Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(2):526-532. doi:10.7150/jca.89271 This issue Cite
Research Paper
ALKBH1 rs2267755 C>T polymorphism decreases neuroblastoma risk in Chinese children
1. Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
2. Department of Pathology, Children's Hospital of Nanjing Medical University, Nanjing 210008, Jiangsu, China.
3. Department of Pathology, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, Guangdong, China.
4. Laboratory Animal Center, School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen 518055, Guangdong, China.
# These authors contributed equally to this work.
Received 2023-8-18; Accepted 2023-11-17; Published 2024-1-1
Abstract
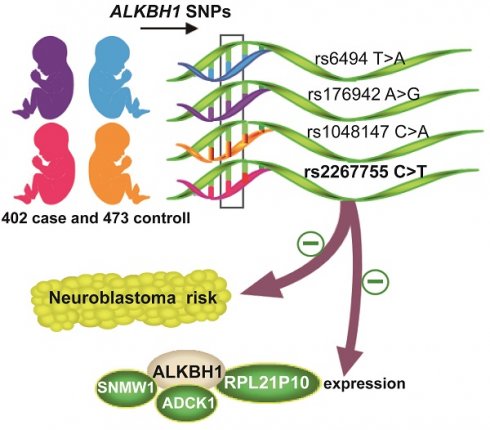
Neuroblastoma is a highly malignant extracranial solid tumor in pediatrics. ALKBH1 as a recently discovered DNA N6-methyldeoxyadenosine (6mA) demethylase closely links to tumorigenesis. Whether the ALKBH1 polymorphism contributes to neuroblastoma risk remains unclear. In the present study, we genotyped the ALKBH1 single nucleotide polymorphisms (SNPs) in 402 neuroblastoma patients and 473 healthy controls by TaqMan assay. Odds ratios (ORs) and 95% confidence intervals (CIs) were also calculated to evaluate the strength of the association. Our result exhibited that the rs2267755 C>T (CT vs. CC, adjusted OR=0.69, 95% CI=0.50-0.94, P=0.019) is significantly associated with reduced neuroblastoma risk. And its protective effect is particularly significant in children with tumors originating from the retroperitoneal. Combined genotype analysis revealed that carriers with 1-2 protective genotypes are more susceptible to neuroblastoma than those with 3-4 protective genotypes (adjusted OR=0.71, 95% CI=0.53-0.97, P=0.028). Moreover, the rs2267755 C>T is significantly associated with messenger RNA (mRNA) expression of ALKBH1 and three of its surrounding genes, including SNWQ, ADCK1, and RPL21P10. These results suggest that the rs2267755 C>T may be a genetic variant to reduce neuroblastoma risk.
Keywords: neuroblastoma, ALKBH1, polymorphism, susceptibility
Introduction
Neuroblastoma is the most frequently extracranial solid tumor in pediatrics, accounting for nearly 15 % of all pediatric cancer deaths [1, 2]. Although previous studies have identified partial genetics aberrations as potential neuroblastoma therapeutic targets, it is necessary to explore new regulators for developing new neuroblastoma therapies [2]. In recent years, methylation has been recognized as playing an important role in neuroblastoma progression [3, 4]. Multiple studies demonstrated that suppressing DNA m5C affects neuroblastoma cell proliferation and colony-forming activity [5, 6]. Targeted inhibition of histone H3K27 demethylation effectively treats high-risk neuroblastoma. Moreover, RNA N6-methyladenosine methylation gene variations are significantly associated with neuroblastoma risk [8-12].
DNA N6-methyladenine (6mA) is a recently identified modification in mammals, which is widely present in the human genome [13]. Like most methylation modifications, 6mA is an invertible DNA modification. The identification of its regulators had achieved definite progress. Studies demonstrated that the METTL4 could catalyze 6mA modification on genomic DNA (gDNA) and be active against mitochondria DNA in vitro [14, 15]. ALKBH1 and ALKBH4 are responsible for DNA 6mA demethylation in mammals [16, 17].
