Impact Factor
ISSN: 1837-9664
J Cancer 2024; 15(2):332-342. doi:10.7150/jca.90149 This issue Cite
Research Paper
Causal relationship between gut microbiota and glioblastoma: a two-sample Mendelian randomization study
1. Department of Radiology, The Second Affiliated Hospital of Xinjiang Medical University, Urumqi, 830011, China.
2. Department of Radiology, The Second Xiangya Hospital of Central South University, Changsha, Hunan, 410011, China.
# Chao Ju and Yanjing Chen contributed equally to this work.
Received 2023-9-14; Accepted 2023-11-8; Published 2024-1-1
Abstract
Background: Observational research and medical trials have suggested a connection between gut microbiota and glioblastoma, but it remains unclear if the relationship is causal.
Method: A two-sample Mendelian randomization (MR) study was conducted by employing data from the MiBioGen consortium's largest genome-wide association study (n=18340) and the FinnGen consortium R8 release information (162 cases and 256,583 controls). Inverse variance weighted (IVW), weighted median estimator (WME), weighted model, MR-Egger, simple mode, and MR-PRESSO were used to determine the causal relationship between gut microbiota and glioblastoma. Reverse MR analysis was also performed on bacteria identified as causally related to glioblastoma.
Results: Seven causal relationships were identified between genetic liability in the gut microbiota and glioblastoma, involving various bacterial families and genera. No significant causal effect was found on gut microbiota from glioblastoma, and no significant heterogeneity of instrumental variables (IVs) or horizontal pleiotropy was observed.
Conclusion: A two-sample MR analysis reveals a causal association between the gut microbiota and glioblastoma, highlighting the need for more investigation to comprehend the processes behind this association.
Keywords: Gut microbiota, glioblastoma, Mendelian randomization, Genetics, SNPs
Introduction
Gut microbiota including bacteria, viruses, and fungi refers to the collection of microorganisms, which reside in the human gastrointestinal tract [1]. They play an essential role in various physiological functions, such as digestion, nutrient absorption, and immune system regulation [2]. Recent studies have shown that gut microbiota is also associated with the risk of several diseases, including metabolic diseases [3], autoimmune diseases [4], rheumatoid arthritis [5] and cancer [6,7].
Glioblastoma is a highly aggressive brain tumor that arises from glial cells in the brain. It is one of the most common and deadly forms of primary brain cancer, with less than 10% five-year survival rate, with a median survival time of less than 15 months [8]. The current treatment options for glioblastoma include radiation therapy, surgery, and chemotherapy, but their efficacy is limited and the prognosis remains poor [9].
Bidirectional communication system “gut-brain axis” connects gastrointestinal tract and central nervous system [10]. Studies have shown that gut microbiota dysbiosis, or an imbalance in gut microbial community, can lead to the development of various cancers, including brain tumors [11]. In addition, preclinical studies demonstrated that certain gut bacteria could modulate immune system and affect the efficacy of cancer therapies, including those used to treat glioblastoma [12].
MiBioGen is a comprehensive database of microbial genomes, which provides a platform for ours to access and analyze microbial genomic data. The database is designed to help ours understand the genetic basis of microbial evolution, ecology, and pathogenesis [1]. MiBioGen contains a vast collection of bacterial and archaeal genomes, as well as tools for comparative genomic analysis, annotation, and visualization [13,14]. The FinnGen study is a large-scale genetic study of various disorders in the Finnish population. The study aims to identify genetic factors that contribute to risk of disorders, as well as to understand the biological mechanisms underlying the disorders [14]. The FinnGen database contains genomic data from over 160,000 individuals, including both cases and controls. The data includes information on genetic variants, as well as clinical and phenotypic data [15].
MR is a popular statistical technique used in observational studies to estimate causal effect of an exposure on an outcome by leveraging genetic variation as IVs [16]. This approach exploits random allocation of genetic variants at conception to determine impact of an exposure on an outcome of interest [17]. Two-sample MR approach involves using separate datasets for the genetic variants and the exposure-outcome data, which allows for increased statistical power and flexibility in the analysis [18,19]. One of the key advantages of two-sample MR is that it enables researchers to estimate causal effects for a wide range of exposures and outcomes, without the need for expensive or time-consuming data collection [20]. Additionally, it can help to overcome some limitations of traditional MR, such as weak instrument bias and pleiotropy (when a single genetic variant influences multiple traits) [13]. Overall, Two-sample MR is a powerful and flexible approach that can provide valuable insights into causal relationships between exposures and outcomes.
Therefore, we used the MibioGen and FinnGen databases for the first time to investigate causal relationship between gut microbiota and glioblastoma using two-sample MR method, eventually to accelerate the pace of discovery in the field of human genetics, and provide new insights into genetic basis for disease.
