 (reference) | p=0.950 |
| Moderate or poor | 114 (83.8%) | 275 (83.6%) | 0.983 (0.572-1.689) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
* p value < 0.05 as statistically significant.
Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of HOTTIP rs2067087 in male oral cancer patients with age > 60.
| Variable | CC (N=160) | CG+GG (N=305) | OR (95% CI) | p value |
|---|---|---|---|---|
| Clinical Stage | ||||
| Stage I+II | 66 (41.3%) | 158 (51.8%) | 1.000 (reference) | p=0.030* |
| Stage III+IV | 94 (58.8%) | 147 (48.2%) | 0.653 (0.444-0.962) | |
| Tumor size | ||||
| ≦ T2 | 76 (47.5%) | 149 (48.9%) | 1.000 (reference) | p=0.782 |
| > T2 | 84 (52.5%) | 156 (51.1%) | 0.947 (0.646-1.389) | |
| Lymph node metastasis | ||||
| No | 107 (66.9%) | 224 (73.4%) | 1.000 (reference) | p=0.137 |
| Yes | 53 (33.1%) | 81 (26.6%) | 0.730 (0.482-1.107) | |
| Metastasis | ||||
| M0 | 159 (99.4%) | 304 (99.7%) | 1.000 (reference) | p=0.642 |
| M1 | 1 (0.6%) | 1 (0.3%) | 0.523 (0.032-8.418) | |
| Cell differentiated grade | ||||
| Well | 29 (18.1%) | 47 (15.4%) | 1.000 (reference) | p=0.452 |
| Moderate or poor | 131 (81.9%) | 258 (84.6%) | 1.215 (0.731-2.020) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
* p value < 0.05 as statistically significant.
Allelic discrimination plot obtained for the HOTTIP SNP. (A) rs3807598, (B) rs17501292, (C) rs2067087, and (D) rs1859168 using a TaqMan assay. The x-and y-axes indicate the fluorescence values of the VIC and FAM dyes, respectively, while the dots are individual sample points.”
We further examined the correlations of the HOTTIP genotypic frequencies to oral cancer susceptibility. Although these HOTTIP SNPs have not been previously investigated in oral cancer specifically, their known associations with cancer-related outcomes in other epithelial malignancies [22-26, 29], and the functional importance of HOTTIP in oral cancer biology suggest a plausible role in oral carcinogenesis. However, in our study, no statistically significant associations were found between the oral cancer patients and the controls, suggesting a limited disease susceptibility and carcinogenic effect of HOTTIP polymorphisms in oral cancer development. Intriguingly, after we analyzed clinical statuses associated with the HOTTIP genotypic frequencies among male oral cancer patients and patients with age ≥ 60, we found that the HOTTIP rs3807598 “GC+CC” polymorphic variants were associated with lower risk of clinical stage and lymph node metastasis, and the HOTTIP rs2067087 “CG+GG” genotype were associated with lower risk of metastasis in male oral cancer patients and lower risk of clinical stage in male oral cancer patients with age ≥ 60, respectively.
Generally, aging was considered as the most important risk factor of malignant disease and was suggested to be the largest risk factor for the development of cancer [43-45]. Most cancers were found to arise in individuals over the age of 60 [46]. The ageing microenvironment may influence tumor progression [46], and molecular alterations in tumors may differ among patients of different ages [47]. Compared with these results, in our study, the clinical statuses associated with genotypic frequencies of HOTTIP rs3807598 and rs2067087 polymorphisms were different in male oral cancer patients and patients with age ≥ 60, suggesting the possible change and influences of ageing microenvironment to tumor progression and molecular alterations with age in these oral cancer patients. For the correlations between the HOTTIP rs3807598 and rs2067087 to disease or cancer, some studies have associated the HOTTIP rs3807598 and rs2067087 with increased disease susceptibility and cancer risk such as CRC [24], and GC [26]. However, in HCC, the HOTTIP rs2067087 was suggested to be associated with increased HCC risk, while the HOTTIP rs3807598 variant genotype was found to show significantly longer survival time in HBV negative subgroup, and no significant associations between the HOTTIP rs3807598 and HCC risk was found in the same study [28]. Another study which focused on Hirschsprung disease has suggested a negative association between the lncRNAs HOTTIP rs3807598 C > G and risk of Hirschsprung disease [48]. Taken together, although the lncRNAs HOTTIP has demonstrated to be associated with oncogenic regulation in cancer progression in various cancers including oral cancer [7, 8, 15, 16, 19, 26-28, 41], and the HOTTIP rs3807598 and rs2067087 were suggested to be associated with cancer risk [24, 26]. However, the inconsistency of HOTTIP rs3807598 and rs2067087 to cancer risk, disease susceptibility, and survival time still observed [28, 48]. One possible mechanism to explain these inconsistencies is that the HOTTIP was suggested to be functioned as a competing endogenous RNA (ceRNA). In renal cell carcinoma (RCC), it was reported that the HOTTIP down-regulation attenuated RCC cell proliferation, migration, and invasion, which could be rescued by miR-506 down-regulation [30]. In acute myocardial infarction (AMI), HOTTIP knockdown markedly promoted cardiomyocyte growth and inhibited cardiomyocyte apoptosis in vitro, and miR-92a-2 overexpression could significantly enhance the protective effect of HOTTIP knockdown against AMI through partially inhibiting c-Met expression [49]. Therefore, it might be the ageing microenvironment and the vital regulatory role of HOTTIP ceRNA which contribute to the better prognosis of HOTTIP rs3807598 and rs2067087 polymorphisms in male oral cancer patients with age ≥ 60 in our study. However, future well-designed studies are required to elucidate the detailed mechanisms and inconsistencies between the HOTTIP polymorphism in oral cancer and other diseases.