Recently, more evidence suggested that 6mA is closely related to tumorigenesis [18, 19]. Thereinto, silencing of ALKBH1 significantly impaired the growth of cancer xenografts in vivo. Quyang L et al. proposed that ALKBH1 is a potential therapeutic target to reduce the burden of vascular calcification. Mechanistically, they found that ALKBH1 activates bone morphogenetic protein 2(BMP2) and aggravates osteogenic reprogramming in chronic kidney disease [20]. A recent study revealed that ALKBH1 is highly expressed in human head and neck squamous cell carcinoma cells and patient tissues, promoting cell proliferation by enhancing DDX18 expression [21]. More importantly, Rashad, S. et al uncovered that ALKBH1 assists tRNA cleavage in demethylation stress in rat neuroblastoma cells [22]. However, it remains unclear whether ALKBH1 is associated with neuroblastoma risk.
The study of genetic variants is one of the most important parts to elucidate the role of key genes in neuroblastoma. Our previous studies clarified that SNPs of multiple genes are tightly related to neuroblastoma risk [23, 24]. Given the crucial role of ALKBH1 in cancer, we conducted a comprehensive analysis to illuminate the relationship between the ALKBH1 gene SNPs and neuroblastoma susceptibility in Chinese children, providing new evidence for predicting the risk of neuroblastoma.
Subjects and Methods
Subjects
In this study, 402 neuroblastoma patients and 473 control samples were recruited from Children's Hospital of Nanjing Medical University in Jiangsu province, China (Table S1) [25, 26]. All neuroblastoma patients were confirmed by histopathology or cytology, and did not receive radiotherapy and/or chemotherapy prior to blood collection. Patients with a history of malignancy in other organs were excluded. Control samples without neuroblastoma were recruited in the same hospital. All subjects' guardians had written informed consent at the start of this research. The present study was approved by the institutional review board of Children's Hospital of Nanjing Medical University (Approval No: 202112141-1).
DNA extraction and selection of ALKBH1 polymorphism
Total genomic DNA was extracted using the TIANamp Blood DNA Kit (TianGen Biotech Co., Ltd., Beijing, China) from EDTA-peripheral blood. As previously, ALKBH1 potentially functional SNPs were chosen from the NCBI dbSNP database (https://www.ncbi.nlm.nih.gov) and SNPinfo (https://manticore.niehs.nih.gov), we selected these SNPs according to the following criteria: 1) located at both sides of ALKBH1 (5'UTR, 5' near gene, 3'UTR or 3' near gene); 2) potential biological functions predicted by snpinfo; 3) the minor allele frequency in Chinese subject is >5%; 4) SNPs showing significant linkage disequilibrium (LD) with each other (R2≥0.8) are excluded. Consequently, we selected the four ALKBH1 potential functional SNPs (rs2267755 C>T, rs1048147 C>A, rs6494 T>A, rs176942 A>G) with R2 lower than 0.5. The Taqman real-time PCR method was used to genotype the four ALKBH1 SNPs under the blind status of neuroblastoma cases and control [27]. To guarantee the reliability of the genotyping of ALKBH1 SNPs, 10% of samples were randomly redetected and the accuracy was 100%.
Statistics analysis
In this study, statistical analysis was conducted with the Chi-square test to evaluate the significant difference for categorical variables, and a t-test was employed for continuous variables. ALKHB1 SNPs were identified with χ2 test to compare the difference between the cases and control samples, which agreed with Hardy-Weinberg equilibrium (HWE). To investigate the association between each ALKBH1 SNP and neuroblastoma risk, odds ratios (ORs) with 95% confidence intervals (CIs) were calculated using unconditional logistic regression. This statistical analysis was also performed based on age, gender, and clinical stages. For all statistical tests on two sides, P-value < 0.05 was considered statistically significant, which was calculated by SAS software (Version 10.0, SAS Institute, Inc., Cary, North Carolina).
Result
Association of ALKBH1 gene polymorphism with neuroblastoma susceptibility
In this study, genotyping of ALKBH1 was performed in 402 neuroblastoma patients and 473 controls, whose clinical characteristics were listed in Table S1. As shown in Table 1, we successfully genotyped four ALKBH1 SNPs (rs2267755 C>T, rs1048147 C>A, rs6494 T>A, rs176942 A>G) in 399 cases and 473 controls samples, which coincided with Hardy-Weinberg equilibrium (HWE) among controls (HWE P > 0.05). Thereinto, the rs2267755C>T variant was significantly associated with neuroblastoma risk (CT versus CC: adjusted OR=0.69, 95% CI=0.50-0.94, P=0.019; CT/TT versus CC: adjusted OR=0.74, 95% CI=0.55-0.98, P=0.039). We then identified rs2267755 CT/TT, rs1048147 CC, rs6494 TA/TT, and rs176942 AG/AA) as protective genotypes following their values of ORs. The combined genotype result showed that individuals with 3-4 protective genotypes experienced a 0.71-fold decrease in the risk of development neuroblastoma when compared with those with 1-2 protective genotypes (95% CI=0.53-0.97, P=0.028).
