Impact Factor
ISSN: 1837-9664
J Cancer 2023; 14(3):480-489. doi:10.7150/jca.79593 This issue Cite
Research Paper
Prognostic MicroRNA Fingerprints Predict Recurrence of Early-Stage Hepatocellular Carcinoma Following Hepatectomy
1. OncoSeek Limited, Hong Kong Science and Technology Parks, Hong Kong Special Administrative Region, People's Republic of China
2. Department of Clinical Oncology, Queen Mary Hospital, LKS Faculty of Medicine, The Hong Kong Special Administrative Region, People's Republic of China
3. Department of Surgery, Queen Mary Hospital, LKS Faculty of Medicine, The Hong Kong Special Administrative Region, People's Republic of China
*Contributed equally to this work.
Received 2022-10-6; Accepted 2022-12-24; Published 2023-2-5
Abstract
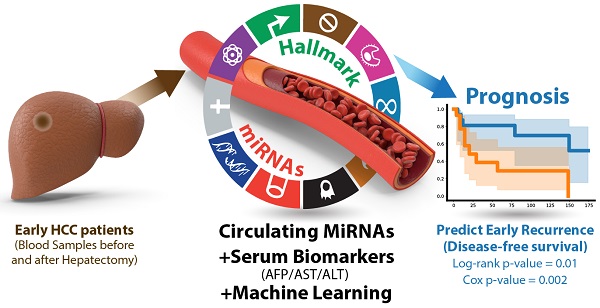
Purpose: This study aims to develop liquid biopsy assays for early HCC diagnosis and prognosis.
Methods: Twenty-three microRNAs were first consolidated as a panel (HCCseek-23 panel) based on their reported functions in HCC development. Serum samples were collected from 103 early-stage HCC patients before and after hepatectomy. Quantitative PCR and machine learning random forest models were applied to develop diagnostic and prognostic models.
Results: For HCC diagnosis, HCCseek-23 panel demonstrated 81% sensitivity and 83% specificity for identifying HCC in the early-stage; it showed 93% sensitivity for identifying alpha-fetoprotein (AFP)-negative HCC. For HCC prognosis, the differential expressions of 8 microRNAs (HCCseek-8 panel: miR-145, miR-148a, miR-150, miR-221, miR-223, miR-23a, miR-374a, and miR-424) were significantly associated with disease-free survival (DFS) (Log-rank test p-value = 0.001). Further model improvement using these HCCseek-8 panel in combination with serum biomarkers (i.e. AFP, ALT, and AST) demonstrated a significant association with DFS (Log-rank p-value = 0.011 and Cox proportional hazards analyses p-value = 0.002).
Conclusion: To the best of our knowledge, this is the first report to integrate circulating miRNAs, AST, ALT, AFP, and machine learning for predicting DFS in early HCC patients undergoing hepatectomy. In this setting, HCCSeek-23 panel is a promising circulating microRNA assay for diagnosis, while HCCSeek-8 panel is promising for prognosis to identify early HCC recurrence.
Keywords: Hepatocellular carcinoma, miRNA fingerprints, Liquid biopsy, Machine learning, HCC diagnosis, HCC prognosis, hepatectomy
Introduction
Hepatocellular carcinoma (HCC) is a highly fatal cancer, accounting for nearly 830,000 deaths every year worldwide [1]. Traditionally, alpha-fetoprotein (AFP) is the most common serological biomarker for HCC detection. However, AFP detection is known to have a low specificity issue and low positive predictive value (PPV) [2-4]. Therefore, more reliable biomarkers for both HCC diagnosis and prognosis are needed.
MicroRNAs are endogenously expressed small non-coding RNA first discovered by Ambros and Ruvkun groups in 1993. Circulating microRNAs have been under the spotlight for diagnostic and prognostic applications because of their functions in various liver diseases such as viral hepatitis, alcoholic or non-alcoholic liver diseases, liver fibrosis, and cirrhosis [5,6]. Since miRNA has been reported to outperform AFP for HCC diagnosis [6-8], circulating microRNA is one of the emerging liquid biopsy technologies to complement AFP for HCC screening and prognosis [7-9].
The advent of machine learning enables us to analyze multi-dimensional data for clinical applications [10,11]. In HCC, Chaudhary et al applied machine learning to analyze the genetics of the tissue biopsies for clinical application [12]. However, whether machine learning can translate the molecular genetic profile from the liquid biopsy for HCC prognosis still needs to be examined. Previously our group compared different machine learning strategies such as regression-based (logistic regression), non-regression-based (neural network), and ensemble machine learning models (random forest and gradient boosting) using liquid biopsy miRNA profile for HCC diagnosis. We identified random forest as a promising model for identifying HCC. In this study, we aim to apply the random forest model for HCC prognosis. To do so, we analyzed multi-dimensional data, including microRNA expressions, dynamic change in miRNA levels before and after hepatectomy, and traditional serum biomarkers.
This study sought to build a liquid biopsy microRNA (miRNA) assay for HCC diagnosis and prognosis. We began by consolidating 23 miRNAs (HCCseek-23 panel) essential for HCC development. Subsequently, we tested the diagnostic power of HCCseek-23 panel for early HCC diagnosis. After that, we narrowed down the miRNA panel to eight signature miRNAs (HCCseek-8 panel) based on their association with patient survival. Ultimately, we developed an integrated prognostic model with the HCCseek-8 panel in combination with the traditional serum biomarkers AST, ALT, and AFP.
Methodology
Patient enrolment criteria
This is a retrospective study with all the data prospectively collected in a patient database. Patients with clinically diagnosed resectable HCC were included. All the patients had CT/MRI typical features of HCC. Patients who were diagnosed with HCC were discussed in the multidisciplinary meeting with oncologists, radiologists, and surgeons. Patients who were considered technically resectable by minimally invasive surgery and with good liver function reserved were included. Patients with the extrahepatic disease, Child C liver cirrhosis, and severe pre-existing medical conditions were excluded. A total of 103 HCC patients treated with hepatectomy at Queen Mary Hospital between January 2006 and October 2012 were included in this study (Table 1). All blood samples were obtained after written informed consent. Eighty-one patients were diagnosed with stage I and 17 patients were diagnosed with stage II based on The Hong Kong liver cancer (HKLC) staging system.