A key limitation of our study is the absence of HOTTIP expression data in cancer versus control samples. Due to the design of the current study, we were unable to collect tissue specimens or preserve whole-blood RNA for expression analysis. Nevertheless, previous studies have consistently shown that HOTTIP is significantly upregulated in oral cancer tissues compared to adjacent normal tissues, supporting its potential oncogenic role in oral cancer [50]. Therefore, validating the clinical relevance of our genetic findings through expression profiling is warranted and will be a focus of future investigations.
In conclusion, our study first revealed the associations of HOTTIP polymorphisms to oral cancer disease susceptibility and clinical statuses. In male oral cancer patients with age ≥ 60, patients who carried the HOTTIP rs3807598 “GC+CC” genotype was associated with lower risk of clinical stage and lymph node metastasis, while carriers with HOTTIP rs2067087 polymorphic “CG + GG” genotype was associated with lower risk of clinical stage, respectively. The HOTTIP 3807598 and rs2067087 polymorphisms may provide as possible predictor to evaluate oral cancer disease progression and prognosis.
Acknowledgements
We thank the Human Biobank of Chung Shan Medical University Hospital, Taichung, Taiwan for specimen preparation. This study was supported by Chung Shan Medical University and Changhua Christian Hospital (CSMU-CCH-113-07). This study was also funded by China Medical University, Taiwan (CMU108-S-42).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Noggle B, Cheng H, Sarkar M. Oral Cancer Incidence Among Adult Males With Current or Former Use of Cigarettes or Smokeless Tobacco: Population-Based Study. JMIR Cancer. 2024;10:e51936
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
3. Yang YH. Current advances of oral cancer in Taiwan. Oral Dis. 2024
4. Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP. et al. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics. 2017;7:1088-99
5. Su CW, Chang YC, Chien MH, Hsieh YH, Chen MK, Lin CW. et al. Loss of TIMP3 by promoter methylation of Sp1 binding site promotes oral cancer metastasis. Cell Death Dis. 2019;10:793
6. Hsieh MJ, Lin CW, Su SC, Reiter RJ, Chen AW, Chen MK. et al. Effects of miR-34b/miR-892a Upregulation and Inhibition of ABCB1/ABCB4 on Melatonin-Induced Apoptosis in VCR-Resistant Oral Cancer Cells. Mol Ther Nucleic Acids. 2020;19:877-89
7. Wang DT, Luo J, Feng HJ, Wang YY. Serum HOTTIP expression is upregulated in nasopharyngeal carcinoma patients and predicts poor prognosis. Braz J Otorhinolaryngol. 2025;91:101471
8. Feng H, Zhao F, Luo J, Xu S, Liang Z, Xu W. et al. Long non-coding RNA HOTTIP exerts an oncogenic function by regulating HOXA13 in nasopharyngeal carcinoma. Mol Biol Rep. 2023;50:6807-18
9. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904-14
10. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120-4
11. Han S, Jin X, Liu Z, Xing F, Han Y, Yu X. et al. The long noncoding RNA HOTTIP promotes breast cancer cell migration, invasiveness, and epithelial-mesenchymal transition via the Wnt-β-catenin signaling pathway. Biochem Cell Biol. 2019;97:655-64
12. Liu J, Hu HB, Liu YM, Li FX, Zhang LP, Liao ZM. LncRNA HOTTIP promotes the proliferation and invasion of ovarian cancer cells by activating the MEK/ERK pathway. Mol Med Rep. 2020;22:3667-76
13. Sun Q, Zhang SY, Zhao JF, Han XG, Wang HB, Sun ML. HIF-1α or HOTTIP/CTCF Promotes Head and Neck Squamous Cell Carcinoma Progression and Drug Resistance by Targeting HOXA9. Mol Ther Nucleic Acids. 2020;20:164-75