Stratification analysis
We next evaluated the correlation between rs2267755 C>T variant with neuroblastoma susceptibility in subgroups divided by age, gender, sites of origins, and INSS stages (Table 2). Stratified analysis results revealed that the rs2267755 C>T was associated with reduced neuroblastoma risk in children with tumors originating from the retroperitoneal (adjusted OR=0.60, 95% CI=0.41-0.87, P=0.007). Besides, individuals with 3-4 protective genotypes significantly decreased neuroblastoma susceptibility compared with those with 1-2 protective genotypes in a subgroup of tumors originating from retroperitoneal (adjusted OR=0.64, 95% CI=0.43-0.95, P=0.026).
Effect of rs2267755C>T on the expression of ALKBH1
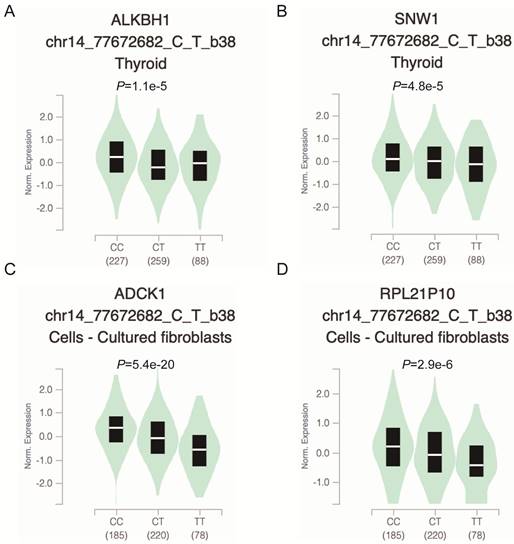
To further explore whether the functional relevance of rs2267755C>T affects the messenger (mRNA) expression of adjacent genes, we used the GTEx (https://www.gtexportal.org/) portal to investigate Cis-expression quantitative trait loci (eQTL) target genes of the rs2267755C>T. Result manifested that the rs2267755 T allele was significantly associated with reduced mRNA expression of ALKBH1 and SNW1 in the thyroid (Figure 1A, B), as well as ADCK1 and RPL21P10 in the cultured fibroblasts cell (Figure 1C, D).
ALKBH1 gene polymorphisms and neuroblastoma susceptibility in Jiangsu children.
| Genotype | Cases (N=399) | Controls (N=473) | P a | Crude OR (95% CI) | P | Adjusted OR (95% CI) b | P b |
|---|---|---|---|---|---|---|---|
| rs2267755 C>T (HWE=0.262) | |||||||
| CC | 131 (32.83) | 125(26.43) | 1.00 | 1.00 | |||
| CT | 179 (44.86) | 248 (52.43) | 0.69 (0.50-0.94) | 0.019 | 0.69 (0.50-0.94) | 0.019 | |
| TT | 89 (22.31) | 100 (21.14) | 0.85 (0.58-1.24) | 0.395 | 0.85 (0.58-1.24) | 0.397 | |
| Additive | 0.278 | 0.90 (0.75-1.09) | 0.278 | 0.90 (0.75-1.09) | 0.279 | ||
| Dominant | 268 (67.17) | 348 (73.57) | 0.039 | 0.74 (0.55-0.98) | 0.039 | 0.74 (0.55-0.98) | 0.039 |
| Recessive | 310 (77.69) | 373 (78.86) | 0.678 | 1.07 (0.78-1.48) | 0.677 | 1.07 (0.78-1.48) | 0.673 |
| rs1048147 C>A (HWE=0.266) | |||||||
| CC | 197 (49.37) | 250 (52.85) | 1.00 | 1.00 | |||