Clinical characteristics of HCC patients treated with hepatectomy
| 98 patients (196 serum samples in total) | ||||
|---|---|---|---|---|
| Sex | Male | Female | ||
| 75 | 23 | |||
| AFP group | AFP positive | AFP negative | ||
| 53 | 45 | |||
| HCC stage | Stage I | Stage II | ||
| 81 | 17 | |||
| Mean | Std | Min | Max | |
| Age(year) | 60.94 | 10.5 | 28 | 82 |
| Body weight(kg) | 63.71 | 12.5 | 31.5 | 107 |
| Body Height(cm) | 162.65 | 7.63 | 144 | 179 |
| Pre-operation AST(u/L) | 40.58 | 18.63 | 16 | 109 |
| Pre-operation ALT (u/L) | 42.47 | 24.49 | 12 | 142 |
| Pre-operation platelet (10^9/L) | 162.83 | 51.31 | 66 | 288 |
| Pre-operation AFP (ng/ml) | 930.46 | 5458.68 | 1 | 53430 |
| Overall survival (month) | 91.91 | 51.01 | 4.14 | 180.04 |
| Disease-free survival (month) | 65.03 | 54.7 | 0.72 | 177.51 |
AFP>= 20ng/mL= AFP positive group; AFP< 20ng/mL= AFP negative group
Random forest model development
Random forest is a modified version of the bagging tree-based machine learning technique. It avoids overfitting by sampling the samples and features randomly at first. Initially, the samples (i.e. HCC patients) are randomly subset to multiple training sets by bootstrapping. Then, in each training set, a decision tree is established by randomly extracting a part of the features from the features (i.e. microRNAs, AST, ALT, AFP) until an optimal solution is reached. The final prediction result is the classification result from the majority vote among all the decision trees in the random forest model. For the first section of developing an HCC diagnostic model, 55 intermediate-stage HCC patients (Barcelona Clinic Liver Cancer system stage B) and 33 healthy individuals reported in our previous paper were used [13]. The microRNA expression qPCR results were split into a training dataset (n= 70) and a test dataset (n= 18), followed by random forest model development. After that, the HCCseek-23 random forest model's diagnostic performance was evaluated using 196 serum samples from the early HCC patients treated with hepatectomy (Table 1). For the second section of developing an HCC prognostic model, as mentioned in the enrolment criteria section, a total of 103 HCC patients treated with hepatectomy at Queen Mary Hospital between January 2006 and October 2012 were included in this study. The samples were collected before (B1) and after the surgical operation (B2). Samples from five patients were removed due to poor quality (i.e. more than 10 miRNAs showed Ct value higher than 35. The clinical information of the subjects is summarized in Table 1. During the model development, we first classified the patients into two groups based on the median survival data (i.e. 86.2 months for overall survival (OS) and 58.81 months for disease-free survival (DFS)). The long and short OS group has a mean OS of 135.9 and 47.3 months, respectively, while the long and short DFS group has a mean DFS of 110.3 and 19.7 months, respectively. After labeling the samples with the survival classification, we split the dataset (n=98) into the training dataset (n=68) and test dataset (n=30). Subsequently, the survival group classifications and the microRNA expression levels were subjected to random forest model development, followed by validation with Log-rank and Univariate Cox proportional hazards analyses.
Sample preparation and miRNA isolation
To ensure the quality of the serum sample, a two-step centrifugation was applied. First, we removed the blood cells with low-speed centrifugation, followed by high-speed centrifugation to remove residual impurities. A volume of 300 uL serum sample was transferred to a new 1.5mL centrifuge tube and the protein debris was precipitated with high-salt buffer for 1 min at room temperature. The supernatant containing RNA was then precipitated by isopropanol, followed by RNA purification using a column-based Silica membrane technology. After washing the columns with ethanol-based buffer, RNase-free H2O was added into the column, followed by 1-minute room temperature incubation and RNA elution. The eluted RNA was subjected to PolyA tailing reaction with PolyA polymerase and ATP and incubated for 60 minutes at 37°C, followed by 5-minute deactivation at 70°C. Subsequently, cDNA synthesis was performed by adding oligo-dt adapter primer, miRNA-specific forward primer, reverse transcriptase, and incubate for 20 minutes at 42°C, followed by 85°C incubation for 5 minutes [13].
MicroRNA analysis
Roche Light-Cycler 480 was applied for qPCR reaction. The Ct values for microRNA were generated with the 2nd derivative maximum of fluorescence curve. For the microRNA with no Ct value after calculation, a Ct value of 45 was filled to facilitate further model development. The Ct values were converted to fold differences by the equation 2^(-ΔCt) normalized to the endogenous expression level of miR-451a.
Statistical analysis and data visualization
Clinicopathologic and genomic variables were tested for their effect on the survival probabilities of patients; the effect of these prognostic factors were estimated by the Kaplan-Meier method and compared between survival subgroups by Log-rank test and univariate Cox proportional hazards analyses. The effect size of univariate survival analysis was estimated based on p-values and hazard ratio with 95% confidence intervals (95% CI); p-values < 0.05 were interpreted as statistically significant associated with survival outcome. A hazard ratio >1 indicates that the variable associated with increased risk of death, while hazard ratio< 1 indicates the variable associated with decreased risk of death; hazard ratio= 1 means that variable have no effect on the length of survival. Lifelines 0.25.7 in python (version 3.8.3) was used for statistical analyses. Student's T test were performed using SciPy package in python environment, the effect size was estimated by the difference between two means divided by pooled standard deviation. Univariate Cox proportional hazards model and the Log-rank test were analyzed using lifelines.statistics package in python. The results were visualized in Kaplan-Meier curves and Cox proportional hazards model hazard ratio plot using matplotlib.pyplot and lifeline package in python.
