Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(7):1195-1201. doi:10.7150/jca.84470 This issue Cite
Research Paper
The impact of FOXP3 polymorphisms on oral cancer progression and clinicopathological characteristics
1. Institute of Medicine, Chung Shan Medical University, Taichung, Taiwan
2. Department of Dentistry, Changhua Christian Hospital, Changhua, Taiwan
3. Department of Post-Baccalaureate Medicine, College of Medicine, National Chung Hsing University, Taichung, Taiwan.
4. Institute of Oral Sciences, Chung Shan Medical University, Taichung, Taiwan
5. Department of Dentistry, Chung Shan Medical University Hospital, Taichung, Taiwan
6. Division of Hematology and Oncology, Department of Internal Medicine, Chung Shan Medical University Hospital, Taichung, Taiwan
7. School of Medicine, Chung Shan Medical University, Taichung, Taiwan
8. Department of Otolaryngology, Chung Shan Medical University Hospital, Taichung, Taiwan
9. Department of Medical Research, Chung Shan Medical University Hospital, Taichung, Taiwan
Received 2023-4-11; Accepted 2023-4-19; Published 2023-5-5
Abstract
Oral cancer is the sixth leading cause of cancer mortality worldwide. Genetic, epigenetic, and epidemiological risk factors were suggested to be correlated with the carcinogenesis of oral cancer. In this study, we focused on the correlations of FOXP3 single-nucleotide polymorphisms (SNPs) to oral cancer susceptibility and clinicopathological characteristics. The FOXP3 SNPs rs3761547, rs3761548, rs3761549, and rs2232365 in 1053 controls and 1175 male patients with oral cancer were analyzed with real-time polymerase chain reaction. The results showed that the betel quid chewer who carried the FOXP3 rs3761548 polymorphic variant “T” were significantly associated with lower risk to develop oral cancer [AOR (95% CI) = 0.649 (0.437-0.964); p = 0.032]. The betel quid chewers with genotypic variant “T” of FOXP3 rs3761548 in male oral cancer patients were associated with lower risk of cell differentiated grade [AOR (95% CI) = 0.592 (0.377-0.930); p = 0.023]. The carriers of FOXP3 rs3761548 polymorphic variant “T” in male oral cancer patients with alcohol consumption were associated with lower risk to develop greater tumor [AOR (95% CI) = 0.609 (0.378-0.983); p = 0.042] and lower risk of cell differentiated grade [AOR (95% CI) = 0.440 (0.248-0.779); p = 0.005]. In conclusion, our results have revealed that the FOXP3 rs3761548 polymorphic variant “T” was associated with lower risk of oral cancer susceptibility, greater tumor size, and cell differentiated grade among betel quid chewers. The FOXP3 rs3761548 polymorphisms may play a role as pivotal biomarkers to predict oral cancer disease development and prognosis.
Keywords: oral cancer, FOXP3, polymorphism, betel quid
Introduction
Oral cancer is the sixth leading cause of cancer mortality worldwide [1, 2]. Genetic, epigenetic, and epidemiological risk factors such as tobacco smoking, alcohol consumption, betel quid chewing, ethnicity, familial and genetic predisposition, immunosuppression, viruses, and radiation were suggested to be correlated with the carcinogenesis of oral cancer [3-7]. Transcription factor forkhead box protein 3 (FOXP3) is a member of the family of forkhead transcription factors and is mainly localized in the nucleus [8]. The FOXP3 was suggested as the main marker of regulatory T cells (Tregs) [8, 9]. It was suggested that the Treg cell differentiation and function was depended on FOXP3 expression [8-11], and the Tregs were suggested to play a critical role in oral squamous cell carcinoms (OSCC) [12, 13]. The tumor microenvironment (TME) is a complicate system composed by cancer cells, immune cells and stromal cell [8, 14, 15]. Tregs in the TME were suggested as checkpoint which blockade immune surveillance of tumors and suppress antitumor immune responses [8, 14, 16].
Previous studies have suggested that the tumor-infiltrating FOXP3 positive T cells were associated with development and poor prognosis in OSCC [13, 17], and the FOXP3 polymorphisms were associated with cancer development and prognosis in various cancers such as cervical cancer [18, 19], gastric adenocarcinoma [20], and colorectal cancer [21]. However, the associations and influences of FOXP3 polymorphisms to OSCC tumor progression and clinicopathological characteristics remained not well-investigated. In this study, we focused on four SNPs of FOXP3 rs3761547, rs3761548, rs3761549, and rs2232365, and try to elucidate their associations to oral cancer susceptibility and clinicopathological characteristics with environmental risk factors.