| CA | 164 (41.10) | 181 (38.27) | 1.15 (0.87-1.52) | 0.332 | 1.15 (0.87-1.52) | 0.335 | |
| AA | 38 (9.52) | 42 (8.88) | 1.15 (0.71-1.85) | 0.570 | 1.15 (0.71-1.85) | 0.572 | |
| Additive | 0.354 | 1.10 (0.90-1.35) | 0.353 | 1.10 (0.90-1.35) | 0.356 | ||
| Dominant | 202 (50.63) | 223 (47.15) | 0.306 | 1.15 (0.88-1.50) | 0.306 | 1.15 (0.88-1.50) | 0.309 |
| Recessive | 361 (90.48) | 431 (91.12) | 0.743 | 1.08 (0.68-1.71) | 0.742 | 1.08 (0.68-1.71) | 0.742 |
| rs6494 T>A (HWE=0.974) | |||||||
| TT | 349 (87.47) | 400 (84.57) | 1.00 | 1.00 | |||
| TA | 45 (11.28) | 70 (14.80) | 0.74 (0.49-1.10) | 0.136 | 0.74 (0.49-1.10) | 0.133 | |
| AA | 5 (1.25) | 3 (0.63) | 1.91 (0.45-8.05) | 0.378 | 1.94 (0.46-8.19) | 0.369 | |
| Additive | 0.380 | 0.85 (0.60-1.22) | 0.380 | 0.85 (0.60-1.21) | 0.380 | ||
| Dominant | 50 (12.53) | 73 (15.43) | 0.220 | 0.79 (0.53-1.16) | 0.221 | 0.78 (0.53-1.16) | 0.219 |
| Recessive | 394 (98.75) | 470 (99.37) | 0.340 | 1.99 (0.47-8.37) | 0.349 | 2.01 (0.47-8.49) | 0.342 |
| rs176942 A>G (HWE=0.788) | |||||||
| AA | 276 (69.17) | 334 (70.61) | 1.00 | 1.00 | |||
| AG | 102 (25.56) | 126 (26.64) | 0.98 (0.72-1.33) | 0.895 | 0.98 (0.72-1.33) | 0.896 | |
| GG | 21 (5.26) | 13 (2.75) | 1.96 (0.96-3.98) | 0.064 | 1.97 (0.97-4.01) | 0.062 | |
| Additive | 0.290 | 1.14 (0.90-1.45) | 0.290 | 1.14 (0.90-1.45) | 0.288 | ||
| Dominant | 123 (30.83) | 139 (29.39) | 0.644 | 1.07 (0.80-1.43) | 0.644 | 1.07 (0.80-1.43) | 0.642 |
| Recessive | 378 (94.74) | 460 (97.25) | 0.056 | 1.97 (0.97-3.98) | 0.060 | 1.98 (0.98-4.01) | 0.059 |
| Combine protective genotypes c | |||||||
| 0-2 | 119 (29.82) | 110 (23.26) | 1.00 | 1.00 | |||
| 3-4 | 280 (70.18) | 363 (76.74) | 0.028 | 0.71 (0.53-0.97) | 0.028 | 0.71 (0.53-0.97) | 0.029 |
OR, odds ratio; CI, confidence interval, HWE, Hardy-Weinberg equilibrium.
a χ2 test for genotype distributions between neuroblastoma patients and cancer-free controls.
b Adjusted for age and sex.
c Protective genotypes were carriers with rs2267755 CT/TT, rs1048147 CC, rs6494 TA/TT, and rs176942 AG/AA genotypes.
Stratification analysis for the association between ALKBH1 gene protective genotypes and neuroblastoma risk in Jiangsu children.
