Impact Factor
ISSN: 1837-9664
J Cancer 2022; 13(9):2717-2726. doi:10.7150/jca.73761 This issue Cite
Research Paper
HMGB1 overexpression promotes a malignant phenotype and radioresistance in ESCC
1. Department of Radiation Oncology, The Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, P. R. China
2. Department of Clinical Laboratory, The Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, P. R. China
3. Research Center, The Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, P. R. China
Received 2022-4-7; Accepted 2022-5-15; Published 2022-5-21
Abstract
Esophageal cancer is a common malignant disease that is generally treated with radiotherapy. High mobility group box 1 (HMGB1) plays an essential role in tumor cell proliferation, migration, and cell cycle progression. Here, we aimed to clarify the effects of HMGB1 on radioresistance in esophageal squamous cell carcinoma (ESCC) cell lines and patient survival. We performed immunohistochemistry for HMGB1 in biopsy samples of 39 stage I-III ESCC patients grouped by HMGB1 expression status. Then, 1‐, 3‐, 5-, and 10‐year overall survival outcomes were calculated by Kaplan-Meier survival analysis. The cellular localization of HMGB1 was examined before and after irradiation by Immunofluorescence staining. Stable cell lines (KYSE30 and KYSE510) with differential HMGB1 expression were constructed using lentiviruses. Furthermore, we examined phosphorylated histone H2AX (γ-H2AX) expression in both HMGB1 overexpression and negative control groups by western blotting. HMGB1-negative expression was associated with superior ESCC patient 10-year survival (P=0.016). HMGB1 overexpression promoted cell migration, proliferation, and radioresistance and mitigated cell cycle arrest at the G0/G1 phase induced by irradiation. This demonstrates that HMGB1-positive expression is correlated with unfavorable clinical outcomes, and HMGB1 overexpression may promote the malignant phenotype of ESCC cells and induce radioresistance by regulating cell cycle distribution in ESCC.
Keywords: ESCC, HMGB1, malignant phenotype, radioresistance, γ-H2AX
Introduction
Esophageal cancer (EC) is one of the gastrointestinal tumors and is the eighth most common malignancy worldwide [1, 2]. Radiotherapy is an important modality for treating EC [3]. The continuous advances in radiotherapy equipment and technology have improved the prognosis of EC patients. However, anti-tumor efficacy remains suboptimal in part because of radioresistance [4]. Radioresistance in patients has become a focal point of EC research, especially in esophageal squamous cell carcinoma (ESCC), because it is the globally predominant pathological type of EC [5]. High mobility group box 1 (HMGB1) is a non-histone chromatin-binding protein in eukaryotes and is an essential protein for life. HMGB1 is involved in the modulation of cancer cell proliferation, invasion, and metastasis [6, 7]. HMGB1 can affect the efficacy of radiotherapy and has an impact on the survival of cancer patients. Jiao et al. proposed that by targeting the HMGB1-RB axis, SEPT9 methylation promoted tumorigenesis and radioresistance of cervical cancer [8], and under chemoradiotherapy or hypoxia, HMGB1 can be translocated [9]. In addition, high levels of HMGB1 protein expression are reportedly associated with poor prognosis of colorectal cancer patients [10].
Radiotherapy not only mediates a therapeutic response by inducing DNA double-strand breaks (DSBs) but also has immunomodulatory effects. Irradiation can induce translocation of HMGB1 to the cytoplasm, and it can regulate cell proliferation and migration by binding to its high‐affinity receptor. Intracellular HMGB1 regulates DNA repair systems and autophagy in tumor cells to induce radioresistance. Extracellular HMGB1 has a complex role in the radiation-related immune response. It not only stimulates the anti-tumor immune response by promoting the recognition of dying tumor cells but also participates in maintaining immunosuppression [11, 12].