Results
Establishing HCCseek-23 microRNA panel
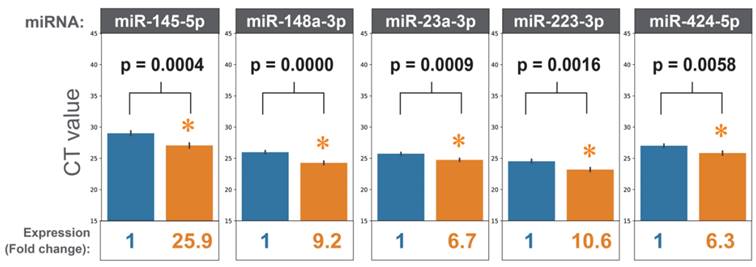
To design an HCC-specific microRNA panel, we reviewed the literature focusing on microRNAs' functions in HCC development. Twenty-three microRNAs were selected based on their reported functions in regulating cancer hallmarks in HCC (Supplementary Table 1). Quantitative PCR (qPCR) was performed to detect the expression levels of these 23 microRNAs (HCCseek-23 panel) in the serum samples taken from 103 stage I and II HCC patients before and after hepatectomy. After quality control filtering, 98 HCC patients were available for analysis. Seventeen microRNAs (miR-122-5p, miR-125a-5p, miR-125b-5p, miR-145-5p, miR-148a-3p, miR-191-5p, miR-192-5p, miR-214-3p, miR-22-5p, miR-223-3p, miR-23a-3p, miR-30c-5p, miR-320d, miR-365a-3p, miR-423-5p, miR-424-5p, miR-574-3p) were significantly up-regulated after surgery (p-value < 0.05, Figure 1, Supplementary Figure 1, Supplementary Table 2-3). As expected, the negative control microRNA miR-451 showed no difference comparing pre- and post-surgical operations (Supplementary Figure 1).
HCCseek-23 miRNA random forest model can identify early HCC
Before we develop an HCC prognostic model, we attempted to test if the HCCseek-23 microRNA panel could identify early-stage HCC patients. To do so, we applied the random forest model we reported previously [13] to this HCCseek-23 miRNA panel (detailed in the methodology section). As expected, the HCCseek-23 random forest model provided good performance for identifying stage I and II HCC patients (n=196, 81% sensitivity, 83% specificity, and 0.79 AUC) and identifying early-stage AFP-negative HCC patients (n= 45, 93% sensitivity and 83% specificity, Supplementary Table 4). Collectively, these results suggested miRNA's clinical utility for early HCC diagnosis.
HCCseek-23 miRNA random forest model is not associated with patient survival
Next, we test if the HCCseek-23 random forest model is associated with patient survival. To do so, we applied the Log-rank test and univariate Cox proportional hazards analyses. In the Log-rank test, no significant association was found between the predicted HCC probability and the OS and DFS of the early-stage HCC patients (n=98) (p-values are 0.09 and 0.25; Supplementary Table 5). In univariate Cox proportional hazards analysis, we separated the patients into two categories (long-term and short-term survival groups) based on the median OS and DFS for evaluation. Again, no significant association was found between the predicted HCC probability and the survivor classification (Supplementary Figure 2-3). To sum up, although the HCCseek-23 random forest model can identify early-stage HCC patients, it is unsuitable for HCC prognosis. It is necessary to narrow down the microRNA panel specifically for prognosis purposes.
Identification of signature miRNAs for OS and DFS prognosis
To tailor-make a microRNA panel for early-stage HCC prognosis, we first narrowed down our microRNA list by the Log-rank and Cox proportional hazards analysis. To do so, we calculated the differential microRNA expression pattern comparing post-operation versus pre-operation time points (B2-B1). These differential expressions, together with the expression data at both pre-operation (B1) and post-operation (B2) time points were subjected to the Log-rank and Univariate Cox proportional hazards tests. Eight microRNAs (HCCseek-8 panel i.e. miR-145, miR-148a, miR-150, miR-221, miR-223, miR-23a, miR-374a, miR-424) showed significant association with DFS in either Log-rank test or univariate Cox proportional hazards analysis (Table 2). When analyzing OS, four microRNAs (HCCseek-4 panel i.e. miR-125a, miR-223-3p, miR125b, miR-150) showed significant association (Table 2). The KM-curves, p-values, and cumulative hazard ratio plots were shown in Supplementary Figures 4-17. In summary, we narrowed down the microRNA panel to 8 microRNAs and 4 microRNAs for further prognostic model development.
MiRNAs expressions before and after hepatectomy. When comparing the miRNA expressions before surgery (blue bars) and after surgery (orange bars), eighteen miRNAs showed statistically significant results. For simplicity, five miRNAs are shown (i.e. miR-145-5p, miR-148a-3p, miR-223-3p, miR-23a-3p, miR-424-5p). Expressions of all the miRNAs can be found in supplementary Figure 1. * indicates p-values<0.05 in Student's t-test.
Eight miRNA panel selected for DFS prognosis and Four miRNA panel selected for OS prognosis
| Eight miRNA panel | |||
|---|---|---|---|
| MiRNA | Univariate Cox analysis (p-value) | Hazard Ratio (HR) | Log-rank (p-value) |
| miR-145-5p (B2-B1) | 0.71 | 0.97 | 0.02* |
| miR-148a-3p (B1) | 0.54 | 0.00 | 0.04* |
| miR-150-5p (B2) | 0.64 | 0.01 | 0.01* |
| miR-150-5p (B2-B1) | 0.47 | 1.00 | 0.01* |
| miR-221-3p (B1) | 0.84 | 0.00 | 0.04* |
| miR-223-3p (B1) | 0.41 | 0.00 | 0.00* |
| miR-23a-3p (B2-B1) | 0.05* | 1.02 | 0.46 |
| miR-374a-5p (B1) | 0.04* | 0.00 | 0.53 |
| miR-374a-5p (B2-B1) | 0.02* | 1.05 | 0.32 |
| miR-424-5p (B2-B1) | 0.96 | 1.00 | 0.01* |
| Four miRNA panel | |||
| miR-125a-5p (B1) | 0.94 | 0.03 | 0.05* |
| miR-125b-5p (B1) | 0.61 | 1.00 | 0.04* |
| miR-150-5p (B1) | 0.33 | 1.00 | 0.00* |
| miR-223-3p (B1) | 0.25 | 0.00 | 0.01* |