Material and Methods
Study design
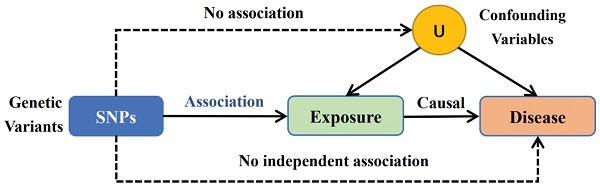
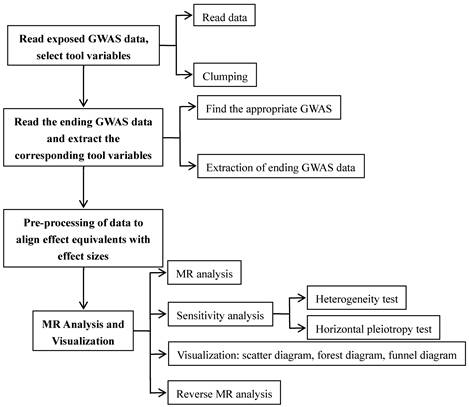
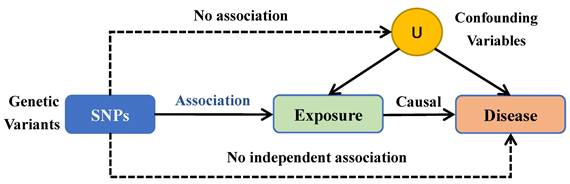
Using a two-sample MR methodology, we assessed the link between the gut microbiota and glioblastoma. To thoroughly study the role played by gut microbiota in the etiology of glioblastoma, we conducted MR studies at five distinct character levels, including phylum, class, order, family, and genus. Figure 1 depicts the research design as well as the fundamental MR assumptions.
Study design and workflow
Exposure data
The aim of the study was to investigate correlations between human genetic variation and gut microbiota, specifically through the use of SNPs linked to composition of human gut microbiota as IVs in a GWAS dataset. The International Consortium MiBioGen conducted a large-scale multi-ethnic GWAS, which analyzed genotyping data and 16S ribosomal RNA gene sequencing from a total of 18,340 participants across 24 cohorts from various countries, including the Germany, United States, Denmark, Canada, Israel, Finland, the United Kingdom, the Netherlands, Belgium, Sweden, and Korea [13,21]. The study identified 211 taxa, including 9 phyla, 16 classes, 20 orders, 35 families, 131 genera [1].
Outcome data
The FinnGen consortium R8 release data [14,22] provided the GWAS summary statistics for glioblastoma. The GWAS involved 256,745 Finnish individuals, of which 162 were cases and 256,583 were controls. To ensure accuracy, the analysis accounted for sex, age, first 10 principal components, and genotyping batch [14].
Instrumental variable selection
IVs is an abbreviation for instrumental variables. The MR method employs genetic variants as IVs to infer the causality of an association. The IVs were selected based on the following criteria: (1) potential IVs were single nucleotide polymorphisms (SNPs) associated with each taxa at the locus-wide significance threshold (P < 5.0 × 10-6) [3]; (2) the linkage disequilibrium (LD) between the SNPs was calculated using the 1000 Genomes project European samples data as the reference panel, and among those SNPs with R2 < 0.001 (clumping window size=10,000 kb), only the SNPs with the lowest P-values were retained; (3) SNPs that have a minor allele frequency (MAF) of ≤ 0.01 were removed; and (4) in the presence of palindromic SNPs existed, forward strand alleles were inferred using allele frequency information.
Statistical analysis
We conducted a study for examining the relationship between features of the microbiome and glioblastoma by employing MR analysis (Figure 2). For features with different IVs, we used six popular MR methods [23], including IVW [20], weighted mode [24], simple mode [24], MR-Egger regression [25], WME [26], and MR-PRESSO [27]. The IVW approach is mentioned to be barely extra effective than the others underneath sure stipulations [26]. Therefore, the consequences with extra than one IV have been usually primarily based on the IVW method, with the other different five methods serving as complements [28].
Three major principles of MR method selection [29]: (1) Preferential use of IVW estimates in the absence of heterogeneity and multi-effects; (2) When there is only heterogeneity and no multi-effects, the results of the WME method are used in preference (the random effects model of IVW can also be used); (3) When there is multiplicity of effects, the results calculated by MR-Egger method are used in preference.
Leave-one-out method refers to gradually eliminate each SNP, calculating meta-effects of remaining SNPs, and observing whether results change after eliminating each SNP, if the results change significantly after eliminating a certain SNP, it means that the presence of a certain SNP has a significant impact on the results [30].
One crucial issue in MR studies is the presence of weak instrumental variable bias [31]. From a traditional empirical perspective, when the F-statistic is blow 10, we typically consider genetic variants as weak instrumental variables. This may introduce some bias into the results, and therefore caution should be exercised in interpreting them at this stage. Ideally, an F-statistic greater than 10 or even greater than 100 would be preferred [32].