Zhang et al. proposed that X-ray irradiation induces dying cells to release HMGB1 promoting CD133- pancreatic cancer cells to maintain stemness. Both in vitro and PDX models showed that inhibiting HMGB1 expression in irradiated, dying cells could attenuate the dedifferentiation stimulation effect on C133- pancreatic cancer cells. X-ray irradiation-induced CD133- pancreatic cancer cells to differentiate into the cancer stem cell (CSC) phenotype, and preventing CSC enrichment and pancreatic carcinoma relapse by inhibiting HMGB1 may be a useful strategy [13]. Similarly, silencing HMGB1 expression could reportedly attenuate the stimulating effects on the migration of irradiated, dying cells, promotion of cancer cell migration via paracrine mechanisms, and enhanced epithelial-mesenchymal transition (EMT) and PI3K/pAkt signaling. During radiotherapy, the dying cells could activate paracrine signaling to facilitate the migration of surviving tumor cells. From these data, Chen et al. suggested that inhibiting HMGB1 could help prevent pancreatic carcinoma relapse and metastasis [14].
It is confirmed that knockdown of HMGB1 could decrease the proliferation and invasive ability of non-small cell lung cancer cells while also increasing the radiosensitivity of the cells [15]. Shrivastava et al. showed a positive correlation between high levels of HMGB1 and radioresistance in bladder cancer cell lines. After HMGB1 knockdown, the radiosensitivity of bladder cancer cells was increased. They found that HMGB1 is possibly a crucial gene in these cells because its loss was related to higher radiation-induced cellular DNA damage levels. These data highlight the potential of HMGB1 to be a radiation marker, and the capability to enhance radiation efficacy [16]. Similarly, Ayoub et al. proposed that HMGB1 might play a role in radioresistance of bladder cancer by promoting pro-tumor immune mechanisms, such as reducing the frequency of pro-tumor myeloid-derived suppressor cells and tumor-associated macrophages [17].
In EC, Matsubara et al. proposed that high levels of HMGB1 in tumor cells or plasma could play a key role in the malignant potential of ESCC [18]. Takahata suggested that a high HMGB1 expression level was a risk factor for severe postoperative pulmonary complications in EC patients [19]. HMGB1 may serve as a new therapeutic target or clinical biomarker for metastatic ESCC [20]. It was confirmed that downregulating HMGB1 expression could promote radiosensitivity in ESCC both in vitro and in vivo [21, 22]. Moreover, activating a DNA damage checkpoint response after irradiation was the chief cause of HMGB1-induced radioresistance [23].
It was previously reported that HMGB1 expression levels were significantly associated with ESCC patient survival [18]. The relationship between knocking down HMGB1 expression in ESCC cells and radiosensitivity has been studied, which reported that knockdown HMGB1 expression in ECA109 and TE13 cell lines increased their radiosensitivity [24]. However, there are few studies focused on HMGB1 overexpression. Therefore, it is necessary to research to verify the relationship between HMGB1 overexpression and radioresistance in ESCC by selecting appropriate cell lines. Better understanding the effect of HMGB1 on radiosensitivity and identifying radioresistance in ESCC patients may provide novel insights. Hence, our study aimed to explore the effect of HMGB1 overexpression on cell proliferation, cell cycle progression, and radiosensitivity of ESCC cell lines and to provide a theoretical basis for the treatment of ESCC.
Material and Methods
Immunohistochemistry (IHC)
This study was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University (IRB approval ID: 2020KY184). All patients gave verbal informed consent to participate in the study. Patient inclusion criteria were 1. age ≥18 years; 2. pathological diagnosis of first primary ESCC; 3. not receiving any treatment at the time of biopsy sample collection; 4. having received radiotherapy alone; 5. complete general and pathological data; and 6. patient informed consent. From January 2008 to November 2013, pretreatment biopsy specimens from 39 stage I-III ESCC patients were retrospectively collected. The patients were treated with radiotherapy alone at the Fourth Hospital of Hebei Medical University. A standard IHC staining procedure was followed. The IHC staining was carried out as described [24, 25]. The localization of HMGB1 cells was observed microscopically, and the IHC results of the sections were assessed by light microscopy by two chief pathologists under independent double-blind conditions. Referring to the assessment method proposed by Soumaoro et al. [26], the entire field of view of the tumor tissue, the intensity of positive cell coloration, and the percentage of positive cells were assessed when the sections were observed under the microscope. The intensity of positive cell coloration was scored as 0, 1, 2, and 3, corresponding to unstained, yellowish, brownish, and tan. The percentage of positive cells was scored as 0, l, 2, 3, and 4, corresponding to 0%, 1-25%, 26-50%, 51-75%, and 76-100% of positive cells in that order. The two scores were summed for evaluation, and the resultant tumor tissue specimens with scores of 0-2 were considered to have negative HMGB1 protein expression Scores greater than or equal to 3 were judged to have positive HMGB1 protein expression.