| Variables | rs2267755 (cases/controls) | AOR (95% CI) a | P a | Protective genotypes (cases/controls) | AOR (95% CI) a | P a | ||
|---|---|---|---|---|---|---|---|---|
| CC | CT/TT | 0-2 | 3-4 | |||||
| Age, month | ||||||||
| ≤18 | 46/33 | 90/106 | 0.61 (0.36-1.03) | 0.065 | 40/28 | 96/111 | 0.61 (0.35-1.06) | 0.076 |
| >18 | 85/92 | 178/242 | 0.80 (0.56-1.13) | 0.205 | 79/82 | 184/252 | 0.76 (0.53-1.09) | 0.135 |
| Sex | ||||||||
| Females | 67/62 | 124/163 | 0.70 (0.46-1.07) | 0.099 | 59/56 | 132/169 | 0.74 (0.48-1.14) | 0.174 |
| Males | 64/63 | 144/185 | 0.77 (0.51-1.16) | 0.209 | 60/54 | 148/194 | 0.69 (0.45-1.05) | 0.086 |
| Sites of origin | ||||||||
| Adrenal gland | 22/125 | 71/348 | 1.16 (0.69-1.95) | 0.577 | 22/110 | 71/363 | 0.98 (0.58-1.66) | 0.940 |
| Retroperitoneal | 62/125 | 103/348 | 0.60 (0.41-0.87) | 0.007 | 53/110 | 112/363 | 0.64 (0.43-0.95) | 0.026 |
| Mediastinum | 39/125 | 80/348 | 0.74 (0.48-1.14) | 0.165 | 37/110 | 82/363 | 0.67 (0.43-1.05) | 0.077 |
| Others | 7/125 | 11/348 | 0.56 (0.21-1.49) | 0.248 | 6/110 | 12/363 | 0.60 (0.22-1.65) | 0.324 |
| INSS stages | ||||||||
| I+II+4s | 54/125 | 119/348 | 0.79 (0.54-1.16) | 0.231 | 51/110 | 122/363 | 0.73 (0.49-1.08) | 0.116 |
| III+IV | 52/125 | 111/348 | 0.76 (0.52-1.13) | 0.174 | 46/110 | 117/363 | 0.77 (0.51-1.15) | 0.199 |
AOR, adjusted odds ratio; CI, confidence interval. aAdjusted for age and sex, omitting the corresponding stratification factor.
Functional relevance of rs2267755 C>T on gene expression in GTEx Database. rs2267755 C>T significantly reduced ALKBH1 (A) and SNW1 (B) mRNA expression in the thyroid (P=1.1e-5, P=4.8e-5) as well as in ADCK1 (C) and RPL21P10 (D) in the cell-cultured fibroblasts (P=5.4e-20, P=2.9e-6).
Discussion
Understanding the role of 6mA modification will provide new insight into the prediction of neuroblastoma and the establishment of new treatment strategies. This study investigated the relationship between 6mA demethylase ALKBH1 and neuroblastoma susceptibility in Chinese children from the perspective of genetic polymorphism. Our research results showed that the ALKBH1 rs2267755C>T variant is significantly associated with reduced neuroblastoma risk. More protective genotypes significantly decreased neuroblastoma susceptibility in Chinese children.
DNA 6mA modification is recently identified in mammals [28]. Its balance is synergistically regulated by methyltransferases (METTL4) and demethylase (ALKBH1, ALKBH4). Methylation-related studies demonstrated that genetic variants of the methylation-related genes could affect tumor susceptibility [12, 29, 30]. Our group previously genotyped 5 N6-methyladenosine (m6A) methylase METTL14 SNPs in 898 neuroblastoma patients and 1734 control samples. We found that 3 SNPs are significantly associated with neuroblastoma risk [8]. Besides, we also found a significant association between m6A methylase METTL3 gene SNP rs1061027 C>A variant and neuroblastoma susceptibility in females and ≤ 18-month children [9]. These results suggested that variation of methylation gene is associated with neuroblastoma risk. However, it is necessary to explore the correlation between 6mA regulator SNPs and neuroblastoma risk.
Recent studies verified that ALKBH1 as DNA 6mA demethylase plays a vital role in human cancer [31, 32]. At present, the function mechanism of ALKBH1 is still at the initial stage. Consistent with FTO and ALKBH5, ALKBH1 is a member of the AlkB family, which catalyzes substrate demethylation in Fe (II)- and α-ketoglutarate-dependent manner [32]. Previous research found that the FTO (ALKBH9) gene SNP rs8047395 A>G variant is significantly correlated with Wilms tumor [29]. And the ALKBH5 gene SNPs had a weak impact on hepatoblastoma risk [33]. Therefore, we predicted ALKBH1 may be associated with neuroblastoma risk. In the present study, we explored the role of the ALKBH1 SNPs in neuroblastoma risk for the first time. Among 4 newly identified ALKBH1 SNPs, the rs2267755 C>T is significantly associated with neuroblastoma risk in Chinese children. Moreover, the protective effect of rs2267755 C>T is significantly increased in children with tumors originating from retroperitoneal.