Mendelian randomization (MR) methods
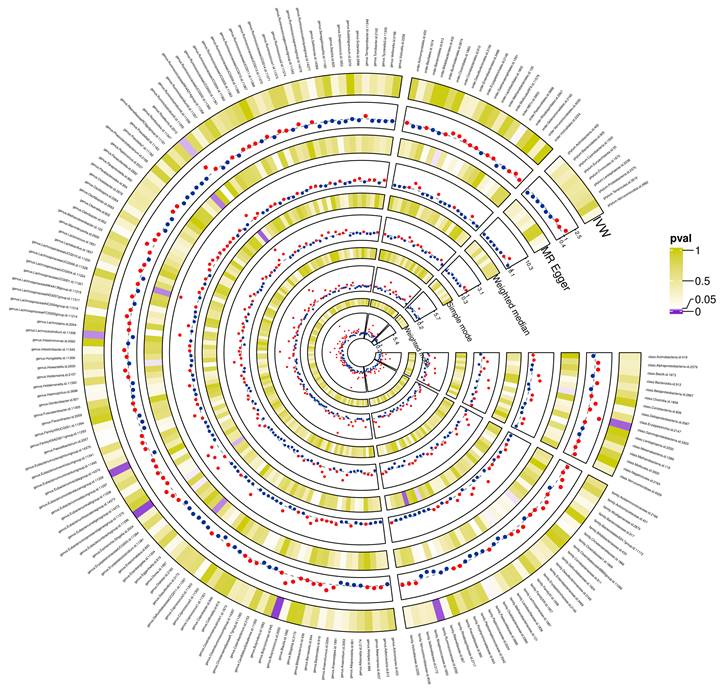
Preliminary correlations between gut microbiota and glioblastoma determined from five popular MR methods. P < 0.05 values were displayed in purple, whereas P > 0.05 estimates were displayed in white or yellow.
Heterogeneity tests were conducted using both Cochran's Q statistic and a two-sample MR package across instruments. Evidence of heterogeneity and invalid instruments was indicated by Q values greater than the number of instruments minus one, while Q statistic values significant at p-values < 0.05 suggested the presence of heterogeneity [33,34].
In order to investigate whether glioblastoma has any causal influence on the noteworthy bacterial, we conducted a reverse MR analysis. In this analysis, glioblastoma was considered as exposure, and identified causal bacterial was treated as outcome. To accomplish this, we employed SNPs associated with glioblastoma as IVs [30]. The settings and procedures used were in line with forward MR.
We performed all statistical analyses using R version 4.2.2 (R Foundation for Statistical Computing, Vienna, Austria). MR analyses had been carried out the usage of the Two-sample MR (version 0.5.6) [35], MR-PRESSO (version 1.0) [27], and qvalue (version 2.30.0) [36] R packages.
Results
SNP selection
Initially, we detected 65, 120, 150, 260, and 902 SNPs related to gut microbiota at the phylum, class, order, family, and genus levels, correspondingly, at a p-value threshold of less than 5 × 10-6. Following several quality control procedures, we handpicked 35 SNPs as instrumental variables (IVs) that met genome-wide statistical significance threshold of p < 5 × 10-6 (Table S1).
All IVs' F statistics exceeded 10, implying the absence of weak instrument bias. Furthermore, MR-PRESSO global test found no evidence of pleiotropic effects (p > 0.05). Finally, after discarding pleiotropic SNPs flagged by MR-PRESSO outlier test and MR-Egger regression, no signs of horizontal pleiotropy were observed in IVs (both MR-PRESSO global test and MR-Egger regression yielded p-values greater than 0.05).
Causal effects of gut microbiota on the development of glioblastoma
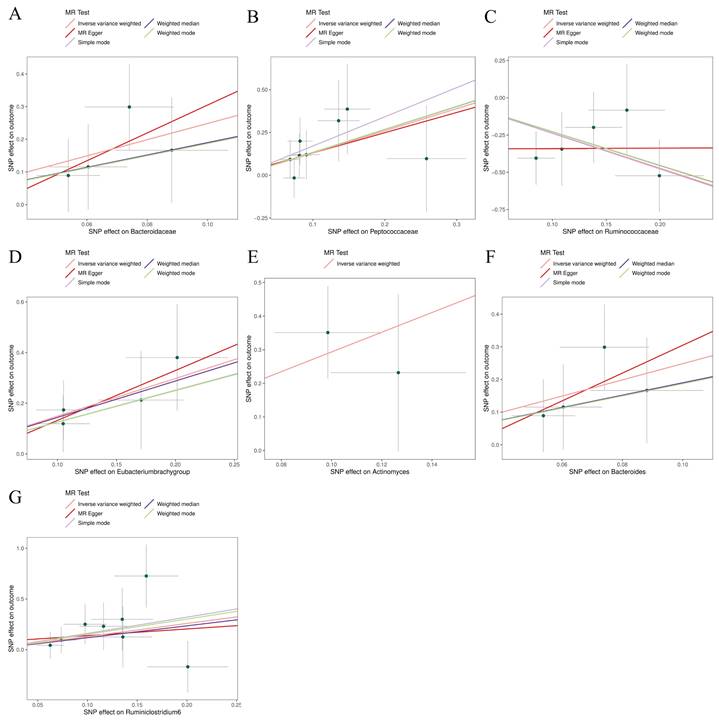
The preliminary associations between bacterial clusters of various levels of classification derived from five popular MR methods and glioblastoma was presented in Figure 3. In the IVs dataset (p < 5 × 10-6), we found a causal relationship between three microbial families and four microbial genera of the gut microbiota and glioblastoma (Table 1). Among the three microbial families, family Ruminococcaceae (OR = 0.094, 95% CI = 0.021-0.417, p = 0.19 × 10-2) was shown to be protective against glioblastoma as assessed by IVW, whereas an increase in the other two microbial families, Bacteroidaceae (OR = 12.003, 95% CI = 1.793-80.32, p = 1.03 × 10-2) and Peptococcaceae (OR = 3.656, 95% CI = 1.233-10.841, p = 1.94 × 10-2), were associated with a high risk of glioblastoma development. In addition, an increase in four microbial genera, Eubacterium (brachy group) (OR = 4.431, 95% CI = 1.529-12.842, p = 0.61 × 10-2), Actinomyces (OR = 18.805, 95% CI = 2.116-167.165, p = 0.85 × 10-2), Bacteroides (OR = 12.003, 95% CI = 1.794-80.320, p = 1.04 × 10-2) and Ruminiclostridium6 (OR = 3.641, 95% CI = 1.009-13.139, p = 4.84 × 10-2) were found to be associated with an increased risk of glioblastoma as assessed by IVW.
