Impact Factor
ISSN: 1837-9664
J Cancer 2025; 16(9):2997-3014. doi:10.7150/jca.111037 This issue Cite
Research Paper
LncRNA HOXB-AS4 Promotes Tumor Malignant Phenotype in Head and Neck Squamous Cell Carcinoma and Serves as a Prognosis Marker
1. Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China.
2. Department of Blood Transfusion, The Third Xiangya Hospital of Central South University, Changsha, China.
3. National Institution of Drug Clinical Trial, Xiangya Hospital, Central South University, Changsha, China.
4. China National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China.
5. Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China.
# Contributed equally.
Received 2025-1-24; Accepted 2025-5-23; Published 2025-7-1
Abstract
Background: Head and neck squamous cell carcinoma (HNSCC) is the most common malignant tumor in the head and neck with a high suicide rate. Numerous studies have indicated that lncRNAs play a significant role in tumor occurrence and development, and that they may serve as promising diagnostic markers and therapeutic targets. The lncRNA HOXB-AS4 is substantially expressed in HNSCC; this work aimed to clarify the role of HOXB-AS4 in HNSCC and to further investigate its potential mode of action.
Methods: In this study, bioinformatics analysis was utilized to identify differentially expressed lncRNAs from RNA-seq data in the TCGA-HNSCC data set. The effect of lncRNA on HNSCC cell function was assessed using cell function tests. The probable downstream target mRNA of lncRNA was discovered after analyzing the differential mRNA and reviewing the literature. Mass spectrometry was utilized to investigate the signaling pathways it may control. RT-qPCR and western blotting were employed to confirm its regulatory action.
Results: HOXB-AS4 was abnormally overexpressed in HNSCC, which was related with poor clinical characteristics and prognosis, as well as promoting HNSCC cell migration, invasion, proliferation, and clone formation. The elevated expression of HOXB-AS4 in HNSCC may play a role in tumor promotion by influencing the HOXB7 gene located on the same chromosome, thereby activating the phosphorylation of AKT.
Conclusions: HOXB-AS4 may promote malignancy in HNSCC by controlling the HOXB7/AKT pathway.
Keywords: Head and neck squamous cell carcinoma, HOXB-AS4, HOXB7, AKT, p-AKT
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the most common malignant tumor in the head and neck, originating from the mucosal epithelium of the oral cavity, nasopharynx, and larynx [1]. According to data from The Global Cancer Observatory (GLOBOCAN) for 2020, the incidence and mortality rate of HNSCC, mainly associated with betel nut chewing, smoking, alcohol consumption, bacterial infections, HPV infection, and ultraviolet radiation, have shown higher in South-Central Asia, Eastern Europe, and Western Europe. Additionally, in all regions, the morbidity and mortality rates for men are consistently higher than those for women [2,3]. HNSCC not only affects the vital physiological functions of breathing, chewing, and swallowing, but it also predisposes patients to psychological issues due to their appearance. As a result, in addition to direct tumor-related deaths, HNSCC patients have a greater suicide incidence than other tumor patients, second only to pancreatic cancer [4]. More than 60% of HNSCC patients are in late stages when they are first diagnosed, with recurrence rates as high as 40-60% after multimodal treatments such as surgery, radiation, and chemotherapy, and patients with distant metastases have an exceedingly low 5-year survival rate [5,6]. Nowadays, it is still necessary to research on the putative molecular processes of HNSCC development and metastasis in order to develop novel strategies for early tumor identification and treatment.
Long non-coding RNAs (lncRNAs) are RNAs longer than 200 nt that do not encode proteins and make up a major component of the genomes of complex organisms. Broadly defined lncRNAs include RNAs transcribed by RNA polymerase I (Pol I), Pol II, and Pol III, as well as RNAs generated from processed introns. Now, it has been proposed that non-coding RNAs be divided into three groups to better describe lncRNAs, which are longer than 500 nt and primarily originate from Pol II [7]. The number of currently identified lncRNAs is gradually increasing and now surpasses 20,000 [8], most of which are functionally unknown, with only a few recognized to regulate a wide range of biological activities via various molecular mechanisms [7]. The majority of lncRNAs with better characterizations are linked to the regulation of gene transcription and have many modes of action on transcriptional regulatory processes [9,10]. Some lncRNAs interact spatially with neighboring mRNA-expressed genes [11]. For example, when released from chromatin-associated transcription sites, the human lncRNA A-ROD enhances the expression of its neighboring protein-coding gene, DKK1 [12]. Some lncRNAs are also found near transcription factors that act as cis-transcriptional stabilizers [11]. For example, the mouse lncRNA Halr1, also known as linc-HOXA1, is located 40kb upstream of the homologous transcription factor gene hoxa1, and its depletion or promoter variation leads to increased expression of hoxa1 or other genes in the hoxa family [13,14]. Furthermore, some lncRNAs can function as transcriptional regulators [11].
As lncRNAs are continually being investigated, numerous studies have discovered their involvement in a range of pathological processes, highlighting research topics including cancer [15-17], cardiovascular disease, etc, and about one-third of the papers related with the keyword "lncRNA" in PubMed contain the phrase "cancer" [18]. Some of these researches have focused on the function of lncRNAs in HNSCC. For instance, the lncRNA RASAL2-AS1 plays a pro-cancer role in HNSCC by boosting METTL14-mediated methylation of m6A and influencing the downstream target gene LIS1 [19]. In addition, some other lncRNAs, including LINC01614, LINC00313, and HOXC13-AS, also have significant functions in HNSCC through various modes of action and can be employed as prognostic and diagnostic indicators for HNSCC [20-22]. However, the majority of lncRNAs involved in HNSCC have yet to be reported.
The Cancer Genome Atlas Program (TCGA) database has the distinct advantage of being able to research gene expression at a large sample level, thereby facilitating the discovery of new markers and therapeutic targets. At present, there have been some research articles by mining HNSCC data in TCGA database [23]. In this study, we employed a large amount of data on lncRNAs from the RNA sequencing data of the TCGA database to identify key molecules in HNSCC through TCGA data mining combined with Lasso regression analysis, and found that HOXB-AS4 is an independent prognostic marker for HNSCC. Further investigation of this molecule may aid in the development of novel diagnostic and therapeutic markers for HNSCC.
Materials and methods
Bioinformatics analysis
The TCGA database (http://cancergenome.nih.gov/) provided the RNA-seq data for the HNSCC, and the data were collated and standardized using R software. Download the other two data sets (GSE41613 and GSE42743) from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) as a validation set. The "Limma" package was used to assess differential lncRNA and mRNA between tumor and normal groups. Genes with |log2FC| > 5 and p.adj <0.05 were identified as differential genes. The "glmnet" package was used for LASSO regression to build the prognostic model, while the "survival", "ROCR", and "rms" packages were used for survival analysis, ROC curve, and nomogram. The BEST online database (https://rookieutopia.com/app_direct/BEST/) examined the relationship between the expression of HOXB-AS4 and the clinical pathological characteristics of HNSCC patients. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were carried out with the "clusterProfiler" program in R software. Mass spectrometry (MS) data were processed using the TOmicsVics program for programmable counter array (PCA) and GSEA software for Gene Set Enrichment Analysis. Immune infiltration analyses were performed using the ASSISTANT for Clinical Bioinformatics online tools (https://www.aclbi.com/static/index.html#/) and TCGA - HNSC data sets. Genomic alteration and pharmacological analyses were performed using the BEST website (https://rookieutopia.com/app_direct/BEST/).
Cell culture and transfection
The Cal-27 cell line was donated by the Department of Stomatology, the second hospital of Xiangya, the Central South University, which had previously been purchased from the American Type Culture Collection cell bank, and it was cultivated in DMEM (Thermo Fisher, gibco) media containing 10% fetal bovine serum (VivaCell, C04001-500) and 1% penicillin-streptomycin (Thermo Fisher, gibco). The SCC-9 cell line was purchased from Wuhan Pricella Biotechnology Co, and it was cultured in DMEM/F12 (Thermo Fisher, gibco) complete medium containing the same serum and antibiotic. Both cell lines were cultured in an incubator with 5% CO2 at 37°C and proved negative for mycoplasma. Transfection was performed when the cell density reached 60%. To complete the transfection, siRNA was combined with the transfection reagent in opti-MEM (Thermo Fisher, gibco) according to the Lipofectamine 3000 (Thermo Fisher Scientific, L3000015) instructions and then dropped into the cell culture dish. All siRNA was designed and synthesized by Guangzhou RiboBio Co. The sequences of siRNAs targeting HOXB-AS4 were as follows: siRNA-HOXB-AS4-1: GGTGACTCCCAAGGCCTGA, siRNA-HOXB-AS4-2: CGTTTATTGCCGGGTACTT, siRNA-HOXB-AS4-3: CCGTCTCCTTATCTCTTTA.
Detection of cell proliferation
After 48 hours of cell transfection, cells were digested with 0.25% trypsin-EDTA (Thermo Fisher, gibco) at 37°C for 2min, and terminated by the addition of complete medium with twice the volume of trypsin. The cells were then collected and counted, and a cell suspension was prepared at a concentration of 1×104 cells per ml, then added 200μl per well of the above cell suspension to 96-well plates, and the cells were incubated for 2 hours after the addition of CCK8 reagent (life-ilab, AC11L054) in every day for the next 0-6 days. The absorbance values were then measured using microplate reader, analyzed with GraphPad Prism 8, and plotted on line charts.
Detection of cell migration and invasion
The transwell assay was used to detect the migration and invasion ability of the cells, we should add the matrix gel (CORNING, 356234) into the chambers (CORNING, 353097) and keep it at 37°C for longer than two hours to make it polymerize into a gel for use. After 48 hours of transfection, cells were prepared into cell suspensions at concentrations of 4×104 cells per ml and 2×104 cells per ml using medium containing 2% FBS, then 200μl per well of above cell suspension was added to the chambers with and without matrix gel respectively to detect cell invasion and migration ability, while 800μl medium containing 15% FBS was added to the lower chamber to create a concentration difference between the upper and lower chambers to enable the cells to better move downward through the chambers. After 24 hours of incubation at 37°C, removed the chambers, washed twice with PBS (Thermo Fisher, gibco), and the cells were fixed with 4% paraformaldehyde (Sangon Biotech, Shanghai) for 15 minutes, followed by staining with crystal violet (beyotime, C0121) for 15 minutes at room temperature, washing off the excess crystal violet, wiping off the stromal gel on the upper surface with a cotton ball, drying and then photographing three fields of view per well under the inverted microscope, observing and counting the number of cells that had passed through the chambers, and analyzing and plotting bar charts using GraphPad Prism 8.