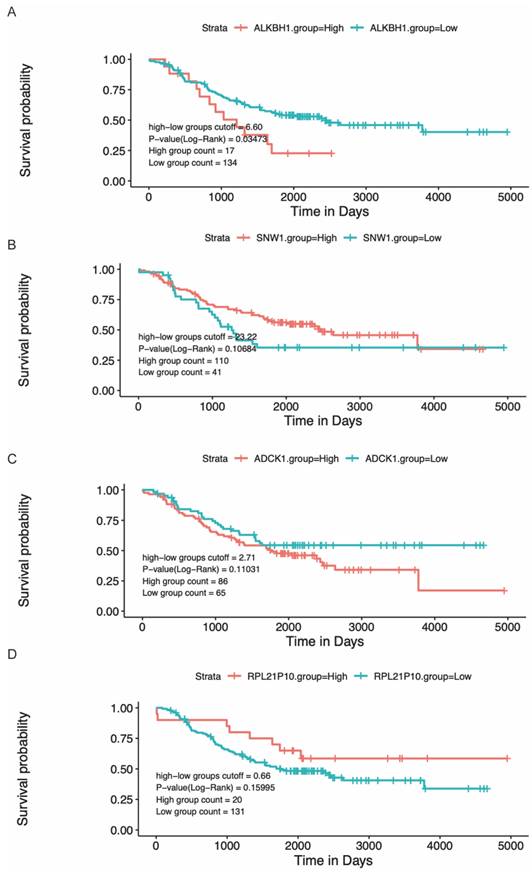
Modulating gene expression is one of the most important ways for regulators to affect tumor development. Consistent with this, lacking ALKBH1 affected the expression of developmentally important miRNAs in embryonic stem cells [36]. Depletion of ALKBH1 inhibits the expression of adipogenic-related genes CEBPA, PPARG, PLIN1, and ADIPOQ in human mesenchymal stem cells [37]. Therefore, we evaluated the effect of rs2267755 C on ALKBH1 and surrounding gene expression. Harnessing the GTEx database (Figure 1), we found that rs2267755 C is significantly correlated with the expression of ALKBH1 in the thyroid, surrounding gene SNW1 in the thyroid, and ADCK1 and RPL21P10 in the cultured fibroblasts cells. Moreover, we explore the survival probability of neuroblastoma patients among these relevant genes through the PCAT (PDX for childhood cancer Therapeutics http://pedtranscriptome.org/?home), in which the statistical analysis (Figure 2) shows that ALKBH1 level is negatively correlated with the survival probability of neuroblastoma patients. The other three gene expression levels are not significant. Therefore, we speculate rs2267755 C>T is associated with increased survival rates of neuroblastoma patients. More experiments are needed to confirm the role of rs2267755C in modulating gene expression.
In general, we elucidated the relationship between ALKBH1 gene SNPs and neuroblastoma susceptibility. Our study revealed a significant correlation between rs2267755C>T variant and neuroblastoma risk in Chinese children. Further studies were needed to illustrate the biological mechanisms.
Abbreviations
6mA: DNA N6-methyldeoxyadenosine; SNP: single nucleotide polymorphism; m6A: RNA N6-methyldeoxyadenosine; mRNA: messenger RNA; HWE: Hardy-Weinberg equilibrium; AOR: adjusted odds ratio; CI: confidence interval; eQTL: Cis-expression quantitative trait loci; gDNA: genomic DNA.
Supplementary Material
Supplementary table.
Acknowledgements
This study was supported by grants from National Natural Science Foundation of China (82002636, 82173593), the Science, Technology and Innovation Commission of Shenzhen (JCYJ20220531093213030), Guangdong Basic and Applied Basic Research Foundation (2021A1515111116).
Data availability statement
The original contributions presented in the study are provided in the article/Supplementary Materials; all data can be directed inquiries from the corresponding authors.
Ethical approval
The research has obtained approval from the institutional review board of Children's Hospital of Nanjing Medical University (Approval No: 202112141-1). According to the Declaration of Helsinki, written informed consent was obtained from all subjects' guardians at the start of this research.