Cell culture
Ethical approval for this study was obtained from the Ethics Committee of The Fourth Hospital of Hebei Medical University. The KYSE180, ECA109, TE1, KYSE150, KYSE510, and KYSE30 ESCC lines were obtained from the Research Center of The Fourth Hospital of Hebei Medical University (Shijiazhuang, China). In addition, the cells were cultured in RPMI 1640 medium (Gibco, Beijing, China), which contained 100 U/mL penicillin-streptomycin solutions (Solarbio, Beijing, China) and 10% fetal bovine serum (FBS, Cellmax, Lanzhou, China). The cultures were stored in an incubator (5% CO2 and saturated humidity) at 37°C.
Immunofluorescence staining
Exponentially proliferating ESCC KYSE30 and KYSE510 cells were seeded in 24-well plates for culture for 12 h. The cells of the 6 Gy groups were irradiated by 6 MV X-rays (dosage rate of 2 Gy/min) and generated by a linear accelerator (Siemens, Buffalo Grove, IL, USA). The controls were not treated. The cells were cultured consecutively for two h and then fixed in 4% paraformaldehyde at room temperature for 20 min. After being rinsed three times with phosphate buffer saline (PBS, Gibco, Beijing, China), the cells were permeabilized with 0.2% Triton X-100 (Vetec, Wuxi, China) in PBS for 10 min and blocked with 10% goat serum (Solarbio, Beijing, China) for 30 min. Then cells underwent incubation with antibody anti-HMGB1 (dilution 1:500; cat. no. 10829-1-AP; Proteintech) at 4°C for 12 h. After three rinses with PBS, the samples underwent incubation for one h with green fluorescent Goat Anti-Rabbit IgG (dilution 1:200; cat. no. SA00013-3; Proteintech), protected from light at room temperature. Following the staining of cell nuclei with DAPI (Solarbio, Beijing, China), cells were viewed under an ECLIPSE Ti2 confocal laser scanning microscope (Nikon, Tokyo, Japan).
Cell transfection
The HMGB1 overexpression lentiviruses were obtained from Gene Chem Co., Ltd (Shanghai, China). The cells were then cultured until they reached 50% confluence in 6-well plates. Next, the lentivirus with HMGB1 overexpression and the negative control (NC) lentiviral particles were added to the cultures for 12 h (MOI=10). Another 36 h later, transfection efficiency was observed under the fluorescence microscope (Nikon, Tokyo, Japan). The cells were then selected and cultured with puromycin (1 μg/mL) for two weeks.
Real-time quantitative polymerase chain reaction (RT-qPCR)
RNA extraction and RT-qPCR were performed as Zhang et al. described (24). And the primer sequences for RT-qPCR included the following:
HMGB1, F: 5'-AATACGAAAAGGATATTGCT-3', R: 5'-GCGCTAGAACCAACTTAT-3'; GAPDH, F: 5'-CGCTGAGTACGTCGTGGAGTC-3', R: 5'-GCTGATGATCTTGAGGCTGTTGTC-3'.