Materials and Methods
Study subjects
A total of 1175 male oral cancer patients and 1053 cancer-free controls were enrolled in our study. All the participants were recruited during 2016 to 2020 at Chung Shan Medical University Hospital in Taichung, Taiwan. For the TNM staging, the oral cancer patients who enrolled in our study were staged clinically at the time of diagnosis according to the American Joint Committee on Cancer (AJCC). The tumor differentiation was examined and rated according to the AJCC classification by pathologist. The demographic data such as age and gender were reported by each participant. Individuals who without oral precancerous disease, including leukoplakia, erythroplakia, and history of cancer of any sites were enrolled as the control group of our study. This project was approved by the institutional review board of Chung Shan Medical University Hospital (IRB number: CS1-21151), and informed written consent was provided to each patient who enrolled in this study.
Sample preparation and DNA extraction
The peripheral blood specimens from oral cancer patients and normal controls who participated in our study were collected for genomic DNA extraction [22]. The genomic DNA extraction assay was performed following the manufacturer's manual of QIAamp DNA blood mini kits to acquire the DNA. The final step of DNA elution was completed with the Tris-EDTA (TE) buffer. The extracted DNA was used as DNA template in further real-time polymerase chain reactions (PCRs) [23].
Selection of FOXP3 SNPs
In the current study, a total of four FOXP3 SNPs rs3761547, rs3761548, rs3761549, and rs2232365 were selected based on the International HapMap Project database [24]. The FOXP3 rs3761547 was selected because the FOXP3 rs3761547 polymorphisms was suggested as a male-specific risk factor in multiple sclerosis [25], or important markers to determine susceptibility and risk of diseases such as Crohn's disease [26] and interstitial lung disease (ILD) [27]. The rs3761548 was selected because it was suggested that the FOXP3 rs3761548 SNPs could affect the susceptibility to gastric adenocarcinoma (GA) and the serum IL-35, IL-10, and TGF-β concentrations [20], and the rs3761548 polymorphism of FOXP3 gene may provide as a risk factor in the development of breast cancer (BC) [28]. The rs3761549 was selected because the rs3761549 variants may play a role to protect against high-risk human papilloma virus (HR-HPV) and cervical cancer malignant lesions by down-regulating FOXP3 [18].
FOXP3 SNPs genotyping
Assessment of allelic discrimination for the FOXP3 rs3761547, rs3761548, rs3761549, and rs2232365 SNP was performed with an ABI StepOne Software v2.3 Real-Time PCR System. The TaqMan assay was applied for the analysis of genotyping. The analysis and calculation of the collected data of genotyping was processed with the SDS 7000 series software (Applied Biosystems, Foster City, CA, USA) [29].
Statistical analysis
To compare the age (years), betel quid chewing, cigarette smoking, alcohol drinking, tumor stage, tumor T status, lymph node status, metastasis, and cell differentiation, the student's t test or Chi-squared test was performed between the patients with oral cancer and the controls. A p < 0.05 was suggested to present statistically significant. To compare the odds ratio (OR) with their 95% confidence intervals (CIs) of the association between the oral cancer risk and genotypic frequencies, and the clinical pathological statuses, the data was assessed and analyzed by multiple logistic regression models. All the data analysis in our study was evaluated with SAS statistical software (Version 9.1, 2005; SAS Institute, Cary, NC).
Results
The distribution of demographical characteristics in 1053 controls and 1175 male patients with oral cancer was listed in Table 1. In the current study, we observed that the distributions of age (years) ≦ 55 was 531 (50.4%) in controls and 601 (51.2%) in oral cancer patients. The distributions of environmental risk factors exposure between the controls and oral cancer patients were 175 (16.6%) and 856 (72.9%) in betel quid chewing (p < 0.001), 559 (53.1%) and 985 (83.8%) in cigarette smoking (p < 0.001), and 213 (20.2%) and 536 (45.6%) in alcohol drinking (p < 0.001), respectively.
The distributions of demographical characteristics in 1053 controls and 1175 male patients with oral cancer
| Variable | Controls (N=1053) | Patients (N=1175) | p value |
|---|---|---|---|
| Age (yrs) | |||
| ≦ 55 | 531 (50.4%) | 601 (51.2%) | p = 0.734 |
| > 55 | 522 (49.6%) | 574 (48.8%) | |
| Betel quid chewing | |||
| No | 878 (83.4%) | 319 (27.1%) | |
| Yes | 175 (16.6%) | 856 (72.9%) | p < 0.001* |
| Cigarette smoking | |||
| No | 494 (46.9%) | 190 (16.2%) | |
| Yes | 559 (53.1%) | 985 (83.8%) | p < 0.001* |
| Alcohol drinking | |||
| No | 840 (79.8%) | 639 (54.4%) | |
| Yes | 213 (20.2%) | 536 (45.6%) | p < 0.001* |
| Stage | |||
| I+II | 554 (47.2%) | ||
| III+IV | 621 (52.8%) | ||
| Tumor T status | |||
| T1+T2 | 584 (49.7%) | ||
| T3+T4 | 591 (50.3%) | ||
| Lymph node status | |||
| N0 | 781 (66.5%) | ||
| N1+N2+N3 | 394 (33.5%) | ||
| Metastasis | |||
| M0 | 1165 (99.2%) | ||
| M1 | 10 (0.8%) | ||
| Cell differentiation | |||
| Well differentiated | 169 (14.4%) | ||
| Moderately or poorly differentiated | 1006 (85.6%) |
Mann-Whitney U test or Fisher's exact test was used between healthy controls and patients with oral cancer. * p value < 0.05 as statistically significant.