Note: * p-value ≤ 0.05; B2-B1: differential microRNA expression pattern comparing post-operation versus pre-operation time points; B1: pre- operation; B2: post-operation
Signature miRNA panel predicts DFS but not OS
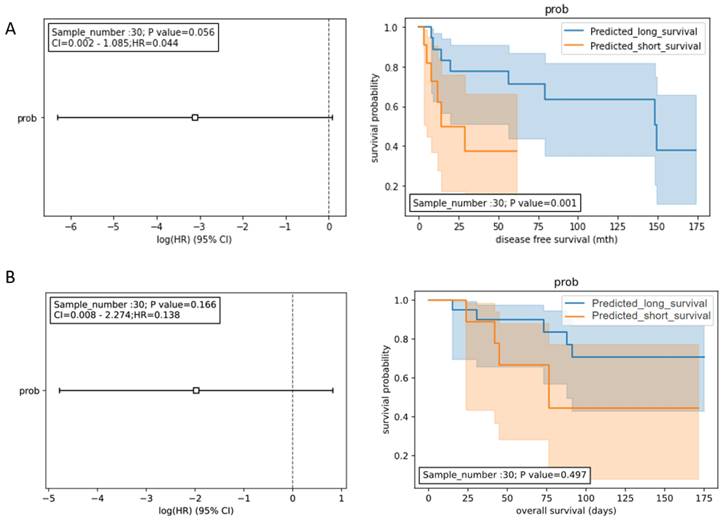
To develop a prognostic model for DFS, the expression data of the 8 miRNA panel (HCCseek-8) shown in Table 2 were input into the Random Forest model. During the model development, the early-stage HCC patients (n=98) were split into the training dataset (n= 68) and the testing dataset (n= 30) using the 70:30 ratio. To evaluate the prognostic performance of the HCCseek-8 miRNA model, we analyzed the predicted survival probability with Log-rank and Univariate Cox proportional hazards tests. A significant association was observed between the predicted survival probability and the patient survival (p-value = 0.001) in Log-rank test, meanwhile, marginal significance was observed in Univariate Cox proportional hazards analysis (p-value = 0.056, Figure 2A). To develop a prognostic model for OS, the expression data of the 4 miRNA panel shown in Table 2 were input into the Random Forest model. However, no association between the predicted survival score and patient survival was observed (Log-rank p-value = 0.497; Cox analysis p-value = 0.138) (Figure 2B). To sum up, the signature miRNAs HCCseek-8 panel is correlated with DFS, suggesting a potential application for predicting DFS. Further model improvement is needed for HCC prognosis.
Survival analyses for the signature miRNA prognostic model. (A) Prognostic models developed from 8 miRNA panel (HCCseek-8). The log-rank test showed significant association between the predicted survival probability score and the DFS (p-value = 0.001), while the univariate COX proportional hazards model showed marginal association (p-value=0.056). (B) Prognostic models developed from 4 microRNA panel (HCCseek-4). The log-rank test and the univariate COX proportional hazards model showed no significant association between the predicted survival probability score and the OS. The hazard ratios are shown in the forest plots (left panel). The orange lines and blue lines in the Kaplan-Meier curve (right panel) shows the survival probabilities of the long-term survival group and the short-term survival group predicted by the prognostic models.
Integrating HCCseek-8 and serum biomarkers for developing HCC prognosis model
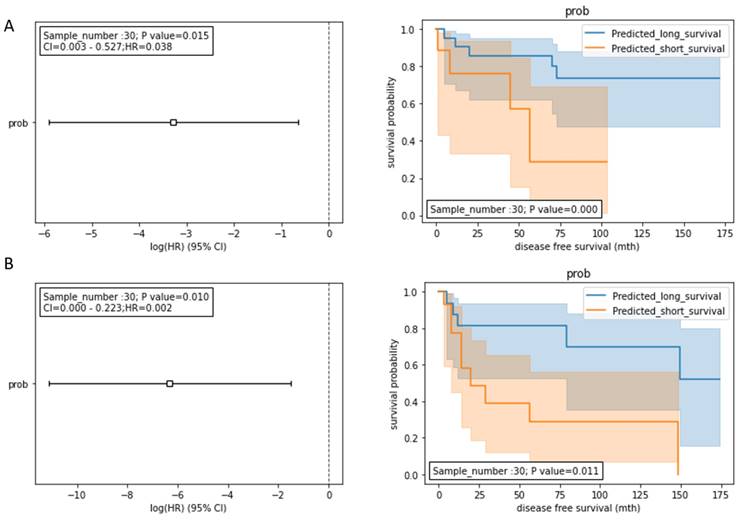
To further improve the prognostic model for predicting DFS, we integrated the model with the traditional serum biomarkers (AST, ALT, and AFP). AST and ALT are liver function-damage indicators, while serum AFP is a serological biomarker for HCC diagnosis [14]. These routinely used biomarkers could be meaningful for introducing an additional data dimension to improve our prognostic model. Therefore, we inputted the miRNA, AST, ALT, and AFP data into the random forest model development. During the model development, the stage I and II HCC patients treated with hepatectomy (n=98) were split into the training dataset (n= 68) and the testing dataset (n= 30). We tested two integration models: 1) HCCseek-8 + AFP + AST; and 2) HCCseek-8 + AFP + AST +ALT. The “HCCseek-8 + AFP + AST +ALT” model showed a more significant association with DFS (Log-rank p-value= 0.011, Cox p-value= 0.010, HR= 0.002, 95% CI: 0.000-0.233), compared to “HCCseek-8 + AFP + AST” model (Log-rank p-value= 0.0003, Cox p-value= 0.015, HR= 0.038, 95% CI: 0.003-0.527) (Table 3, Figure 3A , Figure 3B). Notably, when we remove HCCseek-8 from the model development, the negative control model “AFP+AST” and “AFP+AST+ALT” did not show any significant association with DFS (Table 3). This illustrated the essential role of the microRNA panel in predicting DFS in patients undergoing hepatectomy. Taken together, we provided solid evidence demonstrating the prognostic power of integrating novel HCCseek-8 microRNA panel and traditional serum biomarkers for prognosis in early HCC patients treated with hepatectomy.
Log-rank and Cox analyses for the integrated prognostic models and negative control
| Model | Biomarkers | Cox (p-value) | Log rank (p-value) | HR (95% CI) |
|---|---|---|---|---|
| Integrated Model | HCCseek-8 + AFP + AST | 0.015* | 0.0003* | 0.038 (0.003-0.527) |
| HCCseek-8 + AFP + AST + ALT | 0.010* | 0.011* | 0.002 (0.000-0.233) | |
| Negative control Model | AFP + AST | 0.657 | 0.664 | 0.405 (0.07-2.128) |
| AFP + AST + ALT | 0.716 | 0.475 | 0.292 (0.036-2.354) |
Note: * p-value<0.05
Survival analyses for the integrated models. (A) Prognostic models integrating HCCseek8- panel, AFP, and AST. The log-rank test and the univariate COX proportional hazards model showed significant association between the predicted survival probability score and the DFS (log-rank p-value = 0.0003, Cox p-value = 0.015, HR (95%CI) = 0.038 (0.003-0.527)). (B) Prognostic models integrating HCCseek-8 panel, AFP, AST, and ALT. The log-rank test and the univariate COX proportional hazards model showed significant association between the predicted survival probability score and the DFS (log-rank p-value = 0.011, Cox p-value = 0.010, HR (95%CI) = 0.002 (0.000-0.233)). The orange lines and blue lines in the Kaplan-Meier curve indicate predicted long-term survival group and predicted short-term survival group, respectively.
