3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(11):3481-3494. doi:10.7150/jca.94685 This issue Cite
Research Paper
A Model to Predict Prognosis of Renal Cell Clear Cell Carcinoma Based on 3 Angiogenesis-related Long Non-coding RNAs
1. Department of Urology, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 400042, China.
2. Department of Urology, Mianyang Central Hospital, University of Electronic Science and Technology of China, Sichuan Province, 621099, China.
3. Department of Urology, Three Gorges Hospital of Chongqing University, Chongqing, 404031, China.
4. Department of General Surgery, The Third People's Hospital of Chengdu, Sichuan Province, 610014, China.
5. Department of Vascular Surgery, Yibin First People's Hospital, Sichuan Province, 644000, China.
6. Department of Urology, Xinqiao Hospital of the Army Medical University, Chongqing, 400037, China.
# These authors contributed equally to this work
Abstract
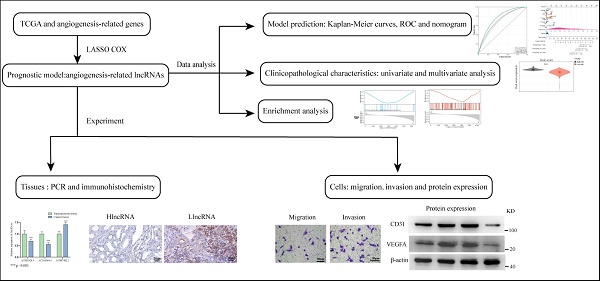
Background: Tumor angiogenesis is closely related to the progression of clear cell renal cell carcinoma (ccRCC). Long non-coding RNAs (lncRNAs) regulating angiogenesis could be potential biomarkers for predicting ccRCC prognosis. With this study, we aimed to construct a prognostic model based on lncRNAs and explore its underlying mechanisms.
Methods: RNA data and clinical information were obtained from The Cancer Genome Atlas (TCGA) database. Angiogenesis-related genes (ARGs) were extracted from the Molecular Signatures database. Pearson correlation and LASSO and COX regression analyses were performed to identify survival-related AR-lncRNAs (sAR-lncRNAs) and construct a prognostic model. The predictive power of the prognostic model was verified according to Kaplan‒Meier curve, receiver operating characteristic (ROC) curve and nomogram analyses. The correlation between the prognostic model and clinicopathological characteristics was assessed via univariate and multivariate analyses. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was subsequently performed to elucidate the mechanisms of the sAR-lncRNAs. In vitro qPCR, immunohistochemistry, migration and invasion assays were conducted to confirm the angiogenic function of sAR-lncRNAs.
Results: Three sAR-lncRNAs were used to construct a prognostic model. The model was moderately accurate in predicting 1- , 3- and 5-year ccRCC prognosis, and the risk score according to this model was closely related to clinicopathological characteristics such as T grade and T stage. A nomogram was constructed to precisely estimate the overall survival of ccRCC patients. KEGG enrichment analysis indicated that the MAPK and Notch pathways were highly enriched in high-risk patients. Additionally, we found that the expression of the lncRNAs AC005324.4 and AC104964.4 in the prognostic model was lower in ccRCC cell lines and cancer tissues than in the HK-2 cell line and paracancerous tissues, while the expression of the lncRNA AC087482.1 showed the opposite trend. In a coculture model, knockdown of lncRNA AC005324.4 and lncRNA AC104964.4 significantly promoted the migration and invasion of human umbilical vein endothelial cells (HUVECs), but siR-AC087482.1 transfection alleviated these effects.
Conclusions: We constructed a prognostic model based on 3 sAR-lncRNAs and validated its value in clinicopathological characteristics and prognostic prediction of ccRCC patients, providing a new perspective for ccRCC treatment decision making.
Keywords: model, prognosis, renal cell clear cell carcinoma, angiogenesis, lncRNA

 Global reach, higher impact
Global reach, higher impact