Sensitivity analyses
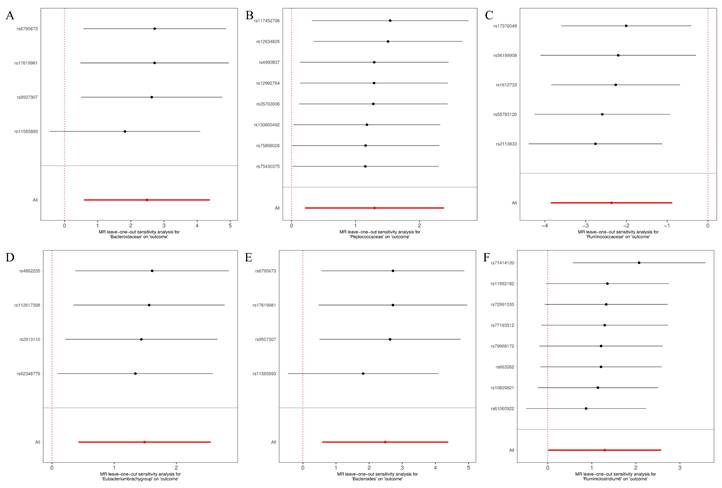
The MR-Egger, weighted mode, simple mode, weighted median, and IVW methods produced comparable causal estimates in both magnitude and direction. Visual inspection revealed probable outliers of the IVs in scatter plots (Figure 4) and leave-one-out plots (Figure 5). Our analysis using the MR-Egger regression intercept approach found no indication of horizontal pleiotropy for gut microbiota in glioblastoma, with a p-value greater than 0.05 (Table S2). Results from MR-PRESSO analysis indicated no outliers in the data (Table S3). Moreover, the Cochrane Q statistics results indicated no significant heterogeneity, with a p-value larger than 0.05 (Table S4).
MR results of causal effects between gut microbiota and glioblastoma (P<5×10-6)
| Gut microbiota(exposure) | method | nSNP | β | SE | p-value | OR | 95%CI |
|---|---|---|---|---|---|---|---|
| Family Bacteroidaceae | MR Egger | 4 | 4.27 | 5.26 | 0.50 | 71.39 | 0.00-2145757 |
| WME | 4 | 1.90 | 1.13 | 0.09 | 6.68 | 0.73-61.61 | |
| IVW | 4 | 2.46 | 0.97 | 0.01 | 12.00 | 1.79-80.32 | |
| Simple mode | 4 | 1.87 | 1.61 | 0.33 | 6.49 | 0.28-151.74 | |
| Weighted mode | 4 | 1.87 | 1.57 | 0.32 | 6.49 | 0.30-140.86 | |
| Family Peptococcaceae | MR Egger | 8 | 1.18 | 1.51 | 0.46 | 3.27 | 0.17-62.92 |
| WME | 8 | 1.34 | 0.75 | 0.07 | 3.82 | 0.88-16.46 | |
| IVW | 8 | 1.30 | 0.55 | 0.02 | 3.66 | 1.23-10.84 | |
| Simple mode | 8 | 1.71 | 1.15 | 0.18 | 5.51 | 0.58-52.60 | |
| Weighted mode | 8 | 1.34 | 1.04 | 0.24 | 3.83 | 0.50-29.20 | |
| Family Ruminococcaceae | MR Egger | 5 | 0.03 | 2.43 | 0.99 | 1.03 | 0.00-119.62 |
| WME | 5 | -2.27 | 1.02 | 0.03 | 0.10 | 0.01-0.76 | |
| IVW | 5 | -2.36 | 0.76 | <0.01 | 0.09 | 0.02-0.42 | |
| Simple mode | 5 | -2.39 | 1.40 | 0.16 | 0.09 | 0.00-1.43 | |
| Weighted mode | 5 | -2.27 | 1.31 | 0.16 | 0.10 | 0.00-1.36 | |
| Genus Eubacteriumbrachygroup | MR Egger | 4 | 1.97 | 2.00 | 0.43 | 7.18 | 0.14-361.09 |
| WME | 4 | 1.44 | 0.67 | 0.032 | 4.21 | 1.13-15.67 | |
| IVW | 4 | 1.49 | 0.54 | <0.01 | 4.43 | 1.53-12.84 | |
| Simple mode | 4 | 1.26 | 0.84 | 0.23 | 3.52 | 0.68-18.09 | |
| Weighted mode | 4 | 1.25 | 0.86 | 0.24 | 3.49 | 0.64-18.95 | |
| Genus Actinomyces | IVW | 2 | 2.93 | 1.11 | <0.01 | 18.80 | 2.12-167.17 |
| Genus Bacteroides | MR Egger | 4 | 4.27 | 5.26 | 0.50 | 71.39 | 0.00-2145757 |
| WME | 4 | 1.90 | 1.16 | 0.10 | 6.68 | 0.69-65.18 | |
| IVW | 4 | 2.49 | 0.97 | 0.01 | 12.00 | 1.79-80.32 | |
| Simple mode | 4 | 1.87 | 1.59 | 0.32 | 6.49 | 0.29-147.15 | |
| Weighted mode | 4 | 1.87 | 1.56 | 0.32 | 6.49 | 0.31-137.60 | |
| Genus Ruminiclostridium6 | MR Egger | 8 | 0.65 | 1.75 | 0.72 | 1.92 | 0.06-59.16 |
| WME | 8 | 1.18 | 0.90 | 0.19 | 3.24 | 0.56-18.78 | |
| IVW | 8 | 1.29 | 0.65 | 0.05 | 3.64 | 1.01-13.14 | |
| Simple mode | 8 | 1.60 | 1.59 | 0.35 | 4.98 | 0.22-111.38 | |
| Weighted mode | 8 | 1.51 | 1.39 | 0.31 | 4.52 | 0.30-69.10 |
CI, confidence interval; IVW, Inverse variance weighted; MR, Mendelian randomization; SNP, single nucleotide polymorphism; SE, standard error; OR, Odds ratio; WME, weighted median estimator.