Clone formation assay
After 48 hours of transfection, cells were prepared into a cell suspension at a concentration of 500 cells per ml using the complete medium, and 2ml of the above cell suspension was added to each well of a six-well plate, i.e., each well contained 1000 cells, and the culture was stopped when the number of individual cell clones reached more than 50 under the microscope. The cells were washed twice with PBS, fixed with 4% paraformaldehyde for 15 minutes, stained with crystal violet staining solution for 15 minutes at room temperature, protected from light, rinsed off the excess crystal violet with running water, dried, and counted using the Adobe Illustrator 2022 APP technology tool, then analyzed and plotted on bar charts using GraphPad Prism 8.
Quantitative Real-time PCR
Tumors and paracancerous tissues were collected from the Department of Head and Neck Surgery, Hunan Cancer Hospital. The conventional trizol method was used to extract RNA from tissue and cell samples. The RNA concentration was then determined and reverse-transcribed into cDNA in accordance with the instructions provided by the Reverse Transcription Kit (Roche). Real-time fluorescence quantitative PCR (RT-qPCR) was used to perform relative quantitative assays. The system consisted of 2 μl of the cDNA template, 1 μl of each upstream and downstream primer, 1 μl of water, and 5 μl of a 2× SYBR GREEN (Roche). RT-qPCR relative quantitative detection was performed using the LightCycler 480 II (Roche). The target gene's relative expression was calculated using β-actin as an internal reference. Q-PCR primers used were all synthesized by Sangon Biotech, and the sequences were: HOXB-AS4: forward 5'-TTTGTCTGCTCTGGACCGTT-3' and reverse 5'-TTTTGGTTCGGAGGCCTAGT-3'; β-actin: forward 5'-TCACCAACTGGGACGACATG-3' and reverse 5'-GTCACCGGAGTCCATCACGAT-3'; HOXB7: forward 5'-AGGAACTGACCGCAAACGAG-3' and reverse 5'-ATCTGTCTTTCCGTGAGGCA-3'; HOXB9: forward 5'-GGAGGCCGTGCTGTCTAATC-3' and reverse 5'-GATCCGGCCTCTCTTTGTCC-3'.
Western blot
After 72 hours of transfection, cell precipitates were collected, proteins were extracted using RIPA lysate (Beyotime, P0013E) supplemented with protease inhibitors, determined the protein concentration in accordance with the guidelines provided by the BCA Protein Concentration Assay Kit (Thermo Fisher, 23225), and computed the sample volume. After SDS-PAGE electrophoresis (Epizyme, PG212), the protein bands were transferred to the PVDF membrane (Merck Millipore, IPVH00010), which was then closed at room temperature for 1 hour with 5% skimmed milk powder, then immersed in the primary antibody at a dilution of 1:1000 and set at 4°C overnight. The PVDF membrane was washed in TBST the next day, and the corresponding secondary antibody was chosen based on the species of the primary antibody for incubation at room temperature for 1 hour, followed by another TBST wash, and color was developed on a chromatograph using the ECL Ultra Sensitive Chromatography Reagent (Thermo Fisher, A38554). Adobe Photoshop 2023 software was used to process and create the photographs. The main primary antibodies employed in the experiment were: AKT (proteintech, 10176-2-AP), Phospho-AKT (proteintech, 28731-1-AP), HOXB7 (ABclonal, A6925) Alpha Tubulin (proteintech, 11224-1-AP), GAPDH (Beijing ComWin Biotech, 01225); and secondary antibody were: Goat Anti-Rabbit IgG, HRP Conjugated (Beijing ComWin Biotech, 01334), Goat Anti-Mouse IgG, HRP Conjugated (Beijing ComWin Biotech, 01123).
Immunofluorescence
After 48 hours of cell transfection, the cells were allowed to form a monolayer of adherent cells on the cell crawler sheet at a density of roughly 60%. The cells on the crawler sheet were washed twice with PBS, fixed with 4% paraformaldehyde at room temperature for 15 minutes, washed, for this experiment is used to detect cytoskeletal molecules, it needs to use the immunostaining permeabilization solution Triton X-100 (Beyotime, P0096) to permeabilize at room temperature for 5 minutes, moisten with PBS for three times, and then block with immunostaining blocking solution (Beyotime, P0102) for 1h after permeabilization, and a primary antibody was added dropwise to the crawler sheet. The usual dilution ratio was 1:50, and the temperature was set to 4°C for overnight treatment. The film was washed twice with PBST the next day after rewarming, and the corresponding fluorescent secondary antibody was applied based on the species of primary antibody. After 1 hour of incubation at room temperature and light protection, the film was dyed with DAPI-containing sealer (Beyotime, P0131) and sealed. Each slide was photographed under the fluorescence microscope in 5 different fields of view at random. The primary antibodies employed in the experiment were: CL594-Phalloidin (Proteintech, PF00003), and fluorescent secondary antibodies were: AF488 donkey anti-mouse IgG (H+L) (Thermo Fisher 1975519).
Statistical analysis
The experimental results were analyzed with GraphPad Prism software (version 8.0; La Jolla, CA, USA). The differences between the two groups were calculated using t-tests. Significant differences between several data sets were calculated using a one-way analysis of variance (ANOVA). p < 0.05 indicated statistical significance. * p <0.05, ** p <0.01, *** p <0.001.
Results
Screening differentially expressed lncRNAs and building a prognostic model for HNSCC-associated lncRNAs
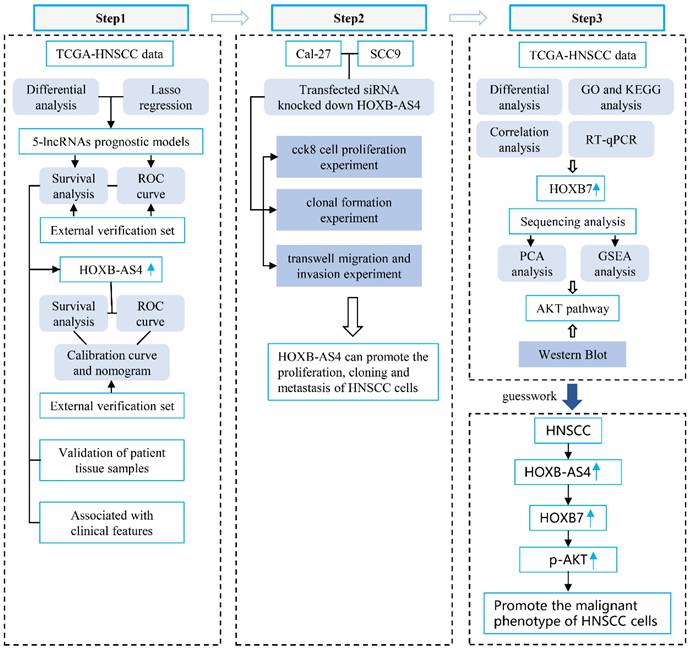
The transcriptome data for HNSCC were obtained from the Cance Genome Atlas (TCGA) database, and the differentially expressed genes between the cancer group and the normal group were obtained by comparing the transcriptome data of these two groups according to the principle of |log2FC| > 5, and all differentially expressed genes were visualized using volcano plots (Figure 1A).
Building a prognostic model for HNSCC-associated lncRNAs. (A) Volcano plots showing differential expression of lncRNAs between HNSCC and normal control groups. (B-C) Lasso regression screening and construction of prognostic model consisting of 5 lncRNAs. (D) Distribution of patient survival status between high- and low-risk groups and risk scores for the 5 lncRNAs. (E) Kaplan-Meier survival analysis between high- and low-risk groups. (F) The ROC curve shows the prediction probability of the prognostic model.
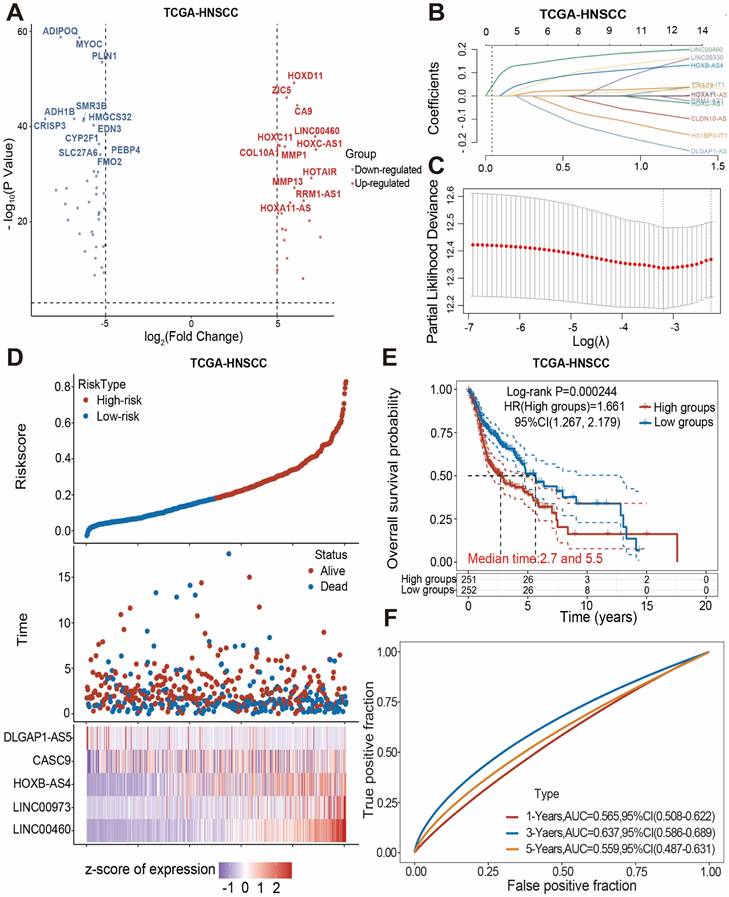
The blue dots on the left side represent differentially down-regulated genes in comparison to the control group, while the red dots on the right side represent differentially up-regulated genes. According to the data that combining the differentially expressed lncRNAs with the clinical prognostic information of patients in the TCGA database, five lncRNAs (DLGAP1-AS5, CASC9, HOXB-AS4, LINC00973, LINC00460) were screened using Lasso regression and constructed the HNSC-associated prognostic model (Figure 1B-C). The HNSCC patients in the TCGA database were divided into high-risk and low-risk subgroups based on the median risk score. As shown in Figure 1D, the high-risk group had a larger proportion of deaths, and HOXB-AS4, LINC00973, and LINC00460 had higher risk ratings in that group. Kaplan-Meier survival analyses revealed that risk ratings were related to HNSCC prognosis, with patients in the high-risk group having a worse prognosis (Figure 1E). ROC curves revealed that the prognostic risk model predicted 1-year, 3-year, and 5-year survival with AUC values of 0.565, 0.637, and 0.559, respectively (Figure 1F). The effect of the above prognostic model was validated using two external datasets, GSE41613 and GSE42743. The Kaplan-Meier survival analysis results confirmed the TCGA database results (Supplementary Figure 1), indicating that the prognostic model was more stable.