Western blotting
Lysis of cells was conducted with radioimmunoprecipitation buffer (Solarbio, Beijing, China), which contained 1% phenylmethanesulfonyl fluoride (Solarbio, Beijing, China) and 1% phosphatase inhibitor (MCE, New Jersey, USA). The samples were centrifuged at 8000 rpm at 4°C for 15 min, and then the supernatant was collected. The concentration of proteins was detected by a BCA protein assay kit (Solarbio, Beijing, China), and Laemmli buffer was added to prepare protein samples. The solution was heated for 5 min at 100°C, followed by analysis with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Subsequently, proteins were transferred to polyvinylidene fluoride (PVDF) membranes. After being blocked with 5% nonfat dry milk for one h, PVDF membranes underwent incubation employing specific primary antibodies at 4°C overnight. Following three rinses with TBS containing 0.1% Tween 20 (TBST) at room temperature, incubation of membranes was performed by utilizing second antibodies for one h. After three more rinses with TBST, the membranes were imaged using the Odyssey system (LI-COR Biosciences, Lincoln, NE, USA). In this process, the antibodies used were as follows: anti-HMGB1 (dilution 1:1,000; cat. no. 10829-1-AP; Proteintech), anti-γ-H2AX (dilution 1:1,000; cat. no. ab81299; Abcam), anti-β-actin (dilution 1:5,000; cat. no. 60008-1-Ig; Proteintech), Goat pAb to Rb (dilution 1:1,0000; cat. no. ab216777; Abcam) and Goat pAb to Ms (dilution 1:1,0000; cat. no. ab216772; Abcam). Image J software was used to quantify autoradiographic bands, then normalize for β-actin levels.
Cell proliferation assay
The cell suspension was seeded into 96‐well plates (100 μL/well; five wells per sample). Once the cells adhered, they were irradiated with X-rays. At 0, 24, 48, and 72 h post-irradiation, 10 μL CCK-8 reagent (Med Chem Express, Princeton, NJ, USA) was added to each well and incubated for two h at 37°C. The absorbance was then measured at 450 nm with a Multiskan ascent (Thermo, Shanghai, China). The experiment was repeated three times.
Colony formation assay
ESCC cells were seeded into 6-well plates and then irradiated at doses of 0, 2, 4, 6, 8, or 10 Gy. After 14 days of culture, the cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Colonies with more than 50 cells were counted. To calculate survival curves (SF), the multitarget single-hit model was fitted to the data using the formula: [SF = 1 - (1 - e-D/D0)N].
Wound-healing assay
The ESCC cells were seeded in 6-well plates. Cells in the 6 Gy groups were irradiated with X-ray, and the 0 Gy groups were not treated. At 90% confluence, a linear scratch wound was generated with a 200 µL pipette tip. After three rinses in PBS, a medium consisting of 1% FBS was added. The images were captured at 0 h, 24 h, and 48 h under a microscope (Nikon, Tokyo, Japan). All data were analyzed using Image J software.
Flow cytometry analysis
Forty-eight h after irradiation, the transfected cells were harvested. Cell cycle distribution was determined via flow cytometry. Propidium iodide (MultiSciences Biotech Co., Ltd., Hangzhou, China) was used to stain cells for cell cycle detection.
Statistical analysis
Non-operated patients diagnosed with stage I-III ESCC from January 2008 to December 2013 were enrolled in this study. Statistical analyses were performed using R software (Version 4.0.3) or GraphPad Prism software 5. We performed Kaplan-Meier (KM) survival analysis to evaluate 1-, 3-, 5-, and 10-year survival among ESCC patients with HMGB1-positive and HMGB1-negative expression and obtained P values. The different levels between the two groups of variables were evaluated by Fisher's exact test. All experimental data were repeated at least three times, and the experimental data were expressed as mean ± standard error. Comparisons of continuous variables between groups were performed by ANOVA. Statistical differences at P<0.05 were considered significant.