Scatter plots for the causal association between gut microbiota and glioblastoma. (A) Bacteroidaceae; (B) Peptococcaceae; (C) Ruminococcaceae; (D) Eubacterium (brachy group); (E); (F) Bacteroides; (G) Ruminiclostridium6.
Bi‑directional causal effects between gut microbiota and glioblastoma risk
We used glioblastoma as exposure and the identified causal bacterial as outcome for evaluating any reverse causation effects. Based on five popular MR methods, we found that glioblastoma was no significance causally associated with the identified causal gut microbiota.
Discussion
Using a two-sample MR study, we investigated manageable causal relationship between gut microbiota and glioblastoma, with summary statistics for gut microbiota from the International Consortium MiBioGen and summary statistics for glioblastoma from the FinnGen consortium R8 release data (2022). The findings supported the hypothesis that the increase in abundance of genetic susceptibility in the family Ruminococcaceae was once defensive towards glioblastoma, while the different two organizations of the family, Bacteroidaceae and Peptococcaceae, and four microbial genera, namely, Eubacterium (brachy group), Actinomyces, Bacteroides, and Ruminiclostridium 6, had been observed to extend the hazard of glioblastoma with growing heritage susceptibility abundance. With the help of reverse MR analysis, no appreciable causal association between glioblastoma and the identified causal gut microbiota was previously identified.
The causal relationship between hereditary susceptibility of gut microbiota and exceptional cancers has been established, but the additional interest focused on the gut microbiota and gastrointestinal tumors because they are in the same ecosystem and it is less complicated to find a conceivable causal relationship between them [37,38], although the doable causal relationships between gut microbiota and different cancers is constantly mentioned in the literature [39]. However, the causal relationship between intestinal plant life and glioblastoma has no longer been reported. Zheng et al. found that composition of the microbiota significantly changed in patients with lung cancer compared with control subjects [40]. Zhu et al. also found a change in the gut microbial neighborhood in breast cancer patients [41]. It is undeniable that connection between gut microbiota and the development of cancer is receiving more and more attention. However, there is nonetheless a dearth of solid proof on the microbial elements of gut microbiota that make contributions to most cancer development. Although some possible causative linkages between the gut microbiota and cancer have been hypothesized in some animal models due to the complicated interplay between the gut microbiota and the human host, the precise causal relationship between the two remains undetermined. The following limitations apply to observational studies: it is impossible to determine the temporal order between exposure and result, and it is impossible to account for the impact of several confounding variables [42]. Gut microbiota is influenced by distinctive factors, including diet [43], BMI [44], medications [45], and different factors [46], all of which contribute to the lack of self-assurance in observational studies. For these reasons, the doable causal relationships between gut microbiota and most cancers nevertheless warrants similar research. Inspired by the application of a massive pattern GWAS database, we have been in a position to use summary-level statistics for causal inference between gut microbiota and glioblastoma, with the hope of exploiting the brain-gut axis for improved interpretation.
Leave-one-out for the causal association between gut microbiota and glioblastoma. (A) Bacteroidaceae; (B) Peptococcaceae; (C) Ruminococcaceae; (D) Eubacterium (brachy group); (E) Bacteroides; (F) Ruminiclostridium6.