Discussion
In summary, this study highlighted four key findings: 1) We developed an HCC diagnostic model using the expression profile of twenty-three miRNAs (HCCseek-23 panel), which demonstrated 81% sensitivity and 83% specificity for identifying early-stage HCC patients; 2) HCCseek-23 panel also exhibited 93% sensitivity for identifying AFP-negative HCC; 3) We further identified eight microRNAs (HCCseek-8 panel) that was significantly associated with DFS in the Log-rank test (p-value = 0.001), suggesting a potential application for HCC prognosis; 4) When integrating HCCseek-8 panel and serum biomarkers, a significant association between the prediction result and DFS was found in both Log-rank test (p-value = 0.011) and Cox proportional hazards analysis, illustrating an outstanding performance for predicting DFS in early-stage HCC patients undergoing surgical operations.
Functional roles of the HCCseek-8 microRNAs in the progression of HCC have been previously documented [15-24]. The miR-145-5p [15], miR-148a-3p [16], miR-150-5p [17], miR-223-3p [19], and miR-424-5p [22] were found to be down-regulated in HCC. The down-regulated expressions appear to potentiate cancer cell migration and invasion in HCC patients. These microRNAs serve as HCC suppressors by targeting transcription factors and oncogenes associated with cancer cell growth, migration, and metastasis. For instance, miR-145-5p targets the ARF6 pathway to inhibit invasion and metastasis. Downregulation of these miRNAs is known to promote HCC invasion and metastasis [15-17,19,22]. Besides regulating invasion and metastasis, the loss of miR-145-5p, miR-150-5p, and miR-223-3p expressions has been identified to promote HCC proliferation [15,17,19]. The miR-148a-3p is critical in controlling hepatic differentiation by regulating c-Met oncogene [16]. In addition, the decreasing level of miR-148a-3p in the blood of HCC patients had an inverse relationship with the profibrogenic cytokine TGF-β, associated with the progression from cirrhosis to HCC, and linked to poorer survival outcomes [24]. MiR-221-3p [18], miR-23a-3p [20], and miR-374a-5p [21] are involved in tumor cell proliferation, genomic stability, and growth suppressor evasion in HCC.
Although some microRNAs in the HCCseek-8 panel have been linked to HCC detection and survival [5,7,18,23,24], analyzing multi-dimensional data could improve the prediction model [10-12]. In this study, we integrated multi-dimensional data such as the microRNA expressions, dynamic change of miRNA levels before and after surgery, and the traditional serum biomarkers for the prognosis of early HCC patients undergoing hepatectomy (Supplementary Figure 18). In addition, we demonstrated the diagnostic performance of the microRNA panel for identifying early-stage HCC patients. Although a large-scale and multi-center investigation is still needed to translate these models into clinical application, this study demonstrated promising results for further model development in the future.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was funded by Innovation and Technology Commission of Hong Kong Government to Tan To Cheung and Victor Chun Lam Wong (SCT/009/20SP).
Author contributions
Conceptualization & Funding acquisition: Victor Chun Lam Wong, Tan To Cheung; Data curation & analysis: Victor Chun Lam Wong, Ming In Wong, Tan To Cheung; Project administration: Victor Chun Lam Wong, Tan To Cheung; Resources: Victor Chun Lam Wong, Ming In Wong, Victor Ho Fun Lee, Kwan Man, Kevin Tak-Pan Ng, Tan To Cheung; Software: Victor Chun Lam Wong, Ming In Wong; Supervision: Victor Chun Lam Wong, Tan To Cheung.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Asafo-Agyei KO, Samant H. Hepatocellular Carcinoma. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. 2022 [cited 22 December 2022]. Available at: http://www.ncbi.nlm.nih.gov/books/NBK559177/
2. Wong MCS, Jiang JY, Goggins WB. et al. International incidence and mortality trends of liver cancer: a global profile. Sci Rep. 2017;7:45846
3. Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002;122:1609-19
4. Ikai I, Arii S, Kojiro M. et al. Reevaluation of prognostic factors for survival after liver resection in patients with hepatocellular carcinoma in a Japanese nationwide survey. Cancer. 2004;101:796-802
5. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne). 2018;9:402
6. Morishita A, Oura K, Tadokoro T, Fujita K, Tani J, Masaki T. MicroRNAs in the Pathogenesis of Hepatocellular Carcinoma: A Review. Cancers (Basel). 2021;13:514
7. Chen L, Chu F, Cao Y, Shao J, Wang F. Serum miR-182 and miR-331-3p as diagnostic and prognostic markers in patients with hepatocellular carcinoma. Tumor Biol. 2015;36:7439-47
8. Zhang Y, Li T, Qiu Y. et al. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore). 2017;96:e5642
9. Long X-R, Zhang Y-J, Zhang M-Y, Chen K, Zheng XFS, Wang H-Y. Identification of an 88-microRNA signature in whole blood for diagnosis of hepatocellular carcinoma and other chronic liver diseases. Aging (Albany NY). 2017;9:1565-76
10. Williams FM. Biomarkers: in combination they may do better. Arthritis Research & Therapy. 2009;11:130
11. Ahn JC, Qureshi TA, Singal AG, Li D, Yang J-D. Deep learning in hepatocellular carcinoma: Current status and future perspectives. World J Hepatol. 2021;13:2039-51
12. Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-59
13. Wong VC-L, Wong M-I, Lam C-T, Lung ML, Lam K-O, Lee VH-F. Hallmark microRNA signature in liquid biopsy identifies hepatocellular carcinoma and differentiates it from liver metastasis. J Cancer. 2021;12:4585-94
14. Tzartzeva K, Obi J, Rich NE. et al. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology. 2018;154:1706-1718.e1
15. Wang S, Wang T, Gu P. microRNA-145-5p Inhibits Migration, Invasion, and Metastasis in Hepatocellular Carcinoma by Inhibiting ARF6. Cancer Manag Res. 2021;13:3473-84