Results
Negative HMGB1 expression correlates with a long-term survival benefit in ESCC patients
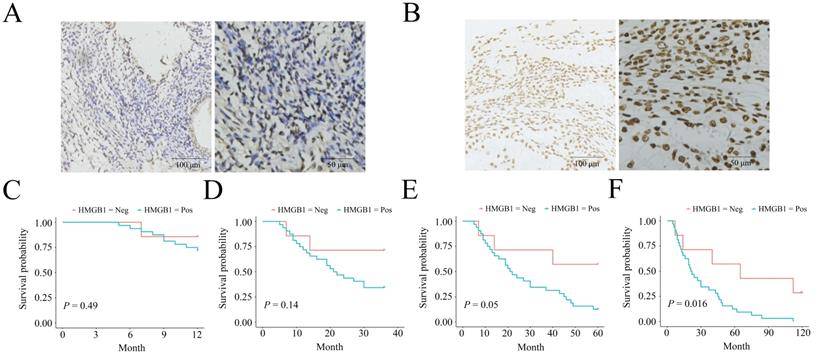
The negative and positive HMGB1 expression profiles in ESCC are listed in Figure 1 A-B. HMGB1 expression was associated with clinical outcomes in ESCC patients. Patient data, including HMGB1 expression status, sex, age, lesion location, lesion length, gross tumor volume (GTV), T stage, N stage, and TNM stage, are shown in Table 1. The two groups had no statistical difference in terms of sex (P=0.686), age (P=1), tumor location (P=0.092), lesion length (P=0.683), GTV (P=0.092), T stage (P=0.075), and N stage (P=0.686), while the two groups statistically differed in the term of TNM stage (P=0.01) between the two groups. IHC confirmed the HMGB1 expression status in each ESCC tissue sample. KM survival analysis indicated no statistical differences in 1-, 3‐, and 5-year survival (P=0.49, P=0.14, and P=0.05, respectively). However, HMGB1 positive expression was negatively associated with 10-year survival of the patients (P=0.016), which suggested that positive HMGB1 expression correlated with poor long-term survival of ESCC patients (Figure 1C-F).
The relationship between HMGB1 protein expression and clinicopathological characteristics of ESCC patients
| Variables | Overall | HMGB1 | P value | ||
|---|---|---|---|---|---|
| (n = 39) | Negative (n = 7) | Positive (n = 32) | |||
| Sex | 0.686 | ||||
| Female | 15 (38.46%) | 2 (5.13%) | 13 (33.33%) | ||
| Male | 24 (61.54%) | 5 (12.82%) | 19 (48.72%) | ||
| Age | 1 | ||||
| < 70-year | 18 (46.15%) | 3 (7.69%) | 15 (38.46%) | ||
| ≥ 70-year | 21 (53.85%) | 4 (10.26%) | 17 (43.59%) | ||
| Lesion location | 0.092 | ||||
| Cervical-Upper | 16 (41.03%) | 6 (15.38%) | 14 (35.9%) | ||
| Middle-Lower | 23 (58.97%) | 1 (2.56%) | 18 (46.15%) | ||
| Lesion length | 0.683 | ||||
| < 5 cm | 18 (46.15%) | 4 (10.26%) | 14 (35.9%) | ||
| ≥ 5 cm | 21 (53.85%) | 3 (7.69%) | 18 (46.15%) | ||
| GTV | 0.092 | ||||
| < 30 cm3 | 20 (51.28%) | 6 (15.38%) | 14 (35.9%) | ||
| ≥ 30 cm3 | 19 (48.72%) | 1 (2.56%) | 18 (46.15%) | ||
| T stagea | 0.075 | ||||
| T1-2 | 14 (35.9%) | 5 (12.82%) | 9 (23.08%) | ||
| T3-4 | 25 (64.1%) | 2 (5.13%) | 23 (58.97%) | ||
| N stagea | 0.686 | ||||
| N0 | 15 (38.46%) | 2 (5.13%) | 13 (33.33%) | ||
| N1-2 | 24 (61.54%) | 5 (12.82%) | 19 (48.72%) | ||
| TNM stagea | 0.01 | ||||
| I | 8 (20.51%) | 2 (5.13) | 6(15.38%) | ||
| II | 13 (33.33%) | 5 (12.82%) | 8 (20.51%) | ||
| III | 18 (46.15%) | 0 (0%) | 18 (46.15%) | ||
a The American Joint Committee on Cancer 6th edition staging system was used to determine tumor stage for patients. HMGB1, high mobility group box 1; ESCC, esophageal squamous cell carcinoma; GTV, gross tumor volume.
HMGB1 expression status in ESCC biopsy specimens and the expression correlated with the survival. A. Negative HMGB1 expression; B. Positive HMGB1 expression; One-y (C), three-y (D), five-y (E), and ten-y (F) survival among HMGB1-negative and HMGB1-positive groups according to Kaplan-Meier survival analysis.