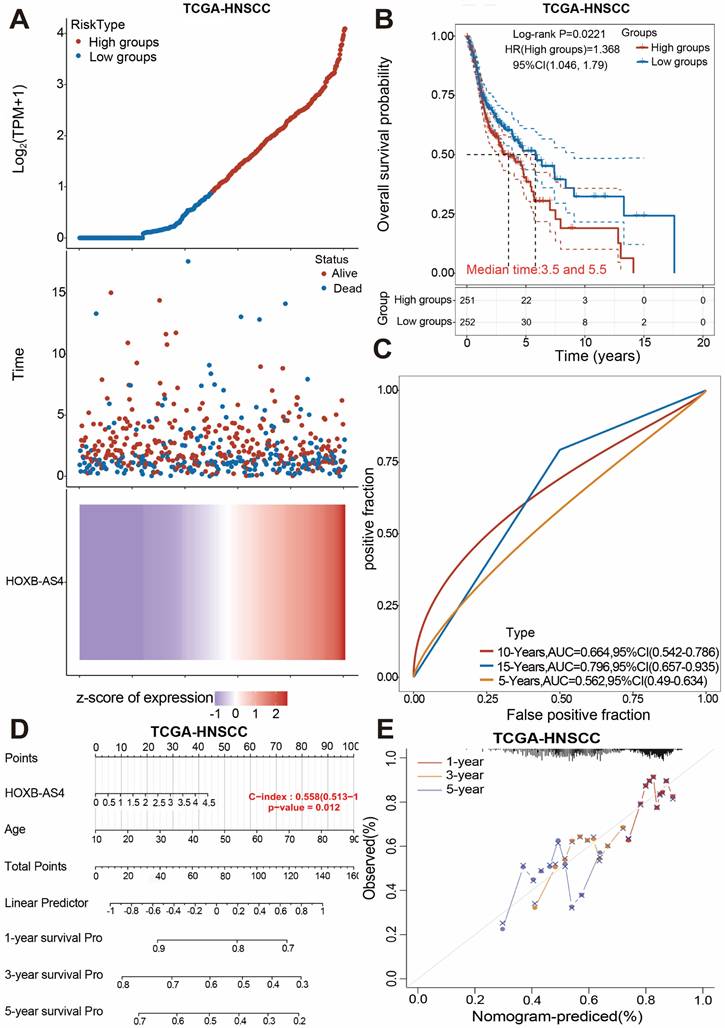
LncRNA HOXB-AS4 has better prognostic marker value in HNSCC
The human HOX family is divided into four subfamilies: A, B, C, and D, which are found on chromosomes 7,17,22,2, with a total of 39 members [24]. HOX genes are overexpressed in a variety of tumors and are strongly related to tumor stage and prognosis [25]. The analysis results in Figure 1 showed that HOXB-AS4 had a higher risk score in the cancer group, so we hypothesized that HOXB-AS4 might play a potential role in HNSCC. HNSCC patients were divided into two groups based on the median value of the HXOB-AS4 risk score, with 251 patients in the high-risk group and 252 patients in the low-risk group, and the survival distribution status of patients in the two groups revealed that the high-risk group had more deaths (Figure 2A). The Kaplan-Meier survival analysis showed that HOXB-AS4 was associated with the prognosis of HNSCC, and patients in the high-risk group had a poorer prognosis (Figure 2B). The ROC curves showed that the AUCs for predicting 5-year, 10-year, and 15-year survival based on HOXB-AS4 were 0.562,0.664, and 0.796, respectively (Figure 2C). A nomogram based on HOXB-AS4 expression level and age was created to assess the survival probability of HNSCC (Figure 2D), while the results of calibration curves showed that the predicted results of the nomogram were more consistent with the actual results (Figure 2E). The predictive effect of HOXB-AS4 in HNSCC was confirmed by Kaplan-Meier survival analyses of external datasets GSE41613 and GSE42743 (Supplementary Figure 2).
HOXB-AS4 is associated with worse clinicopathological features in HNSCC patients
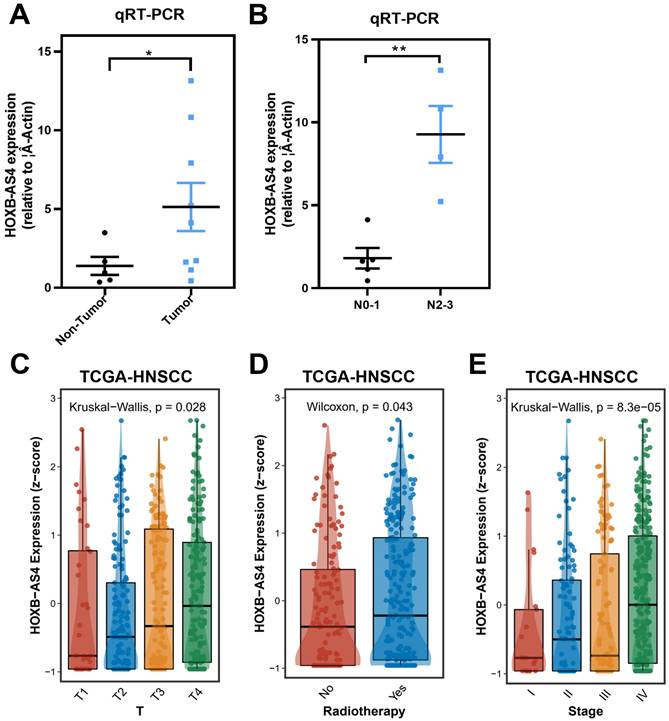
Tumor and paracancerous tissues of HNSCC patients were acquired from Hunan Cancer Hospital, and RT-qPCR validated the expression of HOXB-AS4 after extracting the tissue RNA. The results showed that it was much higher in tumor tissues than paracancerous tissues (Figure 3A). While we had also collected their N stage from some of these patients, and divided them into N0-1 AND N2-3 groups according to their N stage, the comparison result showed that the expression of HOXB-AS4 was higher in the more severe N stage group (Figure 3B).
The clinical information of HNSCC patients was downloaded from the TCGA database and the analytic results of the correlation between HOXB-AS4 expression and pathological features were shown in Figure 3C-E, patients who received radiotherapy had higher HOXB-AS4 expression than those who did not, and in the analysis of the T staging, the level of HOXB-AS4 expression increased gradually with the increase of the T staging, and in total clinical staging it was similarly observed that HOXB-AS4 expression was higher in more advanced tumor stages. These results suggest that HOXB-AS4 expression is up-regulated in HNSCC, which predicts a worse clinical stage and prognosis.
Silencing of HOXB-AS4 inhibits the ability of HNSCC metastasis and proliferation but does not act by regulating the cytoskeleton
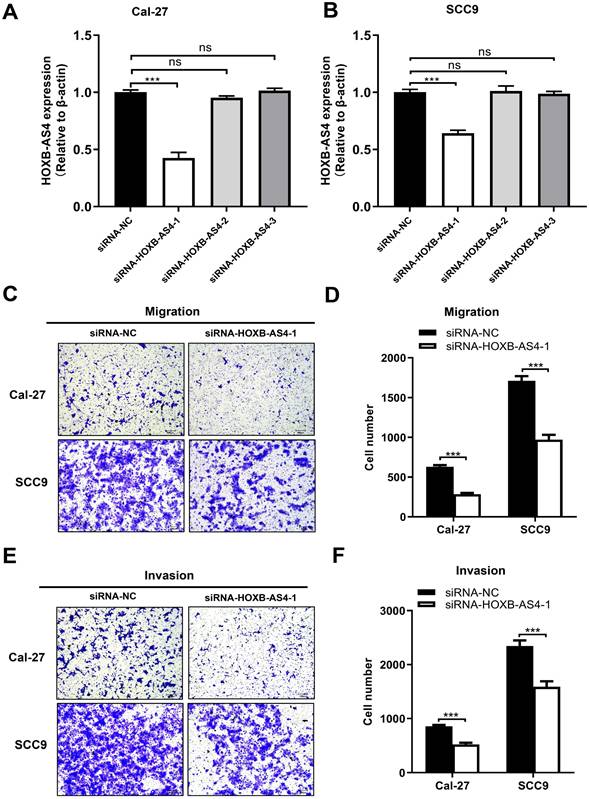
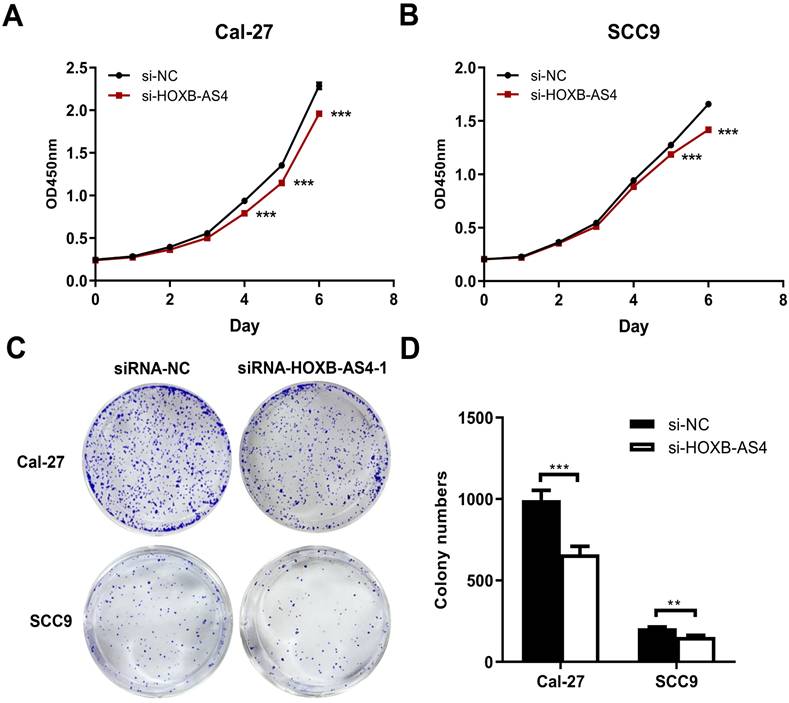
HOXB-AS4 in Cal-27 and SCC-9 cell lines was silenced using siRNA, and the effectiveness of the silencing was verified by RT-qPCR. As demonstrated in Figure 4A-B, hence it was chosen for the following tests. Silencing HOXB-AS4 significantly reduced the migration (Figure 4C-D) and invasion (Figure 4E-F) abilities of Cal-27 and SCC-9 cells, and the results of cck8 and clone formation experiments revealed that the two cell lines' proliferative and clone formation abilities were also reduced (Figure 5A-D). In summary, silencing HOXB-AS4 was able to inhibit the migration, invasion, proliferation, and clone formation ability of HNSCC cells.