14. Tang Q, Li X, Chen Y, Long S, Yu Y, Sheng H. et al. Solamargine inhibits the growth of hepatocellular carcinoma and enhances the anticancer effect of sorafenib by regulating HOTTIP-TUG1/miR-4726-5p/MUC1 pathway. Mol Carcinog. 2022;61:417-32
15. Basavarajappa DS, Padam KSR, Chakrabarty S, An NK, Radhakrishnan R. The regulatory role of HOX interacting lncRNA in oral cancer-An in silico analysis. J Oral Pathol Med. 2022;51:684-93
16. Li N, Dong H, Xia Q, Wang X. Long-chain non-coding RNA HOTTIP enhances oral cancer cell proliferation and migration capacity by down-regulating miR-206. J buon. 2021;26:762-8
17. Xiong L, Tang Y, Tang J, Liu Z, Wang X. Downregulation of lncRNA HOTTIP Suppresses the Proliferation, Migration, and Invasion of Oral Tongue Squamous Cell Carcinoma by Regulation of HMGA2-Mediated Wnt/beta-Catenin Pathway. Cancer Biother Radiopharm. 2020;35:720-30
18. Huang GZ, Wu QQ, Zheng ZN, Shao TR, Lv XZ. Identification of Candidate Biomarkers and Analysis of Prognostic Values in Oral Squamous Cell Carcinoma. Front Oncol. 2019;9:1054
19. Yin X, Yang W, Xie J, Wei Z, Tang C, Song C. et al. HOTTIP Functions as a Key Candidate Biomarker in Head and Neck Squamous Cell Carcinoma by Integrated Bioinformatic Analysis. Biomed Res Int. 2019;2019:5450617
20. Jiang H, Zhou L, Shen N, Ning X, Wu D, Jiang K. et al. M1 macrophage-derived exosomes and their key molecule lncRNA HOTTIP suppress head and neck squamous cell carcinoma progression by upregulating the TLR5/NF-κB pathway. Cell Death Dis. 2022;13:183
21. Xiong L, Tang Y, Tang J, Liu Z, Wang X. Downregulation of lncRNA HOTTIP Suppresses the Proliferation, Migration, and Invasion of Oral Tongue Squamous Cell Carcinoma by Regulation of HMGA2-Mediated Wnt/β-Catenin Pathway. Cancer Biother Radiopharm. 2020;35:720-30
22. Abdelaleem OO, Shaker OG, AbdelHafez MN, Abdelghaffar NK, Eid HM, Zaidan M. et al. The Influence of rs1859168 Polymorphism on Serum Expression of HOTTIP and Its Target miR-615-3p in Egyptian Patients with Breast Cancer. Biomolecules. 2021 11
23. Ali MA, Shaker OG, Ezzat EM, Gaber SN, Hassan EA, Abdelwahed MY. et al. Association Between rs1859168/HOTTIP Expression Level and Colorectal Cancer and Adenomatous Polyposis Risk in Egyptians. J Interferon Cytokine Res. 2020;40:279-91
24. Lv Z, Xu Q, Sun L, Wen J, Fang X, Xing C. et al. Four novel polymorphisms in long non-coding RNA HOTTIP are associated with the risk and prognosis of colorectal cancer. Biosci Rep. 2019 39
25. Abdi E, Latifi-Navid S, Zahri S, Kholghi-Oskooei V, Mostafaiy B, Yazdanbod A. et al. SNP-SNP interactions of oncogenic long non-coding RNAs HOTAIR and HOTTIP on gastric cancer susceptibility. Sci Rep. 2020;10:16763
26. Wang BG, Li YZ, Ding HX, Lv Z, Xu Q, Yuan Y. HOTTIP polymorphism may affect gastric cancer susceptibility by altering HOTTIP expression. Biosci Rep. 2020 40
27. Hamid UZ, Sim MS, Guad RM, Subramaniyan V, Sekar M, Fuloria NK. et al. Molecular Regulatory Roles of Long Non-coding RNA HOTTIP: An Overview in Gastrointestinal Cancers. Curr Mol Med. 2022;22:478-90
28. Wang BG, Xu Q, Lv Z, Fang XX, Ding HX, Wen J. et al. Association of twelve polymorphisms in three onco-lncRNA genes with hepatocellular cancer risk and prognosis: A case-control study. World J Gastroenterol. 2018;24:2482-90
29. Gong WJ, Yin JY, Li XP, Fang C, Xiao D, Zhang W. et al. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37:8349-58
30. Zhang T, Yin H, Guo J, Chang J, Li M, He J. et al. HOTTIP rs1859168 C > A polymorphism reduces neuroblastoma susceptibility in Chinese children. Eur J Pediatr. 2024;184:104