Changes in HMGB1 localization after irradiation and the validation of HMGB1 overexpression
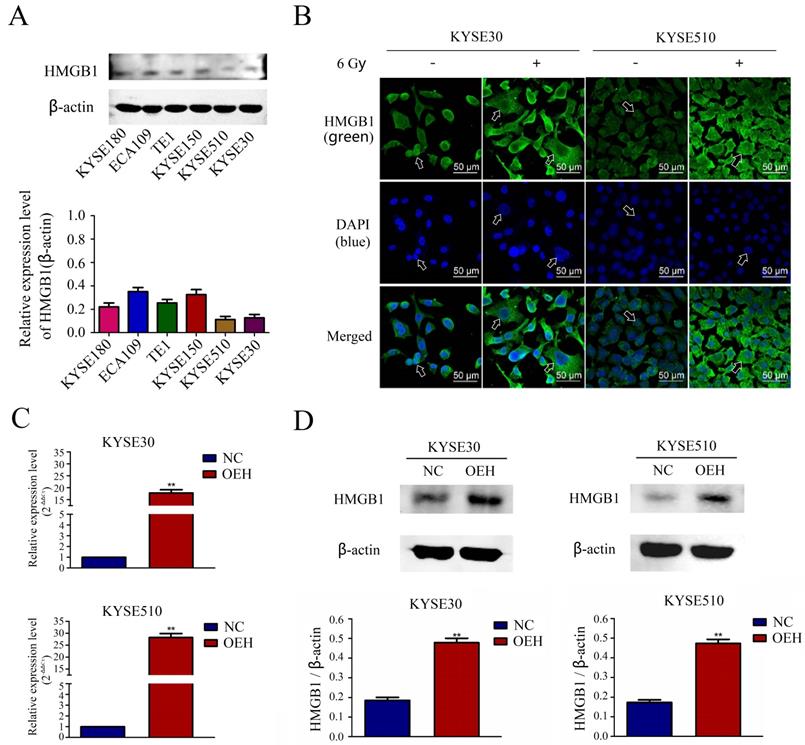
The expression of HMGB1 in the KYSE180, ECA109, TE1, KYSE150, KYSE510, and KYSE30 ESCC lines was determined in vitro by Western blotting, and the results revealed lower expression levels of HMGB1 in the KYSE30, and KYSE510 cell lines compared with the other cell lines (Figure 2A). Therefore, we selected KYSE30 and KYSE510 cells for subsequent experiments. Using the laser confocal microscopy imaging, we investigated the cell-specific localization of HMGB1 in KYSE30 and KYSE510 cells, with or without 6 Gy X-ray treatment. As shown in Figure 2B, the results suggested that compared with the non-irradiated groups, the 6 Gy irradiated cells had an increased distribution of HMGB1 protein in the cytoplasm. The ESCC cell lines KYSE30 and KYSE510 stably overexpressing HMGB1 were successfully constructed by lentiviral transfection. RT-qPCR and Western blotting validated the transfection efficiency, and the validation results for all groups are shown in Figure 2C-D.