To further explore the possible mechanisms by which HOXB-AS4 contributes to the malignant phenotype of HNSCC, we performed GO (S3A) and KEGG (S3B) enrichment analyses on both HOXB-AS4 high- and low-expressing subgroups and found that significant enrichment was observed in the cellular components assembly involved in morphogenesis, as well as regulation of actin cytoskeleton and focal adhesion. However, cellular immunofluorescence staining found no significant difference in the cytoskeleton and nucleus co-staining (S3C) of cells from control and silencing groups. This result suggests that HOXB-AS4 may also not function by regulating the cytoskeleton.
The prognostic value analysis of HOXB-AS4 in HNSCC. HNSCC patients were classified to high and low group according to their median HOXB-AS4 risk score. (A) Grouping, survival status distribution, and the HOXB-AS4 risk score. (B) Kaplan-Meier survival analysis. (C) ROC curve. (D-E) Nomogram and calibration curves.
Correlation analysis between HOXB-AS4 and clinicopathological characteristics of HNSCC patients. (A) RT-qPCR results from HNSCC patient tissue samples. (B) Comparison of HOXB-AS4 expression in patients with different N stages. (C-E) The relationship of HOXB-AS4 to T stage (C), radiotherapy (D), and total patient stage (E).
HOXB-AS4 may affect the AKT signaling pathway by regulating HOXB7 expression leading to a malignant phenotype in HNSCC cells
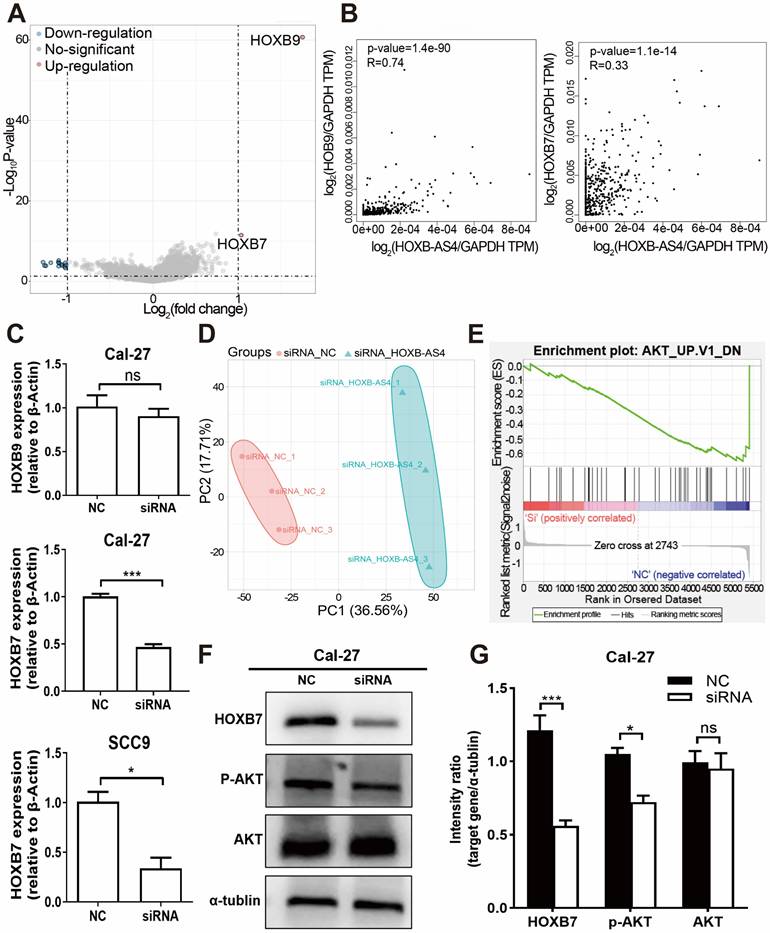
Downloading the mRNA sequencing data from the TCGA database for differential analysis revealed that HOXB7 and HOXB9, which are located at different positions on the same chromosome as HOXB-AS4, were up-regulated in the tumors (Fig. 6A). Additionally, the correlation analysis showed a positive correlation between HOXB-AS4 and the expression of both HOXB7 and HOXB9 (Fig. 6B). After silencing HOXB-AS4 using siRNA transfection of Cal-27 cells, the cells were collected for RNA extraction, reverse transcribed, and then analyzed by RT-qPCR to detect the expression levels of HOXB7 and HOXB9. The results showed that HOXB7 expression was reduced compared with the control group, while HOXB9 expression was not significantly different. In SCC9 cells, the expression of HOXB7 is also reduced after the knockdown of HOXB-AS4 (Figure 6C). Therefore, we hypothesized that HOXB-AS4 may act by affecting the mRNA HOXB7 of the same family on the same chromosome. Cell samples transfected with siRNA were collected and divided into experimental and control groups, each containing three biological replicates, for proteome analysis using liquid chromatography-tandem mass spectrometry (LC-MS). The scores were shown using principal component analysis (Fig. 6D), and it was discovered that the control and experimental samples were clearly separated from each other with large differences. The proteome data were then subjected to GSEA enrichment analysis to search for potential downstream pathways, and the disease was significantly enriched to the differentiated AKT signaling pathway (Figure 6E). Cal-27 cell samples were collected for protein extraction, and the results of western blot experiments showed that silencing of HOXB-AS4 decreased HOXB7 protein expression and AKT phosphorylation level (Figure 6F). Figure 6G is the quantification diagram of the western blot results. Based on the foregoing analysis and experimental data, we hypothesized that lncRNA HOXB-AS4 may alter the AKT signaling pathway by regulating mRNA HOXB7, resulting in the formation of a malignant phenotype and a poor prognosis in HNSCC.