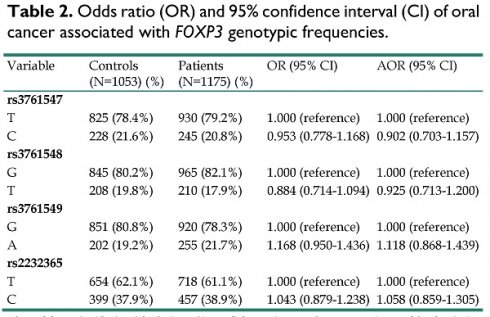
Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with FOXP3 genotypic frequencies.
| Variable | Controls (N=1053) (%) | Patients (N=1175) (%) | OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs3761547 | ||||
| T | 825 (78.4%) | 930 (79.2%) | 1.000 (reference) | 1.000 (reference) |
| C | 228 (21.6%) | 245 (20.8%) | 0.953 (0.778-1.168) | 0.902 (0.703-1.157) |
| rs3761548 | ||||
| G | 845 (80.2%) | 965 (82.1%) | 1.000 (reference) | 1.000 (reference) |
| T | 208 (19.8%) | 210 (17.9%) | 0.884 (0.714-1.094) | 0.925 (0.713-1.200) |
| rs3761549 | ||||
| G | 851 (80.8%) | 920 (78.3%) | 1.000 (reference) | 1.000 (reference) |
| A | 202 (19.2%) | 255 (21.7%) | 1.168 (0.950-1.436) | 1.118 (0.868-1.439) |
| rs2232365 | ||||
| T | 654 (62.1%) | 718 (61.1%) | 1.000 (reference) | 1.000 (reference) |
| C | 399 (37.9%) | 457 (38.9%) | 1.043 (0.879-1.238) | 1.058 (0.859-1.305) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, betel quid chewing, cigarette smoking, and alcohol drinking.
The genotype distributions of FOXP3 gene polymorphisms in 1053 controls and 1175 male patients with oral cancer were listed in Table 2. The highest distribution frequencies in patients with oral cancer of FOXP3 genetic polymorphisms rs3761547, rs3761548, rs3761549, and rs2232365 were polymorphic variant T, polymorphic variant G, polymorphic variant G, and polymorphic variant T, respectively. The odds ratios (ORs) with their 95% confidence intervals (CIs) were estimated by logistic regression models. After adjustment for the effects of age, betel quid chewing, cigarette smoking, and alcohol drinking, no significant associations were found between the oral cancer patients and the controls (Table 2).
We further analyzed the ORs and 95% CIs of oral cancer associated with FOXP3 genotypic frequencies among betel quid chewer. In 142 of total 856 patients, a significant association was found in those individuals who carried the FOXP3 rs3761548 polymorphic variant T, with a lower risk of oral cancer susceptibility [The adjusted odds ratio (AOR) (95% CI):0.649 (0.437-0.964); p = 0.032] (Table 3). Moreover, after we analyzed the AOR and 95% CI of clinical statuses associated with genotypic frequencies of FOXP3 rs3761548 in oral cancer patients, we found that in 856 betel quid chewers among the total 1175 oral cancer patients, carriers who with the rs3761548 polymorphic “T” variant have a lower risk to develop poorer cell differentiated grade [(AOR) (95% CI):0.592 (0.377-0.930); p = 0.023] (Table 4). Besides, in 536 alcohol drinkers among the total 1175 oral cancer patients, carriers who with the rs3761548 genotypic “T” variant have a lower risk to develop greater tumor size [(AOR) (95% CI):0.609 (0.378-0.983); p = 0.042] and poorer cell differentiated grade [(AOR) (95% CI):0.440 (0.248-0.779); p = 0.005] (Table 5).
Discussion
In this study, we revealed the associations between the FOXP3 SNPs and oral cancer. The betel quid chewing, smoking, and alcohol consumption are well-known risk factors responsible for impaired DNA repair capacity in OSCC carcinogenesis, disease development, and progression [30-34]. Most of the oral cancer patients in Taiwan were male who exhibit habits of cigarette smoking as well as betel nut chewing, and over 90% of oral cancer was OSCC [35]. In our study, statistically significant associations of these risk factors were found in 1175 male patients with oral cancer compared with the controls, respectively (p < 0.001, table 1).
Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with FOXP3 genotypic frequencies among betel quid chewer.
| Variable | Controls (N=176) (%) | Patients (N=856) (%) | OR (95% CI) | AOR (95% CI)a |
|---|---|---|---|---|
| rs3761547 | ||||
| T | 132 (75.4%) | 674 (78.7%) | 1.000 (reference) | 1.000 (reference) |
| C | 43 (24.6%) | 182 (21.3%) | 0.829 (0.566-1.213) | 0.845 (0.576-1.239) |
| rs3761548 | ||||
| G | 134 (76.6%) | 714 (83.4%) | 1.000 (reference) | 1.000 (reference) |
| T | 41 (23.4%) | 142 (16.6%) | 0.650 (0.439-0.963)b | 0.649 (0.437-0.964)c |
| rs3761549 | ||||
| G | 140 (80.0%) | 665 (77.7%) | 1.000 (reference) | 1.000 (reference) |
| A | 35 (20.0%) | 191 (22.3%) | 1.149 (0.767-1.720) | 1.161 (0.774-1.742) |
| rs2232365 | ||||
| T | 101 (57.7%) | 530 (61.9%) | 1.000 (reference) | 1.000 (reference) |
| C | 74 (42.3%) | 326 (38.1%) | 0.840 (0.603-1.168) | 0.855 (0.613-1.191) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
a The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, cigarette smoking, and alcohol drinking.
b p = 0.032;c p = 0.032.
Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of FOXP3 rs3761548 in male oral cancer patients.
| Total (N=1175) | Betel Quid Chewers (N=856) | Non-Betel Quid Chewers (N=319) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | G (N=965) | T (N=210) | p value | G (N=714) | T (N=142) | p value | G (N=251) | T (N=68) | p value |
| Clinical Stage | |||||||||
| Stage I+II | 453 (46.9%) | 101 (48.1%) | 0.794 | 336 (47.1%) | 71 (50.0%) | 0.536 | 117 (46.6%) | 30 (44.1%) | 0.544 |
| Stage III+IV | 512 (53.1%) | 109 (51.9%) | 378 (52.9%) | 71 (50.0%) | 134 (53.4%) | 38 (55.9%) | |||
| Tumor size | |||||||||
| ≦ T2 | 475 (49.2%) | 109 (51.9%) | 0.432 | 355 (49.7%) | 76 (53.5%) | 0.402 | 120 (47.8%) | 33 (48.5%) | 0.943 |
| > T2 | 490 (50.8%) | 101 (48.1%) | 359 (50.3%) | 66 (46.5%) | 131 (52.2%) | 35 (51.5%) | |||
| Lymph node metastasis | |||||||||
| No | 637 (66.0%) | 144 (68.6%) | 0.499 | 478 (67.0%) | 100 (70.4%) | 0.434 | 159 (63.4%) | 44 (64.7%) | 0.949 |
| Yes | 328 (34.0%) | 66 (31.4%) | 236 (33.0%) | 42 (29.6%) | 92 (36.6%) | 24 (35.3%) | |||
| Metastasis | |||||||||
| M0 | 958 (99.3%) | 207 (98.6%) | 0.311 | 710 (99.4%) | 139 (97.9%) | 0.087 | 248 (98.8%) | 68 (100.0%) | - |
| M1 | 7 (0.7%) | 3 (1.4%) | 4 (0.6%) | 3 (2.1%) | 3 (1.2%) | 0 (0.0%) | |||
| Cell differentiated grade | |||||||||
| Well | 133 (13.8%) | 36 (17.1%) | 0.193 | 101 (14.1%) | 31 (21.8%) | 0.023a | 32 (12.8%) | 5 (7.4%) | 0.143 |
| Moderate or poor | 832 (86.2%) | 174 (82.9%) | 613 (85.9%) | 111 (78.2%) | 219 (87.2%) | 63 (92.6%) | |||
Cell differentiate grade: grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated.
a The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, cigarette smoking, and alcohol drinking. AOR = 0.592 (0.377-0.930).
Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of FOXP3 rs3761548 in male oral cancer patients with or without alcohol consumption.
| Alcohol Drinkers (N=536) | Non-Alcohol Drinkers (N=639) | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | G (N=451) | T (N=85) | AOR (95% CI)a | p valuea | G (N=514) | T (N=125) | AOR (95% CI) | p value |
| Clinical Stage | ||||||||
| Stage I+II | 204 (45.2%) | 44 (51.8%) | 1.000 (reference) | 0.300 | 249 (48.4%) | 57 (45.6%) | 1.000 (reference) | 0.527 |
| Stage III+IV | 247 (54.8%) | 41 (48.2%) | 0.781 (0.490-1.245) | 265 (51.6%) | 68 (54.4%) | 1.136 (0.765-1.689) | ||
| Tumor size | ||||||||
| ≦ T2 | 227 (50.3%) | 53 (62.4%) | 1.000 (reference) | 0.042 | 248 (48.3%) | 56 (44.8%) | 1.000 (reference) | 0.454 |
| > T2 | 224 (49.7%) | 32 (37.6%) | 0.609 (0.378-0.983) | 266 (51.7%) | 69 (55.2%) | 1.163 (0.783-1.729) | ||
| Lymph node metastasis | ||||||||
| No | 287 (63.6%) | 57 (67.1%) | 1.000 (reference) | 0.597 | 350 (68.1%) | 87 (69.6%) | 1.000 (reference) | 0.695 |
| Yes | 164 (36.4%) | 28 (32.9%) | 0.875 (0.532-1.437) | 164 (31.9%) | 38 (30.4%) | 0.918 (0.598-1.408) | ||
| Metastasis | ||||||||
| M0 | 446 (98.9%) | 83 (97.7%) | 1.000 (reference) | 0.325 | 512 (99.6%) | 124 (99.2%) | 1.000 (reference) | 0.624 |
| M1 | 5 (1.1%) | 2 (2.3%) | 2.312 (0.436-12.261) | 2 (0.4%) | 1 (0.8%) | 1.842 (0.160-21.213) | ||
| Cell differentiated grade | ||||||||
| Well | 56 (12.4%) | 21 (24.7%) | 1.000 (reference) | 0.005 | 77 (15.0%) | 15 (12.0%) | 1.000 (reference) | 0.378 |
| Moderate or poor | 395 (87.6%) | 64 (75.3%) | 0.440 (0.248-0.779) | 437 (85.0%) | 110 (88.0%) | 1.308 (0.721-2.372) | ||
Cell differentiate grade: grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated.