HMGB1 overexpression significantly promoted proliferation and radioresistance of ESCC cells
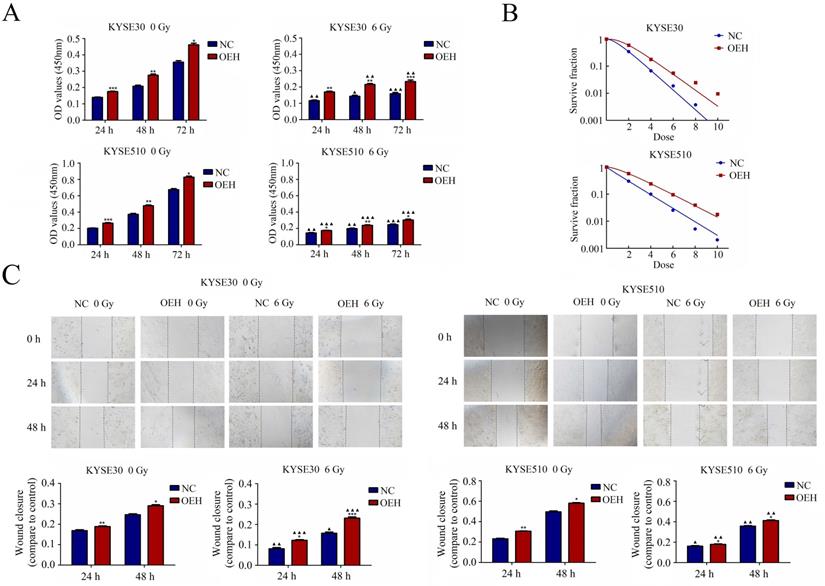
As shown in Figure 3A, the CCK‐8 assay results showed that the cells overexpressing HMGB1 had significantly higher cell proliferation rates than the NC groups with or without irradiation (P<0.05). The rest of the groups showed decreased cell proliferation after irradiation, except for the KYSE30 overexpression HMGB1 group at 24 h, which showed no statistical difference after irradiation compared to its pre-irradiation counterpart (P>0.05). Subsequently, the effect of HMGB1 overexpression on the radioresistance of these cell lines was detected by colony formation assay (Figure 3B). The sensitizer enhancement ratios (SERs) were calculated by a reported method [27]. SER = D0 (HMGB1 overexpression group)/D0 (NC group). Compared with NC groups, the survival curves of KYSE30 and KYSE510 HMGB1 overexpression groups were significantly changed with SERs 0.80 and 0.78. These results suggest that HMGB1 overexpression can promote ESCC cell proliferation and radioresistance.
HMGB1 overexpression enhanced migration of ESCC cells
Cell migration was detected with the use of a wound-healing assay. The cells of HMGB1 overexpression indicated more rapid closure of the wound, which was remarkably different from that in NC cells (P<0.5), indicating increased cell motility. A similar trend was observed after exposure to 6 Gy X-ray in ESCC cells between the NC and the HMGB1 overexpression groups (P<0.5). The results, shown in Figure 3C, suggested that HMGB1 overexpression enhanced the migration of KYSE30 and KYSE510 cells.
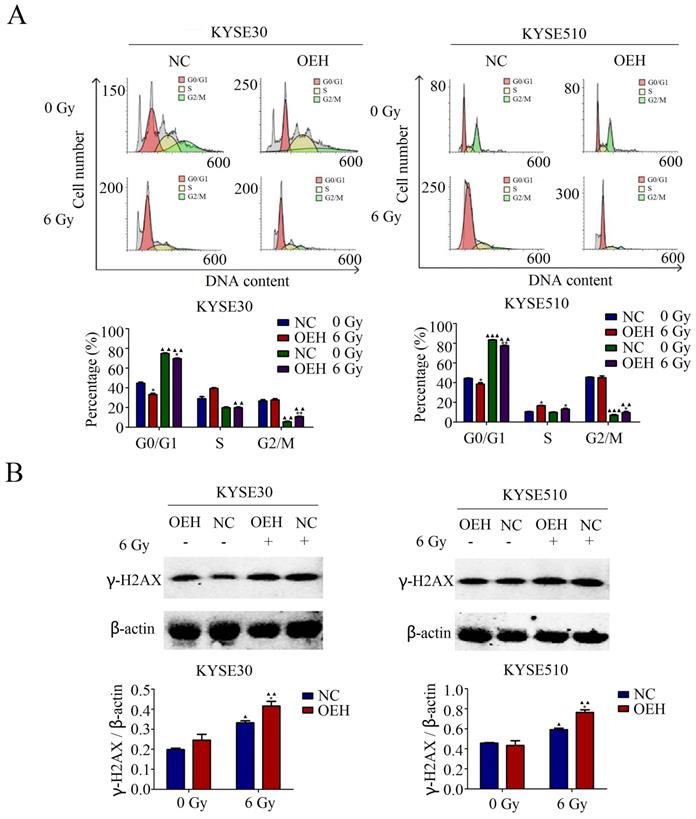
Effects of HMGB1 overexpression on cell cycle distribution
The results of the cell cycle distribution by flow cytometry suggested that the proportion of ESCC cells in the G0/G1 phase was significantly lower in the HMGB1 overexpression groups than in the control groups without 6 Gy X-ray irradiation (P<0.05). Irradiation caused cell cycle arrest at the G0/G1 phase in both control and HMGB1, overexpressing KYSE30 and KYSE510 cells (Figure 4A). Although irradiation arrested the ESCC cells at the G0/G1 phase in all groups, the HMGB1 overexpressing cells showed a weaker ability in G0/G1 phase arrest compared with the NC groups (P<0.05).