31. Hu P, Qiao O, Wang J, Li J, Jin H, Li Z. et al. rs1859168 A > C polymorphism regulates HOTTIP expression and reduces risk of pancreatic cancer in a Chinese population. World J Surg Oncol. 2017;15:155
32. Chen YT, Lin CW, Chou YE, Su SC, Chang LC, Lee CY. et al. Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma. J Cell Mol Med. 2023;27:1144-52
33. International HapMap C. The International HapMap Project. Nature. 2003;426:789-96
34. Yeh JC, Chen YT, Chou YE, Su SC, Chang LC, Chen YL. et al. Interactive effects of CDKN2B-AS1 gene polymorphism and habitual risk factors on oral cancer. J Cell Mol Med. 2023;27:3395-403
35. Lu HJ, Chuang CY, Su CW, Chen MK, Yang WE, Yeh CM. et al. Role of TNFSF15 variants in oral cancer development and clinicopathologic characteristics. J Cell Mol Med. 2022;26:5452-62
36. Wang CP, Yu KJ, Chen TC, Tsai MS, Kang CJ, Chien CY. et al. Multistate oral carcinogenesis-A prospective cohort study and a parallel case-control study in Taiwan. Oral Oncol. 2025;162:107210
37. Rich BJ, Samuels SE, Azzam GA, Kubicek G, Freedman L. Oral Cavity Squamous Cell Carcinoma: Review of Pathology, Diagnosis, and Management. Crit Rev Oncog. 2024;29:5-24
38. Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang CY, Tang CH. et al. A novel melatonin-regulated lncRNA suppresses TPA-induced oral cancer cell motility through replenishing PRUNE2 expression. J Pineal Res. 2021;71:e12760
39. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-63
40. Wu F, Yin Z, Yang L, Fan J, Xu J, Jin Y. et al. Smoking Induced Extracellular Vesicles Release and Their Distinct Properties in Non-Small Cell Lung Cancer. J Cancer. 2019;10:3435-43
41. Navarro A, Moises J, Santasusagna S, Marrades RM, Vinolas N, Castellano JJ. et al. Clinical significance of long non-coding RNA HOTTIP in early-stage non-small-cell lung cancer. BMC Pulm Med. 2019;19:55
42. Padam KSR, Basavarajappa DS, Shenoy US, Chakrabarty S, Kabekkodu SP, Hunter KD. et al. In silico interaction of HOX cluster-embedded microRNAs and long non-coding RNAs in oral cancer. J Oral Pathol Med. 2022;51:18-29
43. Montegut L, Lopez-Otin C, Kroemer G. Aging and cancer. Mol Cancer. 2024;23:106
44. Sedrak MS, Cohen HJ. The Aging-Cancer Cycle: Mechanisms and Opportunities for Intervention. J Gerontol A Biol Sci Med Sci. 2023;78:1234-8
45. The importance of aging in cancer research. Nat Aging. 2022; 2: 365-6.
46. Fane M, Weeraratna AT. How the ageing microenvironment influences tumour progression. Nat Rev Cancer. 2020;20:89-106
47. Chatsirisupachai K, Lagger C, de Magalhaes JP. Age-associated differences in the cancer molecular landscape. Trends Cancer. 2022;8:962-71
48. Zheng Y, Zhuo Z, Xie X, Lu L, He Q, Zhong W. Negative Association Between lncRNA HOTTIP rs3807598 C>G and Hirschsprung Disease. Pharmgenomics Pers Med. 2020;13:151-6
49. Wang B, Ma L, Wang J. LncRNA HOTTIP Knockdown Attenuates Acute Myocardial Infarction via Regulating miR-92a-2/c-Met Axis. Cardiovasc Toxicol. 2022;22:352-64
50. Zhang H, Zhao L, Wang YX, Xi M, Liu SL, Luo LL. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumour Biol. 2015;36:8805-9
Author contact
![]() Corresponding authors: Ying-Erh Chou, PhD. or Cheng-Chen Huang, MD, PhD. School of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: intointo814com (Ying-Erh Chou); E-mail: cshy1319org.tw (Cheng-Chen Huang).
Corresponding authors: Ying-Erh Chou, PhD. or Cheng-Chen Huang, MD, PhD. School of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: intointo814com (Ying-Erh Chou); E-mail: cshy1319org.tw (Cheng-Chen Huang).

 Global reach, higher impact
Global reach, higher impact