HOXB-AS4 is related to HNSCC tumor immunity, stemness, and treatment resistance
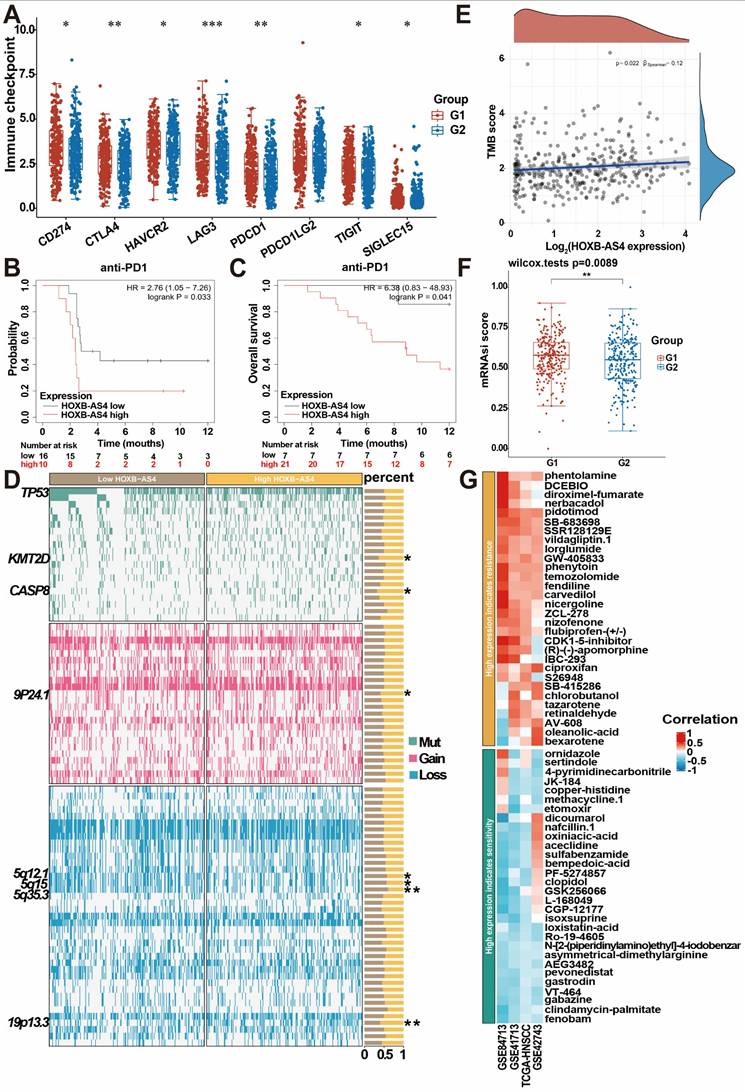
HNSCC patients in the TCGA database were divided into two groups, G1 (high HOXB-AS4 expression) and G2 (low HOXB-AS4 expression), based on the expression of HOXB-AS4, and analyzed potential differences in immune checkpoints between the two groups. The results indicated that the high HOXB-AS4 expression group exhibited elevated levels of immune checkpoints such as CD274, CTLA4, HAVCR2, LAG3, PDCD1, TIGIT, and SIGLEC15 compared to the low expression group (Figure 7A). This suggests that HOXB-AS4 may contribute to a pro-tumor effect by promoting immune escape in tumor cells. However, survival analysis revealed that patients in the high-expression group with anti-PD1 immunotherapy had poorer survival probability and overall survival rates than patients in the low-expression group also with anti-PD1 immunotherapy (Figure 7B-C). It is speculated that the reason for this may be due to the fact that high HOXB-AS4 expression also upregulates other immune checkpoints such as CTLA4, HAVCR2, LAG3, etc., which may play alternative roles and continue to promote tumor immune escape after PD1 blockade on the one hand, and on the other hand, it is possible that the complex tumor microenvironment containing various immune cells and signaling pathways collectively impact immune inhibitor efficacy. Genomic alteration analysis was conducted to compare mutations, amplifications, and deletions of genes in the HOXB-AS4 high-expression group with those in the low-expression group. The results revealed an increase in KMT2D and CASP8 mutations, which are related to the tumor immune microenvironment, as well as an amplification of chromosome 9p24.1, and deletions of chromosomes 5q12.1, 5q15, and 5q35.3 in the HOXB-AS4 high-expression group (Figure 7D). Tumor Mutation Burden (TMB) analysis indicated a positive correlation between HOXB-AS4 and TMB scores to some extent (Figure 7E). Moreover, mRNAsi score results showed that gene expression levels of tumor stem cells were higher in patients with high expression of HOXB-AS4 (Figure 7F), suggesting that HOXB-AS4 may enhance tumor stem cell properties and promote tumor progression, metastasis, and drug resistance in HNSCC. Pharmacological sensitivity analysis of the GDSC drug database for heatmapping demonstrated a significant positive correlation between high expression of HOXB-AS4 and resistance to certain chemotherapeutic drugs such as phentolamine, DCEBIO, diroximel-fumarate, nerbacadol, pidotimod, SBR128129E, vildagliptin, lorglumide, GW405833, phenytoin, temozolomide, fendiline, carvedilol, nigergoline, ZCL278, nizofenone, flurbiprofen-(+/-), CDK1-5-inhibitor, (R)-(-)-apomorphine, IBC293, ciproxifan, S26948, SB415286, chlorobutanol, tazarotene, retinaldehyde, AV-608, oleanolic-acid and bexarotene (Figure 7G).
Discussion
LncRNAs were once thought to be useless 'background noise' in the genome [26], but with continued exploration of lncRNAs, it has been discovered that lncRNAs play important and indispensable roles in the whole process of gene expression, including, but not limited to, chromosome modification, epigenetic regulation, transcriptional regulation, and translational regulation, etc. [27-31]. Through these regulatory functions, lncRNAs are involved in various biological processes including cell proliferation, cell cycle, apoptosis, and immune response [32-35]. Furthermore, numerous studies have shown that abnormal expression of lncRNAs plays a crucial role in the development of various tumors [36]. For example, LINC00853 was significantly up-regulated in gastric cancer patients' tissues and promoted the malignant progression of gastric cancer through the MAP17/PDZK1/AKT signaling pathway [37]; lncRNA VPS9D1-AS1 was highly expressed in tissue samples of patients with endometrial carcinoma (EC) and was demonstrated to play a pro-cancer role in endometrial cancer by negatively regulating miR-187-3p and positively regulating S100A4 endometrial cancer [38]; LINC00313 is highly expressed in HNSCC and affects the progression of HNSCC by influencing the process of epithelial-mesenchymal transition, and the expression of LINC00313 is also negatively correlated with the infiltration of immune cells [20]. In our study, we found that the lncRNA HOXB-AS4 was significantly overexpressed in HNSCC by analyzing the data from the TCGA database along with the tumors and normal tissues of patients with HNSCC, and functional experiments confirmed that its high expression correlated with the progression of the malignant phenotype of HNSCC. HOXB-AS4 might be an important marker for predicting the prognosis of HNSCC.
Effect of HOXB-AS4 on HNSCC cell migration and invasion. (A-B) The silencing impact of HOXB-AS4 siRNAs was detected by using RT-qPCR. (C) Transwell cell migration test to assess the effect of silencing HOXB-AS4 on HNSCC cell movement. (D) A statistical map of HNSCC cell movement following HOXB-AS4 silencing. (E) Transwell cell invasion test to assess the effect of silencing HOXB-AS4 on HNSCC cell invasion. (F) A statistical map of HNSCC cell invasion following HOXB-AS4 silencing.
The action mechanism of lncRNA is extremely complex and has not been completely clarified until now. However, numerous studies on the mechanism of lncRNA have been published. First, lncRNAs can be regulated at the epigenetic level by modifying RNA or DNA methylation and histone modification. For example, the lncRNA HOTAIR, which is expressed in the HOXC locus, suppresses HOXD gene expression by directing the H3K27 methylase Polycomb repressive complex 2 (PRC2) complex to the HOXD locus and then modifying chromatin modification in the region. As a result, it accelerates the malignant progression of metastasis and recurrence during carcinogenesis [39,40]. Second, lncRNAs can play the regulation both during and after transcription. For example, lncRNA ELDR is closely located downstream of epidermal growth factor (EGF), which is highly expressed in various cancers such as HNSCC and ovarian cancer, and activates the downstream molecule STAT3 by stabilizing EGF, thus ELDR plays a carcinogenic role and serves as a prognostic marker for various cancers [41-43]. Furthermore, lncRNAs can compete with miRNA for the mRNA binding site, a process known as competing endogenous RNAs (ceRNAs), inhibiting the regulation of miRNA for mRNA. For example, lncRNA-CDC6 can serve as the ceRNA of miR-215. It also impacts the expression of mRNA CDC6, which promotes breast cancer growth and metastasis, and it might be a predictive biomarker [44]. In our study, through the difference analysis of mRNA data of the HNSCC tumor and normal group in the TCGA database, it was found that HOXB7 and HOXB9, located on the same chromosome of HOXB-AS4, were significantly different. RT-qPCR results showed that the expression of HOXB7 was down-regulated after knocking down the HOXB-AS4, and the correlation analysis between them showed a positive correlation. These results suggest that HOXB-AS4 may play a role in promoting cancer in HNSCC by influencing the expression of HOXB7.
Effect of HOXB-AS4 on HNSCC cell proliferation and clonogenesis. (A-B) The cck8 cell proliferation test to assess the effect of HOXB-AS4 silencing on HNSCC cell proliferation. (C)The clonal formation test to assess the effect of HOXB-AS4 silencing on the clonal ability of HNSCC cells. (D) A statistical map of HNSCC cell clonal formation following HOXB-AS4 silencing.
HOXB-AS4 may activate the AKT signaling pathway by regulating HOXB7, resulting in malignant phenotypes. (A) Comparison of HNSCC mRNA differences in the TCGA database. (B) Correlation analysis of HOXB-AS4 and HOXB7 or HOXB9. (C) RT-qPCR confirmed the expression of HOXB7 and HOXB9 in Cal-27 cells after knocking down HOXB-AS4. (D) PCA score map for proteomic data. (E) GSEA enrichment analysis enhanced the differential AKT signaling pathway. (F) Western blot analysis indicated changes in HOXB7, AKT, and p-AKT levels. (G) The quantification diagram of the western blot results.
HOXB-AS4 plays a role in HNSC tumor immunity, stemness, and drug sensitivity. (A) Differential analysis of major immune checkpoints in the TCGA database. (B-C) Effect of HOXB-AS4 expression on survival probability and overall survival after anti-PD-1 immunotherapy. (D) Genomic alteration analysis. (E) Tumor mutational burden. (F) Tumor cell stemness index. (G) Drug sensitivity analysis.
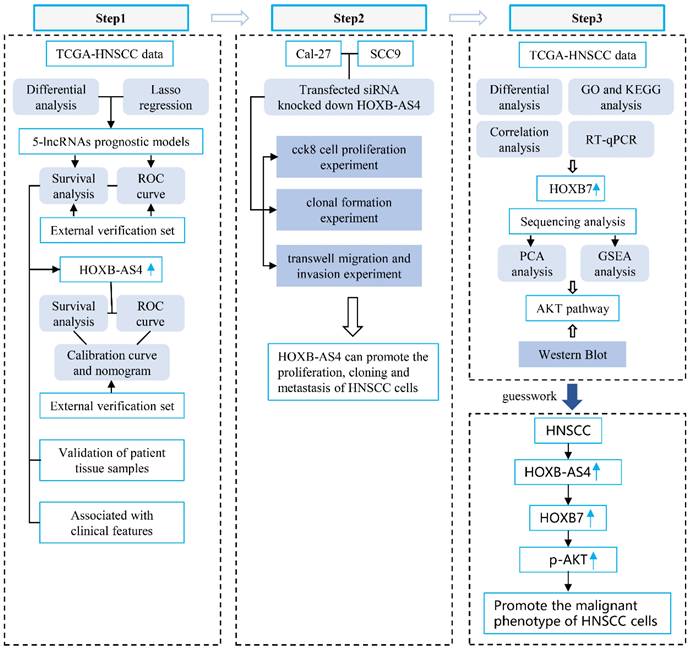
Flowchart.