A growing body of research found possible links between gut microbiota selected for our study and other cancers. For instance, Patients with intrahepatic cholangiocarcinoma (ICC) had a greater abundance of family Peptostreptococcaceae than in sufferers with hepatocellular carcinoma or cirrhosis and in healthy individuals [47]. Abundance of the family of Ruminococcaceae was higher in patients with vascular invasion (VI) than in patients with ICC without VI [48]. Our MR results suggetsted that family Peptostreptococcaceae is a risk factor for glioblastoma. In a study of the association between the gut microbiome and primary liver cancer using a two-sample Mendelian randomization and case-control approach, the family Ruminococcaceae was found as a protective factor against hepatocellular liver cancer and the genus Bacteroidetes as a protective factor for intrahepatic cholangiocarcinoma in a two-sample MR study. In contrast, in case-control studies, healthy controls possessed higher relative abundance of the family Ruminococcaceae and the genus Bacteroidetes than patients with hepatocellular hepatocellular carcinoma [49]. As shown in our results, our MR results suggested that family Ruminococcaceae was also found as a protective factor against glioblastoma, while the genus Bacteroides and the family Bacteroidaceae are risk factors for glioblastoma. One study showed that genus Ruminiclostridium 6 might be potential pathogens with a low malignant potential in plasmacytoid ovarian cancer [6]. In a study of oral microbiota as novel biomarkers for colorectal cancer screening, Eubacterium (brachy group) was ideal for differentiating healthy controls (HCs) from colorectal cancer (CRC) patients [50]. And in the present study, we also found Eubacterium (brachy group) to be a risk factor for glioblastoma. The family Actinomycetaceae of the order Actinomycetales, which also includes the families Mycobacteriaceae (Mycobacterium), Nocardiaceae (Nocardia, Rhodococcus), Corynebacteriaceae (Corynebacterium), and others, contains the genus Actinomyces. All belong to the Actinobacteria phylum [51].
Gut microbiota and various intestinal metabolites influence glioma development and progression through neural signaling, microglia regulation, and energy metabolism [52]. Gut microbiota is involved in regulation of glioma proliferation and immune response. A study comparing the changes that occur in gut microbiota of glioma-bearing mice compared to healthy mice found a significant decrease in ratio of the Firmicutes to the Bacteroidetes and showed significant differences in relative abundance of the Verrucomicrobia and Akkermansia. This shows that there is a correlation between reduced abundance or structural dysregulation of the bacterial flora and glioma progression [53]. The presence of a large number of immune cells and functional lymphatic vessels in the glioma microenvironment and the dysfunction of the lymphatic network constituted by them can promote the progression of glioma [54,55]. Since the gut microbiota itself can participate in regulating development and function of immune cells, and its metabolites can also influence function of the lymphatic network, the flora can be directly or indirectly involved in the regulation of glioma progression [56]. In addition, dysbiosis of gut microbiota can induce a suppressed immune response in the tumor microenvironment, thus increasing the immune escape of glioma cells and accelerating the progression of glioma [56,57]. Loss of flora diversity also leads to a defective immune function in the CNS, which promotes the proliferation of tumor-associated macrophages, mainly abnormal microglia, and ultimately promotes glioma progression [58]. Therefore, we can draw the following inference that modulation or transplantation of bacterial flora is expected to be a new means of treatment for glioma by modulating the immune system.
The main gut microbiota of the organism that produce SCFAs are Bacteroides, Bifidobacterium, Propionibacterium, Lactobacillus, Clostridium, Roseburia, and Pseudomonas spp [59]. Amongst the seven gut microbiomes we explored in this study, Bacteroidaceae is a family of bacteria in the order Bacteroidetes, and the type genus of this family is Bacteroides [51]. In glioma, SCFAs regulate growth and metabolism of glioma cells by affecting immunity, angiogenesis, and epigenetic modifications of the body. In addition to SCFAs, non-SCFAs metabolites produced by gut microbiota metabolism also have a wide range of regulatory effects on organism. Polyamines and nitric oxide are derivatives of spermidine and are also produced by the metabolism of gut microbiota [60]. Nitric oxide, on the other hand, promotes tumor cell growth by inhibiting the JAK3-STAT5 signaling pathway, interfering with T cell function and inducing apoptosis [61].
The gut microbiota is involved in the regulation of multiple systems of the body by directly or indirectly influencing hormone secretion and immune response, and is involved in regulating multiple response responses in the glioma microenvironment. In addition, SCFAs and amino acids in the metabolites of the flora are not only involved in the immune response to glioma, but also in the regulation of gene epigenetic modifications. Therefore, gut microbiota and its metabolites can be used as potential targets for anti-glioma therapy, providing ideas and directions for the discovery of new targets for anti-glioma therapy.
There are advantages of this study in the following points: Compared to traditional observational studies, MR analysis can usually achieve RCT-like results that are less subject to confounding factors and reverse causality. Therefore, this study used a two-sample MR framework using genetic variation to assess and analyze causal relationships between gut microbiota and glioblastoma with reverse causal inference. Genetic variation in the gut microbiota was obtained by maximal GWAS meta-analysis, ensuring strength of instrumentation in the MR analysis. Multiple methods were used to perform sensitivity analyses with consistent results, and the robustness of our findings was demonstrated using MR-PRESSO and MR-Egger regression intercept tests to detect and exclude horizontal pleiotropy.