a The adjusted odds ratio (AOR) with their 95% confidence intervals were estimated by multiple logistic regression models after controlling for age, cigarette smoking, and betel quid chewing.
For the correlations and hazard of these risk factors of OSCC to FOXP3 expression, a previous study of pediatric asthma has suggested that environmental tobacco smoke (ETS) exposure and passive smoking could induce pediatric asthma by affecting the balance of Treg/Th17 cells, and passive smoking was associated with significantly reduced levels of FOXP3 and tumor growth factor-β, which were associated with Treg cells [36]. For the effect of alcohol exposure and FOXP3 expression, it was suggested that the alcohol interferes with the kinetics of FOXP3, and influences the balance of Treg/Th17 cells following lipopolysaccharide (LPS) exposure [37]. In Taiwan, in most cases, more than 80% of the male oral cancer patients were both smokers and betel quid chewers [38]. Previous study has observed that it took two decades on average of betel quid chewing and cigarette smoking before oral cavity cancer diagnosis, and the amount of life-time consumption for these substances was astonishing to these patients [38], suggesting a potentially great impact of these carcinogenic substances to FOXP3 expression in these patients. However, the associations of betel quid chewing to FOXP3 expression remained unclear till date.
We further examined the influence of the FOXP3 genotypic frequencies to oral cancer susceptibility. Previous studies have suggested the controversial role and inconsistency of FOXP3 gene polymorphisms to disease susceptibility in various cancers [21, 39-41]. In our study, no statistical significant associations were found between the oral cancer patients and the controls, suggesting a limited carcinogenic effect and disease susceptibility of FOXP3 polymorphisms to oral cancer (Table 2). Intriguingly, after we analyzed the FOXP3 genotypic frequencies among betel chewers in our study, a statistical significant association was found between the oral cancer patients and the normal controls that carried the genotypic variant “T” of FOXP3 rs3761548, with a lower risk to develop oral cancer [AOR (95% CI) = 0.649 (0.437-0.964), p = 0.032] (Table 3). Moreover, after we analyzed the clinical statuses associated with genotypic frequencies of FOXP3 rs3761548 in oral cancer patients, we found that those betel quid chewers among the oral cancer patients who carried the FOXP3 rs3761548 polymorphic variant “T” were associated with lower risk of moderate or poor cell differentiated grade [AOR (95% CI) = 0.592 (0.377-0.930), p = 0.023] (Table 4). Furthermore, for those alcohol drinkers among the male oral cancer patients in our study, individuals who carried the genotypic variant “T” of FOXP3 rs3761548 were found to be associated with lower risk to develop larger tumor size [AOR (95% CI) = 0.609 (0.378-0.983), p = 0.042] and moderate or poorer cell differentiated grade [AOR (95% CI) = 0.440 (0.248-0.779), p = 0.005] (Table 5). These results have suggested a possible phenomenon that the FOXP3 SNPs (rs3761548, especially) may have greater impact and influence to oral cancer disease progression and development rather than tumor carcinogenesis (Table 3-5).