HOXB7, which is located on human chromosome 17, is a key regulatory gene that can activate a range of carcinogenic pathways by influencing a variety of target molecules [45]. HOXB7 has been found to be overexpressed in a number of tumor tissues and is a valuable early diagnostic and prognostic marker. For example, HOXB7 is a carcinogenic factor that promotes gastric cancer development. Abnormally high HOXB7 expression in gastric cancer is associated with patient clinical characteristics, enhanced the ability of gastric cancer cells to proliferate, migrate, and invade, while inhibiting apoptosis, by activating the PI3KR3/AKT signaling pathway or up-regulating p-AKT and down-regulating PTEN [46]. In addition to inducing AKT signaling pathway, it can also promote the proliferation of gastric cancer cells by inducing the expression of MARKs [47]. HOXB7 is also overexpressed in colorectal cancer, which can activate the PI3K/AKT and MAPK pathways. It also plays a carcinogenic role in colorectal cancer, making it an essential prognostic marker [48]. Furthermore, HOXB7 is overexpressed in solid tumors such as esophageal cancer, intrahepatic cholangiocarcinoma, cutaneous squamous cell carcinoma, osteosarcoma, and pancreatic cancer, suggesting a cancer-promoting role [49-53]. A recent study using pan-cancer computer screening found that HOXB7 has value as a pan-cancer marker, and its expression in malignant tumors is also positively correlated with immune cell infiltration, immune regulatory genes, immune checkpoints, and TMB, which may be a potential immunotherapy indicator [54]. HOX proteins typically function by attaching to cofactors, such as HOXB7 in melanoma, which relies on the cofactor PBX to play a carcinogenic role [55]. Furthermore, HOXB7 can also play an important role in mutual regulation with microRNAs [56,57]. In our work, HOXB7 expression was down-regulated in HNSCC cell lines following the silencing of HOXB-AS4 expression. The results of mass spectrometry revealed that the AKT signaling pathway was significantly enriched, and the western blot experiment confirmed that when HOXB-AS4 was down-regulated, p-AKT was also significantly reduced.
The research on the mechanism of HOXB-AS4 in tumors includes: First, through bioinformatics analysis and the characteristics of lncRNA for model prediction, the prognostic value of HOXB-AS4 has been identified in two types of tumors, namely laryngeal squamous cell carcinoma (LSCC) and clear cell renal cell carcinoma (ccRCC), and in ccRCC, HOXB-AS4 is involved in ferroptosis and oxidative stress processes [58-60]. Second, in colorectal cancer (CRC), HOXB-AS4 competes with HDAC7 to bind miR-140-5p, thereby promoting the proliferation and migration of CRC cells through the ceRNA regulatory network pathway [61]. Therefore, to date, only one study has investigated the role of HOXB-AS4 in HNSCC. However, there are many studies exploring the role of lncRNA in the occurrence and progression of HNSCC. For example, LINC00313 may promote the migration of HNSCC cells by regulating the EMT pathway in HNSCC and is related to immune infiltration [62]; by binding to USF1, LINC00152 can enhance the transcription of MRPL52 mediated by USF1, indirectly up-regulating the expression of MRPL52 and promoting the growth of oral squamous cell carcinoma (OSCC) [63]; lncRNA FOXD1-AS1 can positively regulate the expression of FOXD1 through miR-369-3p and also increase the stability of FOXD1 mRNA by binding to ADAR protein together with FOXD1, thereby promoting the progression of OSCC through these two mechanisms [64]; lncRNA TPRG1-AS1, mediated by CCND1 expression, affects the EMT pathway through the miR-363-3p/MYO1B axis and plays a pro-cancer role in HNSCC [65], and other lncRNAs that act through the ceRNA network pathway in HNSCC include HOXC13-AS and SNHG1 [66,67]. In addition, most studies have mainly constructed prognostic models of HNSCC-related lncRNAs through model prediction [68-73]. In our study, we first discovered that HOXB-AS4 is highly expressed in HNSCC and can promote the malignant progression of HNSCC cells. In terms of mechanism exploration, we found the role of the HOXB-AS4/HOXB7/AKT axis. Thus, HOXB-AS4 may be a valuable prognostic biomarker in HNSCC, and the HOXB-AS4/HOXB7/AKT axis may be a potential therapeutic target for HNSCC.
Immune checkpoints play a crucial role in the self-regulation of the immune system, contributing to autoimmune tolerance under normal physiological conditions. They achieve this process by inhibiting T-cell responses or promoting T-cell depletion through binding to specific ligands, thus effectively terminating the immune response promptly [74]. However, in the tumor microenvironment, there is a potential for overexpression of immune checkpoints, leading to immune escape of tumor cells by inhibiting the immune response and ultimately promoting the proliferation and metastasis of tumor cells [75]. For example, the programmed cell death protein 1 (PD1) receptor, which can be expressed on the surface of tumor-infiltrating lymphocytes, binds to PD1 ligands on the surface of tumor cells (including PD-L1 and PD-L2, usually PD-L1), transmits negative regulatory signals, and causes apoptosis or loss of function of effector T cells that exert anti-tumor effects [76-78]. The cytotoxic T lymphocyte-associated antigen 4 (CTLA4) is the first clinically targeted immune checkpoint receptor expressed only in T cells. When the T-cell receptor (TCR) recognizes an antigen, CD28 amplifies the TCR signal and delivers signals to activate T cells by connecting to the ligands CD80 and CD86. CTLA4, which has the same ligand as CD28, inhibits T-cell activation by competing for binding to the same ligand [79]. The role of immunological checkpoints in tumor immunity is complex and needs further investigation. In addition, Lymphocyte activation gene 3 (LAG3) and T-cell membrane protein 3 (TIM3) are promising targets for inhibitors [80,81]. Currently, drugs developed against critical immune checkpoints are in clinical trials or are already being used in clinics as immune checkpoint inhibitors, which have become a significant tool in immunotherapy for malignancies [82,83]. However, response to single immunosuppressant therapy has been uneven; combinations with multiple immune checkpoint inhibitors or immunotherapy with targeted therapy have proven to be much more effective than single therapies [84,85]. In our study, high expression of HOXB-AS4 in HNSCC was associated with high expression of multiple immune checkpoints; however, there was no significant efficacy observed with anti-PD1 therapy. This suggests that a combination approach using multiple immune checkpoint inhibitors may be more effective.
At present, there are still shortcomings in our study. Due to the limitation of clinical sample size, the results of database analysis and cell experiment could not be well combined with more clinical data, so as to strengthen the potential value of HOXB-AS4 as a prognostic marker.
Conclusion
Based on bioinformatics analysis and experimental verification, our study verified that HOXB-AS4 was overexpressed in HNSCC, which can increase cell proliferation, migration, and invasion. We hypothesized that the function of promoting the malignant progression of HNSCC tumors might work by up-regulating HOXB7 and activating the AKT signaling pathway.
Supplementary Material
Supplementary figures.
Acknowledgements
Thanks to all the authors for their efforts on this article.
Funding
This study was supported by Hunan Provincial Natural Science Foundation of China (2023JJ30879, 2023JJ40411, 2024JJ5524), the natural sciences funding project of Changsha City(kq2208149), the Crucial Science and Technology Project of Hunan Province(2023ZJ1124), and the Hunan Cancer Hospital Climb Plan (ZX2021002-5).
Author contributions
All authors contributed to the study's conception and design. Yan Liu, Hao Tian, and Yanwei Luo designed this study. Yan Liu, Shanshan Lv, and Jie Guo collected samples. Shanshan Lv performed experiments. Lin Du and Jie Guo analyzed the data. Shanshan Lv wrote the original manuscript. Yanwei Luo, Yan Liu, and Tian Hao revised the manuscript. All authors read and approved the final manuscript.
Data availability statement
Data and materials will be provided upon request by contacting YL.
Ethical statement
All human participants in this study were following the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was approved by the Ethics Committee of Hunan Cancer Hospital (KYJJ-2024-060). All patients had agreed to participate in the work.