16. Gailhouste L, Gomez-Santos L, Hagiwara K. et al. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58:1153-65
17. Li T, Xie J, Shen C. et al. miR-150-5p Inhibits Hepatoma Cell Migration and Invasion by Targeting MMP14. PLoS One. 2014;9:e115577
18. Chen Z, Xiang B, Qi L, Zhu S, Li L. miR-221-3p promotes hepatocellular carcinogenesis by downregulating O6-methylguanine-DNA methyltransferase. Cancer Biol Ther. 2020;21:915-26
19. Xu J, Wang B, Liu ZT, Lai MC, Zhang ML, Zheng SS. miR-223-3p regulating the occurrence and development of liver cancer cells by targeting FAT1 gene. Math Biosci Eng. 2019;17:1534-47
20. Xiang Y, Yang Y, Lin C, Wu J, Zhang X. MiR-23a-3p promoted G1/S cell cycle transition by targeting protocadherin17 in hepatocellular carcinoma. J Physiol Biochem. 2020;76:123-34
21. Li H, Chen H, Wang H. et al. MicroRNA-374a Promotes Hepatocellular Carcinoma Cell Proliferation by Targeting Mitogen-Inducible Gene 6 (MIG-6). Oncol Res. 2018;26:557-63
22. Zhao Y, Zhu C, Chang Q. et al. MiR-424-5p regulates cell cycle and inhibits proliferation of hepatocellular carcinoma cells by targeting E2F7. PLOS ONE. 2020;15:e0242179
23. Zhang R, Zhang L-J, Yang M-L, Huang L-S, Chen G, Feng Z-B. Potential role of microRNA-223-3p in the tumorigenesis of hepatocellular carcinoma: A comprehensive study based on data mining and bioinformatics. Mol Med Rep. 2018;17:2211-28
24. Dawood AA, Saleh AA, Elbahr O, Gohar SF, Habieb MS. Inverse relationship between the level of miRNA 148a-3p and both TGF-β1 and FIB-4 in hepatocellular carcinoma. Biochem Biophys Rep. 2021;27:101082
25. Sun W, Zhang Z, Wang J. et al. MicroRNA-150 suppresses cell proliferation and metastasis in hepatocellular carcinoma by inhibiting the GAB1-ERK axis. Oncotarget. 2016;7:11595-608
26. Zha Z, Jia F, Hu P, Mai E, Lei T. MicroRNA-574-3p inhibits the malignant behavior of liver cancer cells by targeting ADAM28. Oncol Lett. 2020;20:3015-23
27. Hua S, Quan Y, Zhan M, Liao H, Li Y, Lu L. miR-125b-5p inhibits cell proliferation, migration, and invasion in hepatocellular carcinoma via targeting TXNRD1. Cancer Cell Int. 2019;19:203
28. Wang Y, Qin X, Guo T, Liu P, Wu P, Liu Z. Up-regulation of CDK16 by multiple mechanisms in hepatocellular carcinoma promotes tumor progression. J Exp Clin Cancer Res. 2017;36:97
29. Gao Y, Luo T, Ouyang X, Zhu C, Zhu J, Qin X. IGF2BP3 and miR191-5p synergistically increase HCC cell invasiveness by altering ZO-1 expression. Oncol Lett. 2020;20:1423-31
30. Liu J, Fan L, Yu H. et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology. 2019;70:241-58
31. Liang H, Sun H, Yang J, Yi C. miR-145-5p reduces proliferation and migration of hepatocellular carcinoma by targeting KLF5. Molecular Medicine Reports. 2018;17:8332-8
32. Dong G, Zhang S, Shen S. et al. SPATS2, negatively regulated by miR-145-5p, promotes hepatocellular carcinoma progression through regulating cell cycle. Cell Death Dis. 2020;11:1-16
33. Lupini L, Pepe F, Ferracin M. et al. Over-expression of the miR-483-3p overcomes the miR-145/TP53 pro-apoptotic loop in hepatocellular carcinoma. Oncotarget. 2016;7:31361-71
34. Tang H, Li R-P, Liang P, Zhou Y-L, Wang G-W. miR-125a inhibits the migration and invasion of liver cancer cells via suppression of the PI3K/AKT/mTOR signaling pathway. Oncol Lett. 2015;10:681-6
35. Ming M, Ying M, Ling M. miRNA-125a-5p inhibits hepatocellular carcinoma cell proliferation and induces apoptosis by targeting TP53 regulated inhibitor of apoptosis 1 and Bcl-2-like-2 protein. Exp Ther Med. 2019;18:1196-202
36. Liu C, Shang Z, Ma Y, Ma J, Song J. HOTAIR/miR-214-3p/FLOT1 axis plays an essential role in the proliferation, migration, and invasion of hepatocellular carcinoma. Int J Clin Exp Pathol. 2019;12:50-63
37. Li Y, Li Y, Chen Y. et al. MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell International. 2017;17:102
38. Gu Y, Wei X, Sun Y. et al. miR-192-5p Silencing by Genetic Aberrations Is a Key Event in Hepatocellular Carcinomas with Cancer Stem Cell Features. Cancer Res. 2019;79:941-53
39. Nielsen KO, Jacobsen KS, Mirza AH. et al. Hepatitis B virus upregulates host microRNAs that target apoptosis-regulatory genes in an in vitro cell model. Experimental Cell Research. 2018;371:92-103
40. Li W, Ding X, Wang S. et al. Downregulation of serum exosomal miR-320d predicts poor prognosis in hepatocellular carcinoma. J Clin Lab Anal. 2020;34:e23239
41. Li M-L, Zhang Y, Ma L-T. LncRNA HCG11 accelerates the progression of hepatocellular carcinoma via miR-26a-5p/ATG12 axis. Eur Rev Med Pharmacol Sci. 2019;23:10708-20
42. Li X, Wang L, Cao X. et al. Casticin inhibits stemness of hepatocellular carcinoma cells via disrupting the reciprocal negative regulation between DNMT1 and miR-148a-3p. Toxicol Appl Pharmacol. 2020;396:114998
43. Deng Y, Wang J, Huang M, Xu G, Wei W, Qin H. Inhibition of miR-148a-3p resists hepatocellular carcinoma progress of hepatitis C virus infection through suppressing c-Jun and MAPK pathway. J Cell Mol Med. 2019;23:1415-26
44. Song SK, Jung WY, Park S-K, Chung C-W, Park Y. Significantly different expression levels of microRNAs associated with vascular invasion in hepatocellular carcinoma and their prognostic significance after surgical resection. PLoS One. 2019;14:e0216847
45. Lin J, Huang S, Wu S. et al. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011;32:1641-7
46. de Oliveira ARCP, Castanhole-Nunes MMU, Biselli-Chicote PM. et al. Differential expression of angiogenesis-related miRNAs and VEGFA in cirrhosis and hepatocellular carcinoma. Arch Med Sci. 2020;16:1150-7
47. He J, Xiao B, Li X, He Y, Li L, Sun Z. MiR-486-5p Suppresses Proliferation and Migration of Hepatocellular Carcinoma Cells through Downregulation of the E3 Ubiquitin Ligase CBL. Biomed Res Int. 2019;2019:2732057