The role of FOXP3 expression in OSCC remained controversial. Some studies have associated the tumor-infiltrating FOXP3+ T cells and FOXP3 overexpression with poor prognosis in OSCC [17, 42], whereas some studies have indicated that high numbers of FOXP3+ T cells were significantly associated with prolonged overall survival [43, 44], and the levels of FOXP3+ Tregs were significantly higher in surviving oral cancer patients [43, 45]. For the associations between the FOXP3 rs3761548 polymorphisms and cancer risk, most studies have associated the A allele or the AA genotype of rs3761548 with increased cancer susceptibility, disease progression, and poor prognosis [20, 41, 46-50]. However, in contrast, a previous study which focused on human papillomavirus infection and cervical cancer precursor lesions has suggested that the homozygous genotype of the rs3761548 variants (A/A) may exert a protective role against HPV infection in women, and the rs3761548 variants (A/A) was observed to be related to decreased FOXP3 expression [19]. Compared with this result, in a study of Triple negative breast cancer (TNBC), it was observed that a positive association for FOXP3 rs3761548 homozygous AA was related to TNBC susceptibility, and most of the TNBC patients (83%) have showed a strong staining for FOXP3 protein in the tumor cells, suggesting a positive association of FOXP3 expression with TNBC susceptibility and prognosis [50]. One possible mechanism to explain these discriminations and inconsistencies was that it is not the overall expression of FOXP3 but its intracellular localization which really represents a prognostic parameter [51]. The elevated levels of the cytoplasmicFOXP3 (cFOXP3)/nuclear FOXP3 (nFOXP3) ratio within tumor infiltrating CD4(+) T cells was proposed as a predictor of OSCC recurrence [51], and alternative splicing of FOXP3 (FOXP3Δ2, FOXP3Δ7, and FOXP3Δ2Δ7 isoforms) may play a critical mechanism which enables different aspects of FOXP3 function in T cells [52]. The limitations to our study is that we lack of the data of FOXP3 expression levels and the exact cellular localization of FOXP3 to these oral cancer patients, so more detailed analysis could not be performed. Future well-designed studies are required to evaluate the mechanisms of FOXP3 rs3761548 SNPs to oral cancer, especially the associations between the FOXP3 SNPs to FOXP3 expression levels and their shifting balance of cytoplasmic and nuclear localization during OSCC progression, which ultimately impact and influence the oral cancer disease prognosis.
In conclusion, our study first demonstrated the associations of FOXP3 polymorphisms to oral cancer disease susceptibility and clinical statuses. The betel quid chewers in male oral cancer patients who carried the FOXP3 rs3761548 polymorphic variant “T” were significantly associated with lower risk to develop oral cancer and moderate or poorer cell differentiated grade. The carriers of the FOXP3 rs3761548 polymorphic variant “T” in male oral cancer patients who with alcohol consumption were associated with lower risk to develop greater tumor size and poorer cell differentiated grade. The FOXP3 rs3761548 may play a role as pivotal tumor marker to predict and evaluate oral cancer disease susceptibility, tumor progression, and prognosis.
Acknowledgements
This study was supported by Chung Shan Medical University and Changhua Christian Hospital (CSMU-CCH-111-02).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Motlokwa PK, Tsima BM, Martei YM, Ralefala T, Galebole F, Stephens-Shields AJ. et al. Disparities in Oral Cancer Stage at Presentation in a High HIV Prevalence Setting In Sub-Saharan Africa. JCO Glob Oncol. 2022;8:e2100439
2. Lu HJ, Peng CY, Tseng HC, Hsin CH, Chuang CY, Chen CC. et al. Preoperative prediction model to evaluate salvage surgery in patients with recurrent or second primary oral cavity squamous cell carcinoma. Oral Oncol. 2022;131:105951
3. Su SC, Chang LC, Huang HD, Peng CY, Chuang CY, Chen YT. et al. Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis. 2021;42:127-135
4. Su CW, Chang YC, Chien MH, Hsieh YH, Chen MK, Lin CW. et al. Loss of TIMP3 by promoter methylation of Sp1 binding site promotes oral cancer metastasis. Cell Death Dis. 2019;10:793
5. Yang SF, Yang WE, Kuo WH, Chang HR, Chu SC, Hsieh YS. Antimetastatic potentials of flavones on oral cancer cell via an inhibition of matrix-degrading proteases. Arch Oral Biol. 2008;53:287-294