Informed consent
All patients had signed a written informed consent to participate in the work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jiménez-Labaig P, Rullan A, Braña I, Hernando-Calvo A, Moreno V, Doger B. et al. Intratumoral therapies in head and neck squamous cell carcinoma: A systematic review and future perspectives. Cancer Treat Rev. 2024;127:102746 doi: 10.1016/j.ctrv.2024.102746
2. Li Z, Liu Y, Huang X, Wang Q, Fu R, Wen X. et al. F. Nucleatum enhances oral squamous cell carcinoma proliferation via E-cadherin/β-Catenin pathway. BMC Oral Health. 2024;24:518 doi: 10.1186/s12903-024-04252-3
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49 doi: 10.3322/caac.21660
4. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:1-22 doi: 10.1038/s41572-020-00224-3
5. Liu J, Sun Q, Zhao J, Qin X, Gao T, Bai G. et al. Early death in supraglottic laryngeal squamous cell carcinoma: A population-based study. Ear Nose Throat J. 2024;103:650-8 doi: 10.1177/01455613221078184
6. Wise-Draper TM, Bahig H, Tonneau M, Karivedu V, Burtness B. Current Therapy for Metastatic Head and Neck Cancer: Evidence, Opportunities, and Challenges. Am Soc Clin Oncol Educ Book. 2022;42:1-14 doi: 10.1200/EDBK_350442
7. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24:430-47 doi: 10.1038/s41580-022-00566-8
8. Frankish A, Carbonell-Sala S, Diekhans M, Jungreis I, Loveland JE, Mudge JM. et al. GENCODE: reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res. 2023;51:D942-949 doi: 10.1093/nar/gkac1071
9. Rinn JL, Chang HY. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu Rev Biochem. 2020;89:283-308 doi: 10.1146/annurev-biochem-062917-01270
10. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2020;21:102-117 doi: 10.1038/s41576-019-0184-5
11. Ferrer J, Dimitrova N. Transcription regulation by long non-coding RNAs: mechanisms and disease relevance. Nat Rev Mol Cell Biol. 2024;25:396-415 doi: 10.1038/s41580-023-00694-9
12. Ntini E, Louloupi A, Liz J, Muino JM, Marsico A, Ørom UAV. Long ncRNA A-ROD activates its target gene DKK1 at its release from chromatin. Nat Commun. 2018;9:1636 doi: 10.1038/s41467-018-04100-3
13. Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z. et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16:504-16 doi: 10.1016/j.stem.2015.03.007
14. Su G, Wang W, Zhao X, Chen J, Zheng J, Liu M. et al. Enhancer architecture-dependent multilayered transcriptional regulation orchestrates RA signaling-induced early lineage differentiation of ESCs. Nucleic Acids Res. 2021;49:11575-11595 doi: 10.1093/nar/gkab1001
15. Yu Z, Tang H, Chen S, Xie Y, Shi L, Xia S. et al. Exosomal LOC85009 inhibits docetaxel resistance in lung adenocarcinoma through regulating ATG5-induced autophagy. Drug Resist Updat. 2023;67:100915 doi: 10.1016/j.drup.2022.100915
16. Tang Y, Tian W, Zheng S, Zou Y, Xie J, Zhang J. et al. Dissection of FOXO1-Induced LYPLAL1-DT Impeding Triple-Negative Breast Cancer Progression via Mediating hnRNPK/β-Catenin Complex. Research (Wash D C). 2023;6:0289 doi: 10.34133/research.0289
17. Ghasemian A, Omear HA, Mansoori Y, Mansouri P, Deng X, Darbeheshti F. et al. Long non-coding RNAs and JAK/STAT signaling pathway regulation in colorectal cancer development. Front Genet. 2023;14:1297093 doi: 10.3389/fgene.2023.1297093
18. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491-509 doi: 10.1007/s00018-016-2174-5
19. Rong M, Zhang M, Dong F, Wu K, Cai B, Niu J. et al. LncRNA RASAL2-AS1 promotes METTL14-mediated m6A methylation in the proliferation and progression of head and neck squamous cell carcinoma. Cancer Cell Int. 2024;24:113 doi: 10.1186/s12935-024-03302-8
20. Ju Y, Zhu F, Fang B. Biomarker Potential of LINC00313 in Head and Neck Squamous Cell Carcinoma: Correlation with Epithelial-Mesenchymal Transition and Immune Cell Infiltration. Int J Med Sci. 2024;21:921-936 doi: 10.7150/ijms.93044
21. Tian X, Hu D, Wang N, Zhang L, Wang X. LINC01614 is a promising diagnostic and prognostic marker in HNSC linked to the tumor microenvironment and oncogenic function. Front Genet. 2024;15:1337525 doi: 10.3389/fgene.2024.1337525
22. Li W, Zhu Q, Zhang S, Liu L, Zhang H, Zhu D. HOXC13-AS accelerates cell proliferation and migration in oral squamous cell carcinoma via miR-378g/HOXC13 axis. Oral Oncol. 2020;111:104946 doi: 10.1016/j.oraloncology.2020.104946
23. Liu J, Zhao J, Xu J, Sun Q, Qin X, Chen G. et al. SPINK5 is a Prognostic Biomarker Associated With the Progression and Prognosis of Laryngeal Squamous Cell Carcinoma Identified by Weighted Gene Co-Expression Network Analysis. Evol Bioinform Online. 2022;18:11769343221077118 doi: 10.1177/11769343221077118
24. Jonkers J, Pai P, Sukumar S. Multiple roles of HOX proteins in Metastasis: Let me count the ways. Cancer Metastasis Rev. 2020;39:661-679 doi: 10.1007/s10555-020-09908-4
25. Li L, Zhang X, Liu Q, Yin H, Diao Y, Zhang Z. et al. Emerging role of HOX genes and their related long noncoding RNAs in lung cancer. Crit Rev Oncol Hematol. 2019;139:1-6 doi: 10.1016/j.critrevonc.2019.04.019
26. Ivanov KI, Samuilova OV, Zamyatnin AA. The emerging roles of long noncoding RNAs in lymphatic vascular development and disease. Cell Mol Life Sci. 2023;80:197 doi: 10.1007/s00018-023-04842-4
27. McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P. et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232-6 doi: 10.1038/nature14443
28. Mumbach MR, Granja JM, Flynn RA, Roake CM, Satpathy AT, Rubin AJ. et al. HiChIRP reveals RNA-associated chromosome conformation. Nat Methods. 2019;16:489-492 doi: 10.1038/s41592-019-0407-x
29. Yang X, Liu M, Li M, Zhang S, Hiju H, Sun J. et al. Epigenetic modulations of noncoding RNA: a novel dimension of Cancer biology. Mol Cancer. 2020;19:64 doi: 10.1186/s12943-020-01159-9
30. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96-118 doi: 10.1038/s41580-020-00315-9
31. Ohyama T, Takahashi H, Sharma H, Yamazaki T, Gustincich S, Ishii Y. et al. An NMR-based approach reveals the core structure of the functional domain of SINEUP lncRNAs. Nucleic Acids Res. 2020;48:9346-9360 doi: 10.1093/nar/gkaa598
32. Li P, Ma X, Huang D, Gu X. Exploring the roles of non-coding RNAs in liver regeneration. Noncoding RNA Res. 2024;9:945-953 doi: 10.1016/j.ncrna.2024.04.003
33. Sharma S, Houfani AA, Foster LJ. Pivotal functions and impact of long con-coding RNAs on cellular processes and genome integrity. J Biomed Sci. 2024;31:52 doi: 10.1186/s12929-024-01038-1
34. Alharbi KS. Noncoding RNAs in hepatitis: Unraveling the apoptotic pathways. Pathol Res Pract. 2024;255:155170 doi: 10.1016/j.prp.2024.155170
35. Liu X, Li Y, Jiang X, Deng Y, Ma C, Yu Q. et al. Long non-coding RNA: Multiple effects on the differentiation, maturity and cell function of dendritic cells. Clin Immunol. 2022;245:109167 doi: 10.1016/j.clim.2022.109167
36. Liu Y, Liu Y, Ye S, Feng H, Ma L. A new ferroptosis-related signature model including messenger RNAs and long non-coding RNAs predicts the prognosis of gastric cancer patients. J Transl Int Med. 2023;11:145-155 doi: 10.2478/jtim-2023-0089
37. Yoon JH, Byun HJ, Kim SY, Jung DH, Lee SK. Exosomal LINC00853 promotes progression of gastric cancer via the MAP17/PDZK1/AKT signaling pathway. Noncoding RNA Res. 2024;9:876-886 doi: 10.1016/j.ncrna.2024.03.011
38. Ren W, Ouyang L. Long noncoding RNA VPS9D1-AS1 promotes the progression of endometrial cancer via regulation of the miR-187-3p/S100A4 axis. Environ Toxicol. 2024;39:4447-4458 doi: 10.1002/tox.24351
39. Poliseno L, Lanza M, Pandolfi PP. Coding, or non-coding, that is the question. Cell Res. 2024;34:609-629 doi: 10.1038/s41422-024-00975-8
40. Zhou L, Jiang J, Huang Z, Jin P, Peng L, Luo M. et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m6A-mediated degradation of STEAP3 mRNA. Mol Cancer. 2022;21:168 doi: 10.1186/s12943-022-01638-1
41. Sur S, Ray RB. Emerging role of lncRNA ELDR in development and cancer. FEBS J. 2022;289:3011-3023 doi: 10.1111/febs.15876
42. Sur S, Nakanishi H, Steele R, Zhang D, Varvares MA, Ray RB. Long non-coding RNA ELDR enhances oral cancer growth by promoting ILF3-cyclin E1 signaling. EMBO Rep. 2020;21:e51042 doi: 10.15252/embr.202051042
43. Sur S, Steele R, Ko BCB, Zhang J, Ray RB. Long noncoding RNA ELDR promotes cell cycle progression in normal oral keratinocytes through induction of a CTCF-FOXM1-AURKA signaling axis. The Journal of biological chemistry J Biol Chem. 2022;298:101895 doi: 10.1016/j.jbc.2022.101895
44. Kong X, Duan Y, Sang Y, Li Y, Zhang H, Liang Y. et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234:9105-9117 doi: 10.1002/jcp.27587
45. Morgan R, Hunter K, Pandha HS. Downstream of the HOX genes: Explaining conflicting tumour suppressor and oncogenic functions in cancer. Int J Cancer. 2022;150:1919-1932 doi: 10.1002/ijc.33949
46. Cai JQ, Xu XW, Mou YP, Chen K, Pan Y, Wu D. Upregulation of HOXB7 promotes the tumorigenesis and progression of gastric cancer and correlates with clinical characteristics. Tumor Biol. 2016;37:1641-50 doi: 10.1007/s13277-015-3948-3
47. He X, Liu Z, Xia Y, Xu J, Lv G, Wang L. et al. HOXB7 overexpression promotes cell proliferation and correlates with poor prognosis in gastric cancer patients by inducing expression of both AKT and MARKs. Oncotarget. 2017;8:1247-1261 doi: 10.18632/oncotarget.13604