Author contributions
X.Z., Y.Z., C.D, Z.Z, and J.H. designed and performed the study and wrote the manuscript; C.Z, and H.W collected the samples and information. X.Z., Z.Z, and J.H analyzed the data, prepared the Tables and Figures, and coordinated the study. All authors contribute significantly to this work and approved the final manuscript submitted for publication.
The correlation between gene levels and survival probability of neuroblastoma patients.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ishola TA, Chung DH. Neuroblastoma. Surg Oncol. 2007;16:149-56
2. Zafar A, Wang W, Liu G, Wang X, Xian W, McKeon F. et al. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med Res Rev. 2021;41:961-1021
3. Durinck K, Speleman F. Epigenetic regulation of neuroblastoma development. Cell Tissue Res. 2018;372:309-24
4. Fetahu IS, Taschner-Mandl S. Neuroblastoma and the epigenome. Cancer Metastasis Rev. 2021;40:173-89
5. Bartolucci S, Estenoz M, Longo A, Santoro B, Momparler RL, Rossi M. et al. 5-Aza-2'-deoxycytidine as inducer of differentiation and growth inhibition in mouse neuroblastoma cells. Cell Differ Dev. 1989;27:47-55
6. Carpinelli P, Granata F, Augusti-Tocco G, Rossi M, Bartolucci S. Antiproliferative effects and DNA hypomethylation by 5-aza-2'-deoxycytidine in human neuroblastoma cell lines. Anticancer Drugs. 1993;4:629-35
7. Lochmann TL, Powell KM, Ham J, Floros KV, Heisey DAR, Kurupi RIJ. et al. Targeted inhibition of histone H3K27 demethylation is effective in high-risk neuroblastoma. Sci Transl Med. 2018;10:eaao4680
8. Zhuo Z, Lu H, Zhu J, Hua RX, Li Y, Yang Z. et al. METTL14 Gene Polymorphisms Confer Neuroblastoma Susceptibility: An Eight-Center Case-Control Study. Mol Ther Nucleic Acids. 2020;22:17-26
9. Bian J, Zhuo Z, Zhu J, Yang Z, Jiao Z, Li Y. et al. Association between METTL3 gene polymorphisms and neuroblastoma susceptibility: A nine-centre case-control study. J Cell Mol Med. 2020;24:9280-6
10. Li Y, Lu T, Wang J, Zhuo Z, Miao L, Yang Z. et al. YTHDC1 gene polymorphisms and neuroblastoma susceptibility in Chinese children. Aging (Albany NY). 2021;13:25426-39
11. Zeng H, Li M, Liu J, Zhu J, Cheng J, Li Y. et al. YTHDF2 Gene rs3738067 A>G Polymorphism Decreases Neuroblastoma Risk in Chinese Children: Evidence From an Eight-Center Case-Control Study. Front Med (Lausanne). 2021;8:797195
12. Guan Q, Lin H, Hua W, Lin L, Liu J, Deng L. et al. Variant rs8400 enhances ALKBH5 expression through disrupting miR-186 binding and promotes neuroblastoma progression. Chin J Cancer Res. 2023;35:140-62
13. Shen C, Wang K, Deng X, Chen J. DNA N(6)-methyldeoxyadenosine in mammals and human disease. Trends Genet. 2022;38:454-67
14. Hao Z, Wu T, Cui X, Zhu P, Tan C, Dou X. et al. N(6)-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol Cell. 2020;78:382-95.e8
15. Kweon SM, Chen Y, Moon E, Kvederaviciute K, Klimasauskas S, Feldman DE. An Adversarial DNA N(6)-Methyladenine-Sensor Network Preserves Polycomb Silencing. Mol Cell. 2019;74:1138-47 e6
16. Yao B, Li Y, Wang Z, Chen L, Poidevin M, Zhang C. et al. Active N(6)-Methyladenine Demethylation by DMAD Regulates Gene Expression by Coordinating with Polycomb Protein in Neurons. Mol Cell. 2018;71:848-57 e6
17. Boulias K, Greer EL. Means, mechanisms and consequences of adenine methylation in DNA. Nat Rev Genet. 2022;23:411-28
18. Xiao CL, Zhu S, He M, Chen D, Zhang Q, Chen Y. et al. N(6)-Methyladenine DNA Modification in the Human Genome. Mol Cell. 2018;71:306-18.e7
19. Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q. et al. N(6)-methyladenine DNA Modification in Glioblastoma. Cell. 2018;175:1228-43 e20