6. Chang YA, Weng SL, Yang SF, Chou CH, Huang WC, Tu SJ. et al. A Three-MicroRNA Signature as a Potential Biomarker for the Early Detection of Oral Cancer. Int J Mol Sci. 2018;19:758
7. Su SC, Hsieh MJ, Lin CW, Chuang CY, Liu YF, Yeh CM. et al. Impact of HOTAIR Gene Polymorphism and Environmental Risk on Oral Cancer. J Dent Res. 2018;97:717-724
8. Wang J, Gong R, Zhao C, Lei K, Sun X, Ren H. Human FOXP3 and tumour microenvironment. Immunology. 2023;168:248-255
9. Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490-500
10. Hori S, Sakaguchi S. Foxp3: a critical regulator of the development and function of regulatory T cells. Microbes Infect. 2004;6:745-751
11. Raugh A, Allard D, Bettini M. Nature vs. nurture: FOXP3, genetics, and tissue environment shape Treg function. Front Immunol. 2022;13:911151
12. Chao JL, Korzinkin M, Zhavoronkov A, Ozerov IV, Walker MT, Higgins K. et al. Effector T cell responses unleashed by regulatory T cell ablation exacerbate oral squamous cell carcinoma. Cell Rep Med. 2021;2:100399
13. Aggarwal S, Sharma SC, S ND. Dynamics of regulatory T cells (Tregs ) in patients with oral squamous cell carcinoma. J Surg Oncol. 2017;116:1103-1113
14. McRitchie BR, Akkaya B. Exhaust the exhausters: Targeting regulatory T cells in the tumor microenvironment. Front Immunol. 2022;13:940052
15. Su SC, Hsieh MJ, Yang WE, Chung WH, Reiter RJ, Yang SF. Cancer metastasis: Mechanisms of inhibition by melatonin. J Pineal Res. 2017;62:e12370
16. Bergerhoff K, Pedersen M. Isolation and Analysis of Tumor-Infiltrating Treg. Methods Mol Biol. 2023;2559:51-63
17. Hayashi T, Yoshikawa K, Suzuki S, Gosho M, Ueda R, Kazaoka Y. Tumor-infiltrating FoxP3+ T cells are associated with poor prognosis in oral squamous cell carcinoma. Clin Exp Dent Res. 2022;8:152-159
18. Shi F, Pang XX, Li GJ, Chen ZH, Dong MY, Wang JL. Genetic association study of intron variants in the forkhead box protein P3 gene in Chinese patients diagnosed with cervical cancer. J Cell Mol Med. 2022;26:2658-2672
19. Cezar-Dos-Santos F, Ferreira RS, Okuyama NCM, Trugilo KP, Sena MM, Pereira ER. et al. FOXP3 immunoregulatory gene variants are independent predictors of human papillomavirus infection and cervical cancer precursor lesions. J Cancer Res Clin Oncol. 2019;145:2013-2025
20. Ezzeddini R, Somi MH, Taghikhani M, Moaddab SY, Masnadi Shirazi K, Shirmohammadi M. et al. Association of Foxp3 rs3761548 polymorphism with cytokines concentration in gastric adenocarcinoma patients. Cytokine. 2021;138:155351
21. Lou Y, Fan M, Shuai R, Yao C. FOXP3 rs2280883 polymorphism confers susceptibility to colorectal cancer in a Chinese Han population. Gene. 2020;734:144395
22. Chen MK, Chiou HL, Su SC, Chung TT, Tseng HC, Tsai HT. et al. The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol. 2009;45:e222-226
23. Chen YT, Lin CW, Chou YE, Su SC, Chang LC, Lee CY. et al. Potential impact of ADAM-10 genetic variants with the clinical features of oral squamous cell carcinoma. J Cell Mol Med. 2023;27:1144-1152
24. International HapMap C. The International HapMap Project. Nature. 2003;426:789-796
25. Wawrusiewicz-Kurylonek N, Chorazy M, Posmyk R, Zajkowska O, Zajkowska A, Kretowski AJ. et al. The FOXP3 rs3761547 Gene Polymorphism in Multiple Sclerosis as a Male-Specific Risk Factor. Neuromolecular Med. 2018;20:537-543
26. Xia S, Zhang D, Zheng S, Wu C, Lin Q, Ying S. et al. Association of Crohn's disease with Foxp3 gene polymorphisms and its colonic expression in Chinese patients. J Clin Lab Anal. 2019;33:e22835
27. Yao J, Zhang T, Zhang L, Han K, Zhang L. FOXP3 polymorphisms in interstitial lung disease among Chinese Han population: A genetic association study. Clin Respir J. 2018;12:1182-1190
28. Arabpour F, Shafizad A, Rahimzadeh M, Norouzan M, Naderi N. FoxP3gene polymorphism is associated with breast cancer in Iranian patients. Exp Oncol. 2018;40:309-314
29. Su S, Chien M, Lin C, Chen M, Yang S. RAGE gene polymorphism and environmental factor in the risk of oral cancer. J Dent Res. 2015;94:403-411
30. Su SC, Chang LC, Lin CW, Chen MK, Yu CP, Chung WH. et al. Mutational signatures and mutagenic impacts associated with betel quid chewing in oral squamous cell carcinoma. Hum Genet. 2019;138:1379-1389
31. Su SC, Lin CW, Ju PC, Chang LC, Chuang CY, Liu YF. et al. Association of LINC00673 Genetic Variants with Progression of Oral Cancer. J Pers Med. 2021;11:468
32. Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP. et al. Exome Sequencing of Oral Squamous Cell Carcinoma Reveals Molecular Subgroups and Novel Therapeutic Opportunities. Theranostics. 2017;7:1088-1099