48. Martinou E, Falgari G, Bagwan I, Angelidi AM. A Systematic Review on HOX Genes as Potential Biomarkers in Colorectal Cancer: An Emerging Role of HOXB9. Int J Mol Sci. 2021;22:13429 doi: 10.3390/ijms222413429
49. Cheng W, Shi X, Lin M, Yao Q, Ma J, Li J. LncRNA MAGI2-AS3 Overexpression Sensitizes Esophageal Cancer Cells to Irradiation Through Down-Regulation of HOXB7 via EZH2. Front Cell Dev Biol. 2020;8:552822 doi: 10.3389/fcell.2020.552822
50. Dai L, Hu W, Yang Z, Chen D, He B, Chen Y. et al. Upregulated expression of HOXB7 in intrahepatic cholangiocarcinoma is associated with tumor cell metastasis and poor prognosis. Lab Invest. 2019;99:736-748 doi: 10.1038/s41374-018-0150-4
51. Gao D, Chen H-Q. Specific knockdown of HOXB7 inhibits cutaneous squamous cell carcinoma cell migration and invasion while inducing apoptosis via the Wnt/β-catenin signaling pathway. Am J Physiol Cell Physiol. 2018;315:C675-C686 doi: 10.1152/ajpcell.00291.2017
52. Yang Y, Chen J, Chen Q. Upregulation of HOXB7 promotes proliferation and metastasis of osteosarcoma cells. Mol Med Rep. 2017;16:2773-2778 doi: 10.3892/mmr.2017.6906
53. Tsuboi M, Taniuchi K, Shimizu T, Saito M, Saibara T. The transcription factor HOXB7 regulates ERK kinase activity and thereby stimulates the motility and invasiveness of pancreatic cancer cells. J Biol Chem. 2017;292:17681-17702 doi: 10.1074/jbc.M116.772780
54. Li C, Mao X, Song L, Sheng J, Yang L, Huang X. et al. Unveiling HOXB7 as a novel diagnostic and prognostic biomarker through pan-cancer computer screening. Comput Biol Med. 2024;176:108562 doi: 10.1016/j.compbiomed.2024.108562
55. Errico MC, Felicetti F, Bottero L, Mattia G, Boe A, Felli N. et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. Int J Cancer. 2013;133:879-92 doi: 10.1002/ijc.28097
56. Hong Z, Fu W, Wang Q, Zeng Y, Qi L. MicroRNA-384 is lowly expressed in human prostate cancer cells and has anti-tumor functions by acting on HOXB7. Biomed Pharmacother. 2019;114:108822 doi: 10.1016/j.biopha.2019.108822
57. Stiegelbauer V, Vychytilova-Faltejskova P, Karbiener M, Pehserl AM, Reicher A, Resel M. et al. miR-196b-5p Regulates Colorectal Cancer Cell Migration and Metastases through Interaction with HOXB7 and GALNT5. Clin Cancer Res. 2017;23:5255-5266 doi: 10.1158/1078-0432.CCR-17-0023
58. Zheng X, Gao W, Zhang Z, Xue X, Mijiti M, Guo Q. et al. Identification of a seven-lncRNAs panel that serves as a prognosis predictor and contributes to the malignant progression of laryngeal squamous cell carcinoma. Front Oncol. 2023;13:1106249 doi: 10.3389/fonc.2023.1106249
59. Zhang Y, Zhou G, Shi W, Shi W, Hu M, Kong D. et al. A novel oxidative-stress related lncRNA signature predicts the prognosis of clear cell renal cell carcinoma. Sci Rep. 2023;13:5740 doi: 10.1038/s41598-023-32891-z
60. Zhou Z, Yang Z, Cui Y, Lu S, Huang Y, Che X. et al. Identification and Validation of a Ferroptosis-Related Long Non-Coding RNA (FRlncRNA) Signature to Predict Survival Outcomes and the Immune Microenvironment in Patients With Clear Cell Renal Cell Carcinoma. Front Genet. 2022;13:787884 doi: 10.3389/fgene.2022.787884
61. Deng Q, Yang J, Chen Y, Chen Z, Li J, Fu Z. LncRNA HOXB-AS4 promotes proliferation and migration of colorectal cancer via the miR-140-5p/hdac7 axis. Biotechnol Genet Eng Rev. 2024;40:1262-1280 doi: 10.1080/02648725.2023.2193465
62. Xu L, Liu S, Yang Y, Shu L, Sun Y. LINC00313 suppresses autophagy and promotes stemness of nasopharyngeal carcinoma cells through PTBP1/STIM1 axis. Radiother Oncol. 2024;196:110310 doi: 10.1016/j.radonc.2024.110310
63. Zou X, Hu X, He F, Zhang M, Kong X, Rui S. et al. LncRNA LINC00152 promotes oral squamous cell carcinoma growth via enhancing Upstream Transcription Factor 1 mediated Mitochondrial Ribosomal Protein L52 transcription. J Oral Pathol Med. 2022;51:454-463 doi: 10.1111/jop.13253
64. Ma Y, Han J, Luo X. FOXD1-AS1 upregulates FOXD1 to promote oral squamous cell carcinoma progression. Oral Dis. 2023;29:604-614 doi: 10.1111/odi.14002
65. Li Z, Qiu X, He Q, Fu X, Ji F, Tian X. CCND1-associated ceRNA network reveal the critical pathway of TPRG1-AS1-hsa-miR-363-3p-MYO1B as a prognostic marker for head and neck squamous cell carcinoma. Sci Rep. 2023;13:11831 doi: 10.1038/s41598-023-38847-7
66. Yamaguchi K, Yamamoto T, Chikuda J, Shirota T, Yamamoto Y. Impact of Non-Coding RNAs on Chemotherapeutic Resistance in Oral Cancer. Biomolecules. 2022;12:284 doi: 10.3390/biom12020284
67. Jia Y, Chen X, Zhao D, Ma S. SNHG1/miR-194-5p/MTFR1 Axis Promotes TGFβ1-Induced EMT, Migration and Invasion of Tongue Squamous Cell Carcinoma Cells. Mol Biotechnol. 2022;64:780-790 doi: 10.1007/s12033-021-00445-1
68. Zhang M, Sun Q, Han Z, Qin X, Gao T, Xu Y. et al. Construction of a novel disulfidptosis-related lncRNAs signature for prognosis prediction and anti-tumor immunity in laryngeal squamous cell carcinoma. Heliyon. 2024;10:e30877 doi: 10.1016/j.heliyon.2024.e30877
69. Li Z, Wang Q, Huang X, Fu R, Wen X, Zhang L. Multi-omics analysis reveals that ferroptosis-related gene CISD2 is a prognostic biomarker of head and neck squamous cell carcinoma. J Gene Med. 2024;26:e3580 doi: 10.1002/jgm.3580
70. Xu Z, Zhang M, Guo Z, Chen L, Yang X, Li X. et al. Stemness-related lncRNAs signature as a biologic prognostic model for head and neck squamous cell carcinoma. Apoptosis. 2023;28:860-880 doi: 10.1007/s10495-023-01832-6
71. Lan T, Yan Y, Zheng D, Ding L. Investigating diagnostic potential of long non-coding RNAs in head and neck squamous cell carcinoma using TCGA database and clinical specimens. Sci Rep. 2024;14:7500 doi: 10.1038/s41598-024-57987-y
72. Jin Y, Wang Z, Huang S, Liu C, Wu X, Wang H. Identify and validate circadian regulators as potential prognostic markers and immune infiltrates in head and neck squamous cell carcinoma. Sci Rep. 2023;13:19939 doi: 10.1038/s41598-023-46560-8
73. Huang HH, You GR, Tang SJ, Chang JT, Cheng AJ. Molecular Signature of Long Non-Coding RNA Associated with Areca Nut-Induced Head and Neck Cancer. Cells. 2023;12:873 doi: 10.3390/cells12060873
74. Van Mol P, Donders E, Lambrechts D, Wauters E. Immune checkpoint biology in health & disease: Immune checkpoint biology and autoimmunity in cancer patients. Int Rev Cell Mol Biol. 2024;382:181-206 doi: 10.1016/bs.ircmb.2023.04.003
75. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275-87 doi: 10.1038/nrc.2016.36
76. Moura LI, Malfanti A, Matos AI, Peres C, Armiñán A, Duro-Castaño A. et al. Off-The-Shelf Multivalent Nanoconjugate Cancer Vaccine Rescues Host Immune Response against Melanoma. Adv Mater. 2025;37:e2417348 doi: 10.1002/adma.202417348
77. Feng S, Shen Y, Zhang H, Liu W, Feng W, Chen X. et al. Human cytomegalovirus tegument protein UL23 promotes gastric cancer immune evasion by facilitating PD-L1 transcription. Mol Med. 2025;31:57 doi: 10.1186/s10020-025-01114-8
78. Tang Q, Zhao S, Zhou N, He J, Zu L, Liu T, Song Z, Chen J, Peng L, Xu S. PD-1/PD-L1 immune checkpoint inhibitors in neoadjuvant therapy for solid tumors. Int J Oncol. 2023;62:49 doi: 10.3892/ijo.2023.5497
79. Mok S, Ağaç Çobanoğlu D, Liu H, Mancuso JJ, Allison JP. Post-immunotherapy CTLA-4 Ig treatment improves antitumor efficacy. Proc Natl Acad Sci U S A. 2024;121:e2404661121 doi: 10.1073/pnas.2404661121
80. Adam K, Butler SC, Workman CJ, Vignali DAA. Advances in LAG3 cancer immunotherapeutics. Trends Cancer. 2025;11:37-48 doi: 10.1016/j.trecan.2024.10.009
81. Tripathi S, Najem H, Dussold C, Pacheco S, Du R, Sooreshjani M. et al. Pediatric glioma immune profiling identifies TIM3 as a therapeutic target in BRAF fusion pilocytic astrocytoma. J Clin Invest. 2024;134:e177413 doi: 10.1172/JCI177413
82. Alsina M, Fleitas-Kanonnikoff T. Immune checkpoint inhibitors for first-line treatment of advanced esophageal squamous cell carcinoma. Med. 2024;5:1038-1040 doi: 10.1016/j.medj.2024.05.016
83. Wang Y, Safi M, Hirsch FR, Lu S, Peters S, Govindan R. et al. Immunotherapy for advanced-stage squamous cell lung cancer: the state of the art and outstanding questions. Nat Rev Clin Oncol. 2025;22:200-214 doi: 10.1038/s41571-024-00979-8
84. Shi H, Yu TK, Johnson B, Selvamani SP, Zhuang L, Lee K. et al. A combination of PD-1 and TIGIT immune checkpoint inhibitors elicits a strong anti-tumour response in mesothelioma. J Exp Clin Cancer Res. 2025;44:51 doi: 10.1186/s13046-025-03314-w
85. Zhou Z, Guan B, Lin J, Zheng R, Xu B. Will personalized ultrafractionated stereotactic adaptive radiotherapy (PULSAR) or split-course SBRT based on systemic therapy (3S) be future directions in the Field of SBRT? Int Immunopharmacol. 2025;146:113689 doi: 10.1016/j.intimp.2024.113689
Author contact
![]() Corresponding authors: Dr. Yan Liu, Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China. E-mail: liuyan2758org.cn. Hao Tian, Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China. E-mail: tianhao01262023com.
Corresponding authors: Dr. Yan Liu, Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China. E-mail: liuyan2758org.cn. Hao Tian, Department of Head and Neck Surgery, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, 410013, Hunan, China. E-mail: tianhao01262023com.

 Global reach, higher impact
Global reach, higher impact