20. Ouyang L, Su X, Li W, Tang L, Zhang M, Zhu Y. et al. ALKBH1-demethylated DNA N6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J Clin Invest. 2021;131:e146985
21. Guo C, Liu Z, Zhang H. DNA 6mA demethylase ALKBH1 regulates DDX18 expression to promote proliferation of human head and neck squamous cell carcinoma. Cell Oncol (Dordr). 2023;46:1097-111
22. Rashad S, Han X, Sato K, Mishima E, Abe T, Tominaga T. et al. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 2020;17:1092-103
23. Lin L, Deng C, Zhou C, Zhang X, Zhu J, Liu J. et al. NSUN2 gene rs13181449 C>T polymorphism reduces neuroblastoma risk. Gene. 2023;854:147120
24. Chang J, Lin L, Zhou C, Zhang X, Yang T, Wu H. et al. Functional polymorphisms of the TET1 gene increase the risk of neuroblastoma in Chinese children. J Cell Mol Med. 2023;27:2239-48
25. Li M, Zhang X, Liu J, Zhou C, Miao L, He J. et al. Association between GPC2 polymorphisms and neuroblastoma risk in Chinese children. J Clin Lab Anal. 2023;37:e24866
26. Jiang S, Sun X, Zhang X, Zhou C, Wu H, He J. et al. MIR938 rs2505901 T>C polymorphism is associated with increased neuroblastoma risk in Chinese children. Biosci Rep. 2023;43:BSR20231223
27. He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP. et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235-44
28. Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K. et al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329-33
29. Hua RX, Fu W, Lin A, Zhou H, Cheng J, Zhang J. et al. Role of FTO gene polymorphisms in Wilms tumor predisposition: A five-center case-control study. J Gene Med. 2021;23:e3348
30. Zhuo Z, Hua RX, Zhang H, Lin H, Fu W, Zhu J. et al. METTL14 gene polymorphisms decrease Wilms tumor susceptibility in Chinese children. BMC Cancer. 2021;21:1294
31. Tian LF, Liu YP, Chen L, Tang Q, Wu W, Sun W. et al. Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res. 2020;30:272-5
32. Zhang M, Yang S, Nelakanti R, Zhao W, Liu G, Li Z. et al. Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res. 2020;30:197-210
33. Ren H, Zhuo ZJ, Duan F, Li Y, Yang Z, Zhang J. et al. ALKBH5 Gene Polymorphisms and Hepatoblastoma Susceptibility in Chinese Children. J Oncol. 2021;2021:6658480
34. Lu H, Li Z, Liu L, Tao Y, Zhou Y, Mao X. et al. A Pan-Cancer Analysis of the Oncogenic Roles of RAD51 in Human Tumors. Advanced Gut & Microbiome Research. 2022;2022:1591377
35. Wei J, Yu X, Yang L, Liu X, Gao B, Huang B. et al. FTO mediates LINE1 m(6)A demethylation and chromatin regulation in mESCs and mouse development. Science. 2022: eabe9582.
36. Ougland R, Jonson I, Moen MN, Nesse G, Asker G, Klungland A. et al. Role of ALKBH1 in the Core Transcriptional Network of Embryonic Stem Cells. Cell Physiol Biochem. 2016;38:173-84
37. Liu Y, Chen Y, Wang Y, Jiang S, Lin W, Wu Y. et al. DNA demethylase ALKBH1 promotes adipogenic differentiation via regulation of HIF-1 signaling. J Biol Chem. 2022;298:101499
Author contact
![]() Corresponding authors: Jing He or Zhenjian Zhuo, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (86-020) 38076560, Email: hejing198374com or hejingorg (Jing He) or Email: zhenjianzhuocom (Zhenjian Zhuo).
Corresponding authors: Jing He or Zhenjian Zhuo, Department of Pediatric Surgery, Guangzhou Institute of Pediatrics, Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, 9 Jinsui Road, Guangzhou 510623, Guangdong, China, Tel./Fax: (86-020) 38076560, Email: hejing198374com or hejingorg (Jing He) or Email: zhenjianzhuocom (Zhenjian Zhuo).

 Global reach, higher impact
Global reach, higher impact