33. Su SC, Yeh CM, Lin CW, Hsieh YH, Chuang CY, Tang CH. et al. A novel melatonin-regulated lncRNA suppresses TPA-induced oral cancer cell motility through replenishing PRUNE2 expression. J Pineal Res. 2021;71:e12760
34. Yang SF, Huang HD, Fan WL, Jong YJ, Chen MK, Huang CN. et al. Compositional and functional variations of oral microbiota associated with the mutational changes in oral cancer. Oral Oncol. 2018;77:1-8
35. Lin CW, Yang WE, Lee WJ, Hua KT, Hsieh FK, Hsiao M. et al. Lipocalin 2 prevents oral cancer metastasis through carbonic anhydrase IX inhibition and is associated with favourable prognosis. Carcinogenesis. 2016;37:712-722
36. Jing W, Wang W, Liu Q. Passive smoking induces pediatric asthma by affecting the balance of Treg/Th17 cells. Pediatr Res. 2019;85:469-476
37. von Haefen C, Mei W, Menk M, Klemz R, Jones A, Wernecke KD. et al. Ethanol changes gene expression of transcription factors and cytokine production of CD4+ T-cell subsets in PBMCs stimulated with LPS. Alcohol Clin Exp Res. 2011;35:621-631
38. Tsai KY, Su CC, Lin YY, Chung JA, Lian Ie B. Quantification of betel quid chewing and cigarette smoking in oral cancer patients. Community Dent Oral Epidemiol. 2009;37:555-561
39. Chen Y, Qi X, Bian C, Ling C, Yi T, Mu X. et al. The association of FOXP3 gene polymorphisms with cancer susceptibility: a comprehensive systemic review and meta-analysis. Biosci Rep. 2019;39:BSR20181809
40. You D, Wang Y, Zhang Y, Li Q, Yu X, Yuan M. et al. Association of Foxp3 promoter polymorphisms with susceptibility to endometrial cancer in the Chinese Han women. Medicine (Baltimore). 2018;97:e0582
41. Cheng Z, Guo Y, Ming L. Functional Foxp3 polymorphisms and the susceptibility to cancer: An update meta-analysis. Medicine (Baltimore). 2018;97:e11927
42. Song JJ, Zhao SJ, Fang J, Ma D, Liu XQ, Chen XB. et al. Foxp3 overexpression in tumor cells predicts poor survival in oral squamous cell carcinoma. BMC Cancer. 2016;16:530
43. Bron L, Jandus C, Andrejevic-Blant S, Speiser DE, Monnier P, Romero P. et al. Prognostic value of arginase-II expression and regulatory T-cell infiltration in head and neck squamous cell carcinoma. Int J Cancer. 2013;132:E85-93
44. Liu S, Liu D, Li J, Zhang D, Chen Q. Regulatory T cells in oral squamous cell carcinoma. J Oral Pathol Med. 2016;45:635-639
45. Wolf GT, Chepeha DB, Bellile E, Nguyen A, Thomas D, McHugh J. et al. Tumor infiltrating lymphocytes (TIL) and prognosis in oral cavity squamous carcinoma: a preliminary study. Oral Oncol. 2015;51:90-95
46. Jahan P, Ramachander VR, Maruthi G, Nalini S, Latha KP, Murthy TS. Foxp3 promoter polymorphism (rs3761548) in breast cancer progression: a study from India. Tumour Biol. 2014;35:3785-3791
47. He YQ, Bo Q, Yong W, Qiu ZX, Li YL, Li WM. FoxP3 genetic variants and risk of non-small cell lung cancer in the Chinese Han population. Gene. 2013;531:422-425
48. Banin Hirata BK, Losi Guembarovski R, Vitiello GAF, Guembarovski AL, Brajao de Oliveira K, Watanabe MAE. FOXP3 Allelic Variants and Haplotype Structures Are Associated with Aggressive Breast Cancer Subtypes. Dis Markers. 2017;2017:6359603
49. Jiang W, Zheng L, Xu L, Zhang Y, Liu X, Hu L. et al. Association between FOXP3 gene polymorphisms and risk of differentiated thyroid cancer in Chinese Han population. J Clin Lab Anal. 2017;31:e22104
50. Lopes LF, Guembarovski RL, Guembarovski AL, Kishima MO, Campos CZ, Oda JM. et al. FOXP3 transcription factor: a candidate marker for susceptibility and prognosis in triple negative breast cancer. Biomed Res Int. 2014;2014:341654
51. Weed DT, Walker G, De La Fuente AC, Nazarian R, Vella JL, Gomez-Fernandez CR. et al. FOXP3 subcellular localization predicts recurrence in oral squamous cell carcinoma. PLoS One. 2013;8:e71908
52. Magg T, Mannert J, Ellwart JW, Schmid I, Albert MH. Subcellular localization of FOXP3 in human regulatory and nonregulatory T cells. Eur J Immunol. 2012;42:1627-1638
Author contact
![]() Corresponding authors: Ying-Erh Chou, PhD. or Shun-Fa Yang, PhD. School of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: intointo814com (Ying-Erh Chou); ysfedu.tw (Shun-Fa Yang)
Corresponding authors: Ying-Erh Chou, PhD. or Shun-Fa Yang, PhD. School of Medicine, Chung Shan Medical University, 110, Section 1, Chien-Kuo N. Road, Taichung, Taiwan, ROC. Fax: 886-4-24723229. E-mail: intointo814com (Ying-Erh Chou); ysfedu.tw (Shun-Fa Yang)

 Global reach, higher impact
Global reach, higher impact