There are certain limitations in this study that need to be noted when we interpret the results. First, GWAS of gut microbiota was obtained from the International Consortium MiBioGen, which included populations from different countries, mainly European populations, while GWAS of GBM was obtained from the FinnGen consortium R8 release data, which included populations of Finnish individuals. Due to the different exposure and outcome GWAS populations, demographic heterogeneity may have biased the results, while the generalizability of MR results in other populations warrants future investigation. Second, the limited number of GBM cases in the FinnGen data and the lack of specific typing of glioblastoma subtypes in the GWAS database may reduce the persuasiveness of this study and lead to a poor use of gut flora to explain the treatment response and prognosis of different subtypes of glioblastoma. Therefore, a further increase in the number of glioblastoma cases and subtyping of glioblastoma is needed to investigate potential causal relationships between gut microbiota and different subtypes of glioblastoma in more depth. Third, we lowered the P threshold between exposure and instrumental variables, which may increase the risk of violating the first hypothesis of MR. However, we performed an F-statistic test for each SNP and did not find SNPs with F-statistic values less than 10, indicating absence of weak SNPs in MR estimates. To better investigate disease pathogenesis, recent studies have proposed the use of multiple histological platforms for an integrated understanding analysis of disease pathogenesis in the context of complex interactions of genetic and environmental factors over time [62].
Conclusions
In summary, our study comprehensively assessed the causal relationship between gut microbiota and glioblastoma. Our results suggest that there are one positive causal direction and six negative directions with glioblastoma. This study may provide new insights into mechanisms and drug-targets of gut microbiota-mediated cancer development.
Abbreviations
AA: acetic acid; BA: butyric acid; CRC: colorectal cancer; MR: Mendelian randomization; MAF: minor allele frequency; HC: healthy control; IVW: Inverse variance weighted; IV: instrumental variable; ICC: intrahepatic cholangiocarcinoma; IGF-1: insulin-like growth factor-1; LD: linkage disequilibrium; PA: propionic acid; SNP: single nucleotide polymorphism; SCFAs: short-chain fatty acids; VI: vascular invasion; WME: weighted median estimator.
Supplementary Material
Supplementary tables.
Acknowledgements
Funding
This study was supported by the Clinical Research Center for Medical Imaging in Hunan Province (2020SK4001), Key Emergency Project of Pneumonia Epidemic of Novel Coronavirus Infection in Hunan Province (2020SK3006), National Natural Science Foundation of China (61971451), Innovative Province Special Construction Foundation of Hunan Province (2019SK2131), the Science and Technology Innovation Program of Hunan Province (2021RC4016), the National Defense Science and Technology Collaborative Innovation Major Project of Central South University (2021gfcx05) and Scientific Research Program of Hunan Provincial Health Commission (202209044797).
Data availability statement
The datasets analyzed during the current study are available in the MiBioGen repository, https:// mibiogen.gcc.rug.nl/, and the FinnGen repository, https:// r8.finngen.fi/.
Author contributions
Conceptualisation, Chao Ju; methodology, Chao Ju, Yanjing Chen and Yijie Huang; verification of the underlying data, Yanjing Chen and Longtao Yang; writing original draft, Chao Ju; writing review and editing, Jun Liu. All the authors participated in planning, execution, and analysis and have read and approved the final submitted version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dragioti E, Li H, Tsitsas G. et al. A large-scale meta-analytic atlas of mental health problems prevalence during the COVID-19 early pandemic. J Med Virol. 2022;94(5):1935-1949
2. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20(2):145-155
3. Sanna S, van Zuydam NR, Mahajan A. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51(4):600-605
4. Xu Q, Ni J-J, Han B-X. et al. Causal Relationship Between Gut Microbiota and Autoimmune Diseases: A Two-Sample Mendelian Randomization Study. Front Immunol. 2021;12:746998
5. Inamo J. Non-causal association of gut microbiome on the risk of rheumatoid arthritis: a Mendelian randomisation study. Ann Rheum Dis. 2021;80(7):e103
6. Long Y, Tang L, Zhou Y. et al. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. 2023;21(1):66
7. Wei Z, Yang B, Tang T. et al. Gut microbiota and risk of five common cancers: A univariable and multivariable Mendelian randomization study. Cancer Med. 2023;12(9):10393-10405
8. Venkataramani V, Yang Y, Schubert MC. et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 2022;185(16):2899-2917.e31
9. Rong L, Li N, Zhang Z. Emerging therapies for glioblastoma: current state and future directions. J Exp Clin Cancer Res. CR. 2022;41(1):142
10. Liu L, Huh JR, Shah K. Microbiota and the gut-brain-axis: Implications for new therapeutic design in the CNS. EBioMedicine. 2022;77:103908
11. Mehrian-Shai R, Reichardt JKV, Harris CC. et al. The Gut-Brain Axis, Paving the Way to Brain Cancer. Trends Cancer. 2019;5(4):200-207
12. Caspani G, Swann J. Small talk: microbial metabolites involved in the signaling from microbiota to brain. Curr Opin Pharmacol. 2019;48:99-106
13. Wang J, Kurilshikov A, Radjabzadeh D. et al. Meta-analysis of human genome-microbiome association studies: the MiBioGen consortium initiative. Microbiome. 2018;6(1):101
14. Kurki MI. et al. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv. 2022 03.03.22271360