48. Luo L-J, Zhang L-P, Duan C-Y. et al. The inhibition role of miR-22 in hepatocellular carcinoma cell migration and invasion via targeting CD147. Cancer Cell Int. 2017;17:17
49. Wan L, Yuan X, Liu M, Xue B. miRNA-223-3p regulates NLRP3 to promote apoptosis and inhibit proliferation of hep3B cells. Exp Ther Med. 2018;15:2429-35
50. Yu Z, Zhao H, Feng X. et al. Long Non-coding RNA FENDRR Acts as a miR-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids. 2019;17:516-29
51. Stiuso P, Potenza N, Lombardi A. et al. MicroRNA-423-5p Promotes Autophagy in Cancer Cells and Is Increased in Serum From Hepatocarcinoma Patients Treated With Sorafenib. Mol Ther Nucleic Acids. 2015;4:e233
52. Lin Q, Zhou C-R, Bai M-J. et al. Exosome-mediated miRNA delivery promotes liver cancer EMT and metastasis. Am J Transl Res. 2020;12:1080-95
53. Yang J, Cui R, Liu Y. MicroRNA-212-3p inhibits paclitaxel resistance through regulating epithelial-mesenchymal transition, migration and invasion by targeting ZEB2 in human hepatocellular carcinoma. Oncol Lett. 2020;20:23
54. Budhu A, Jia H-L, Forgues M. et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907
55. Du H, Xu Q, Xiao S. et al. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. 2019;224:1-11
56. Li D, Tang X, Li M, Zheng Y. Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. J Cell Biochem. 2019;120:12290-9
57. Yang Z, Zi Q, Xu K, Wang C, Chi Q. Development of a macrophages-related 4-gene signature and nomogram for the overall survival prediction of hepatocellular carcinoma based on WGCNA and LASSO algorithm. Int Immunopharmacol. 2021;90:107238
58. Teng F, Zhang J-X, Chang Q-M. et al. LncRNA MYLK-AS1 facilitates tumor progression and angiogenesis by targeting miR-424-5p/E2F7 axis and activating VEGFR-2 signaling pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39:235
59. Gao J, Yin X, Yu X, Dai C, Zhou F. Long noncoding LINC01551 promotes hepatocellular carcinoma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of microRNA-122-5p to regulate ADAM10 expression. J Cell Biochem. 2019;120:16393-407
60. Wei G-Y, Hu M, Zhao L, Guo W-S. MiR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:5158-67
Author Biographies
Dr. Victor Chun-Lam Wong is the Chief Scientific Officer at OncoSeek Limited. He holds a Ph.D. in Clinical Oncology from the University of Hong Kong and has completed postgraduate training at Brown University and Memorial Sloan Kettering Cancer Center in the United States. In addition, he has obtained a postgraduate diploma in Machine Learning Artificial Intelligence through the Emeritus Institute in collaboration with Columbia University. His research focuses on the integration of circulating tumor cells, DNA, microRNA, and machine learning to develop innovative liquid biopsy technologies for cancer surveillance and monitoring. Dr. Wong's work has been recognized with numerous awards, including the Hong Kong Science Park Incubator Program Award in 2014, the Technology Start-up Support Scheme Award in 2015 and 2016, the Best Pitching Award in 2017, and the Public Sector Trial Scheme Award in 2021. His publications have received over 300 citations in cancer research. He also serves as an ad-hoc reviewer for international journals, including Frontiers of Oncology, Aging, Oncotarget, Cancer Letters, Cancer Biology & Therapy, Journal of Experimental & Clinical Cancer Research, Plos One, Oncology Research and Review, Trends in Cancer Updates, Cell Death Discovery, Protein & Cell, International Journal of Biological Sciences, and AAAS Science Advances.
Ming-In Wong received her Bachelor's degree of Science in Biochemistry and Cell Biology at The Hong Kong University of Science and Technology. Currently working as an R&D assistant scientist in OncoSeek Ltd, her R&D interests include machine learning, biomedical science, and genetics.
Prof. Victor Ho-Fun Lee is currently Clinical Associate Professor of the Department of Clinical Oncology, Li Ka Shing Faculty of Medicine, The University of Hong Kong. He graduated in the University of Hong Kong in 2002. He joined the Department of Clinical Oncology, Li Ka Shing Faculty of Medicine, The University of Hong Kong as Clinical Assistant Professor in 2008. He obtained his fellowship in Royal College of Radiologists in Clinical Oncology in 2010. Afterwards, he received further specialist training in interstitial brachytherapy for head and neck cancers and sarcoma in Institut Gustave Roussy in Paris, France and novel radiation techniques like stereotactic radiosurgery and stereotactic ablative radiotherapy in Stanford University USA. In 2013, he received further training on stereotactic body radiation therapy for liver tumors at Princess Margaret Hospital, Toronto, Canada. More recently in 2015 he was awarded HKCR 15A Traveling Fellowship and pursued subspecialty training in image-guided brachytherapy for cervical cancer and pediatric oncology in Addenbrooke's Hospital in Cambridge. He has obtained a Doctor of Medicine (MD) in 2015 for his research on nasopharyngeal carcinoma. He was promoted to Clinical Associate Professor in 2017 and appointed as Assistant Dean (Assessment) of Li Ka Shing Faculty of Medicine, The University of Hong Kong since August 2018. He was awarded the Outstanding Young Researcher Award of The University of Hong Kong in December 2018. His current interests include clinical and genetic studies on nasopharyngeal cancer, head and neck cancers, lung cancers, liver cancers and gastrointestinal cancers and has published extensively in these respects. In addition, he has special interest in preclinical and clinical dosimetric studies on intensity-modulated radiation therapy, stereotactic body radiation therapy (SBRT), stereotactic radiosurgery (SRS) and selective internal radiation therapy (SIRT) with Yttrium-90 microspheres for liver tumors.