15. Kurki MI, Karjalainen J, Palta P. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508-518
16. Sekula P, Del Greco M F, Pattaro C. et al. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J Am Soc. Nephrol. JASN. 2016;27(11):3253-3265
17. Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA. 2017;318(19):1925-1926
18. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89-98
19. Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1-22
20. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658-665
21. MiBioGen consortium. MiBioGen. https://mibiogen.gcc.rug.nl/. Accessed 16 Sep. 2022
22. FinnGen. FinnGen R8 release. https://r8.finngen.fi/. Accessed 1 Dec. 2022
23. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333-2355
24. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985-1998
25. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525
26. Bowden J, Davey Smith G, Haycock PC. et al. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304-314
27. Verbanck M, Chen C-Y, Neale B. et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698
28. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880-1906
29. Cho Y, Haycock PC, Sanderson E. et al. Exploiting horizontal pleiotropy to search for causal pathways within a Mendelian randomization framework. Nat Commun. 2020;11(1):1010
30. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081
31. Burgess S, Thompson SG, CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755-764
32. Staiger D. JHSR. Instrumental variables regression with weak instruments. Econometrica. 1997;65:557-86
33. Greco M FD, Minelli C, Sheehan NA. et al. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34(21):2926-2940
34. Bowden J, Del Greco M F, Minelli C. et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol. 2019;48(3):728-742
35. Hemani G, Zheng J, Elsworth B. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408
36. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440-9445
37. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690-704
38. Tong Y, Gao H, Qi Q. et al. High fat diet, gut microbiome and gastrointestinal cancer. Theranostics. 2021;11(12):5889-5910
39. Garrett WS. The gut microbiota and colon cancer. Science. 2019;364(6446):1133-1135
40. Zheng Y, Fang Z, Xue Y. et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. 2020;11(4):1030-1042
41. Zhu J, Liao M, Yao Z. et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. 2018;6(1):136
42. Colditz GA. Overview of the epidemiology methods and applications: strengths and limitations of observational study designs. Crit Rev Food Sci Nutr. 2010 50 Suppl 1(s1):10-12
43. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(1):35-56
44. Deehan EC, Zhang Z, Riva A. et al. Elucidating the role of the gut microbiota in the physiological effects of dietary fiber. Microbiome. 2022;10(1):77
45. Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69(8):1510-1519
46. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823-1836
47. Solé C, Guilly S, Da Silva K. et al. Alterations in Gut Microbiome in Cirrhosis as Assessed by Quantitative Metagenomics: Relationship With Acute-on-Chronic Liver Failure and Prognosis. Gastroenterology. 2021;160(1):206-218.e13
48. Jia X, Lu S, Zeng Z. et al. Characterization of Gut Microbiota, Bile Acid Metabolism, and Cytokines in Intrahepatic Cholangiocarcinoma. Hepatol Baltim Md. 2020;71(3):893-906
49. Ma J, Li J, Jin C. et al. Association of gut microbiome and primary liver cancer: A two-sample Mendelian randomization and case-control study. Liver Int Off J Int Assoc Study Liver. 2023;43(1):221-233
50. Rezasoltani S, Aghdaei HA, Jasemi S. et al. Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers. 2022;15(1):192
51. Parte AC, Sardà Carbasse J, Meier-Kolthoff JP. et al. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70(11):5607-5612
52. Rutsch A, Kantsjö JB, Ronchi F. The Gut-Brain Axis: How Microbiota and Host Inflammasome Influence Brain Physiology and Pathology. Front Immunol. 2020;11:604179
53. Patrizz A, Dono A, Zorofchian S. et al. Glioma and temozolomide induced alterations in gut microbiome. Sci Rep. 2020;10(1):21002
54. Song E, Mao T, Dong H. et al. VEGF-C-driven lymphatic drainage enables immunosurveillance of brain tumours. Nature. 2020;577(7792):689-694
55. Hu X, Deng Q, Ma L. et al. Meningeal lymphatic vessels regulate brain tumor drainage and immunity. Cell Res. 2020;30(3):229-243
56. Lyu Y, Yang H, Chen L. Metabolic regulation on the immune environment of glioma through gut microbiota. Semin Cancer Biol. 2022;86(Pt 2):990-997
57. Qiu Q, Lin Y, Ma Y. et al. Exploring the Emerging Role of the Gut Microbiota and Tumor Microenvironment in Cancer Immunotherapy. Front Immunol. 2020;11:612202
58. Ma Q, Xing C, Long W. et al. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflammation. 2019;16(1):53
59. Layden BT, Angueira AR, Brodsky M. et al. Short chain fatty acids and their receptors: new metabolic targets. Transl Res J Lab Clin Med. 2013;161(3):131-140
60. Dai Z, Wu Z, Hang S. et al. Amino acid metabolism in intestinal bacteria and its potential implications for mammalian reproduction. Mol Hum Reprod. 2015;21(5):389-409
61. Dehhaghi M, Kazemi Shariat Panahi H, Heng B. et al. The Gut Microbiota, Kynurenine Pathway, and Immune System Interaction in the Development of Brain Cancer. Front Cell Dev Biol. 2020;8:562812
62. Agustí A, Melén E, DeMeo DL. et al. Pathogenesis of chronic obstructive pulmonary disease: understanding the contributions of gene-environment interactions across the lifespan. Lancet Respir Med. 2022;10(5):512-524
Author contact
![]() Corresponding author: Jun Liu, 410011, junliu123edu.cn.
Corresponding author: Jun Liu, 410011, junliu123edu.cn.

 Global reach, higher impact
Global reach, higher impact