Prof. Nancy Kwan Man is currently the Director of Liver Transplantation and Liver Cancer Research in Department of Surgery, The University of Hong Kong. She is well-recognized for the advancements of liver transplantation research. Her innovative development and successful application of integrated clinical, translational and basic research for liver graft injury and cancer recurrence after transplantation, has resulted in major advances and impact on transplant oncology and immunology. She has published more than 200 original articles in international journals including Transplantation, Liver Transplantation, Annals of Surgery, Clinical Cancer Research, Cancer Research, Journal of Hepatology, Hepatology, Gastroenterology, Nature Medicine & New England Journal of Medicine etc with H index 68 (total citation 14187). She is one of the Top 1% Scholars at University of Hong Kong according to ISI's Essential Science Indicators since 2013. Nancy has made great efforts in the serviced to international, national and local academic societies, particular for International Liver Transplantation Society (ILTS). She was the President of ILTS (2018-19) and Program Chair of ILTS2018. She was the member of Basic Science Committee/Transplant Science Committee of ILTS and The Transplantation Society (TTS), Chair of Scholarship Committee, member of Education Committee, Publication Committee of ILTS. She is also the Founder member of Women Leaders in Transplantation (WLIL), International Mentor of WLIL and a Key Opinion Leader of TTS. Being the President of Hong Kong Scientist Association (HKSA). Nancy has made a valued contribution to the journal of “Transplantation” as the Regional Associate Editor and Deputy Editor for her extensive experience for the translational and basic research in liver transplantation. She is also the editorial board member of “Annals of Surgery”, especially for the liver related research. She and her research team have obtained more than 60 international awards including, numbers of “Rising Star Awards”, “Young Investigator Awards” in annual congress of ILTS, and “Mentee-Mentor Basic Science Awards”, “Young Investigator Awards” In TTS/TSS meetings over the past 15 years. She has also got the “First Class Award of 2013 Higher Education Outstanding Scientific Research Output Award from Ministry of Education (MOE), China” and “First Class Award for Science and Technology of Chinese Medical Association (CAE) 2014” for her achievement is liver transplantation research, particular for the mechanisms and therapeutics of cancer recurrence after liver transplantation for liver cancer patients. She was recently got the first prestigious award for liver transplant research entitles with “2022 Basic Science Established Investigator Award of International Liver Transplantation Society” for her outstanding accomplishment in liver transplantation research.
Dr. Kevin Tak-Pan Ng obtained his PhD degree in the department of Surgery, the University of Hong Kong, in 2008. He is currently appointed as a Scientific Officer in the same department. Dr. Ng is expertized in molecular and cell biology in the area of liver cancer, liver injury and liver transplantation. He has published 70 international peer-review articles and more than 100 abstracts in the international conferences. Dr. Ng is currently focusing on clinical, basic and translational researches on liver cancer and liver transplantation to tackle hepatocellular carcinoma (HCC) recurrence after liver transplantation and liver resection.
Prof. Tan To Cheung is the Chief of the Division of Hepatobiliary and Pancreatic Surgery within the Department of Surgery, Queen Mary Hospital, the University of Hong Kong, and President of the Hong Kong Society of Hepatobiliary and Pancreatic Surgery, Vice president (External Affairs) of College of Surgeons on Hong Kong and Council member of Liver Foundation. He is currently Clinical Professor of the University of Hong Kong.He is listed as top 1% HKU researchers ranked by Clarivate Analytics in the worldwide by citations in 2021. Prof. Cheung is an experienced academic surgeons and his research interest was focused in disease and cancers in the liver, biliary tract and pancreas. He is the chairman of Hong Kong pancreatic cystic lesion management consensus meeting 2022, Hong Kong consensus recommendation on the management of HCC 2018 and 2021, and Asia pacific consensus meeting for minimally invasive hepatectomy for primary liver cancer (Hong Kong Statement) in 2016. Prof. Cheung is a world-renowned leading experts in the field of laparoscopic liver resection for primary liver cancer. He is the pioneer of laparoscopic liver hepatectomies in Hong Kong with good outcome. He completed the degree of Doctor of Medicine in 2018. His thesis was the “Application on Minimally invasive Hepatectomy for liver cancer.” Prof. Cheung is very active and visionary in this field internationally. He was the founding member of International Laparosocpic liver Surgery Society and was elected as Advisory board member in 2019. Chairman of radiology committee of International liver transplant society 2019-2022. Prof. Cheung is an active researcher with a successful grant record including principle investigator and co-investigators of various grants from UGC HK and China with at total amount of more than 80 Million. Prof. Cheung was a recipient of the GB Ong Travelling Fellowship Award 2013 and James IV travelling Fellowship Award 2017. He has authored more than 310 peer-reviewed articles and other publications, and is frequently invited to lecture internationally and to present in plenary sessions. He has mentored a large number of trainees, including post graduate degree and oversea academia.
![]() Corresponding author: Department of Surgery, The University of Hong Kong, 21 Sassoon Road, Pokfulam, Hong Kong Special Administrative Region, People's Republic of China; OncoSeek Limited, Hong Kong Science and Technology Parks, Hong Kong Special Administrative Region, People's Republic of China. E-mail addresses: cheung68hk (TC), victorcom (VW); Phone: (+852) 2255 3025 (TC); (+852) 3188 9335 (VW)
Corresponding author: Department of Surgery, The University of Hong Kong, 21 Sassoon Road, Pokfulam, Hong Kong Special Administrative Region, People's Republic of China; OncoSeek Limited, Hong Kong Science and Technology Parks, Hong Kong Special Administrative Region, People's Republic of China. E-mail addresses: cheung68hk (TC), victorcom (VW); Phone: (+852) 2255 3025 (TC); (+852) 3188 9335 (VW)

 Global reach, higher impact
Global reach, higher impact