HMGB1 affects the expression level of γ-H2AX after irradiation
γ-H2AX is the DNA damage marker and key signal for initiating DNA damage repair [28]. It has been reported that in neurons, HMGB1 knockdown led to a weakened DNA damage response, which was reflected in the decreased expression of γ-H2AX [29]. Therefore, we performed western blotting to confirm the relationship between HMGB1 and γ-H2AX in ESCC cells. As revealed in Figure 4B, the protein expression levels of γ-H2AX were examined in both the HMGB1 overexpression and NC groups with or without exposure to 6 Gy irradiation. Interestingly, all groups showed a significant enhancement in γ-H2AX expression levels after 6 Gy irradiation. In addition, γ-H2AX protein expression was higher in HMGB1 overexpressing cells than in the control groups (P<0.05).
The expression of HMGB1 in ESCC cell lines and the validation of HMGB1 overexpression efficiency. A. The comparison of the HMGB1 protein expression levels in six esophageal cancer cell lines was performed using Western blotting; B. Immunofluorescence checking was shown and verified the specific localization of HMGB1 before and after irradiation. Green fluorescence, HMGB1 staining; blue fluorescence, nucleus staining; C. The mRNA expression level of HMGB1 was higher in OEH groups than that in NC groups; D. Overexpression of HMGB1 was validated using Western blotting. OEH, overexpression of HMGB1; NC, negative control.
HMGB1 protein overexpression increases the proliferation radioresistance and migration of ESCC cells. A. Cell proliferation was detected by the CCK-8 assay; B. Colony formation assay was used to evaluate the radioresistance of KYSE30 and KYSE510 cells; C. Migration was detected by wound healing assays (magnification: 40×). The wound closure images were obtained at 0 h, 24 h, and 48 h. The migration capacities of irradiated groups at 24 h and 48 h were compared to the corresponding non-irradiated groups. Each experiment was replicated three times. The comparison with NC group is indicated by asterisks (*P<0.05, **P<0.01 and ***P<0.001), and the comparison with the corresponding non-irradiated group is indicated by triangles (▲P<0.05, ▲▲P<0.01 and ▲▲▲P<0.001). OEH, overexpression of HMGB1; NC, negative control.
Discussion
EC is one of the most common malignant tumors in the digestive tract. Comprehensive treatment, mainly with radiotherapy, is the standard nonsurgical therapy for EC patients [30]. Despite the significant progress in ESCC diagnosis and treatment, the 5-year survival rate of ESCC patients is still low [31]. Local recurrence was the most common reason for treatment failure from radioresistance [32]. Therefore, it is necessary to identify ESCC patients with resistance to radiotherapy and adjust their treatment plans accordingly to improve the radiotherapy efficacy.
HMGB1 is a DNA‐binding protein released during radiotherapy, and it is related to tumor radioresistance based on its DNA damage repair and autophagy functions [11]. HMGB1 is involved in mechanisms that regulate resistance to radiotherapy in pancreatic [13, 14], cervical [8], breast [33], bladder [16, 34], and lung cancers [15].
In addition, Ma et al. found that HMGB1 expression was associated with ESCC recurrence after postoperative radiotherapy. Knocking down HMGB1 expression in ESCC cells could increase the radiosensitivity of these cells both in vitro and in vivo. Interestingly, HMGB1 could also inhibit the reduced autophagy levels in these cells, and activating autophagy restored the radioresistance [21]. A similar finding was reported previously, where inhibiting HMGB1 could significantly increase the radiosensitivity of ESCC cells through G0/G1 phase arrest. Additionally, HMGB1 negative expression was correlated with the long-term survival of ESCC patients who received radiochemotherapy [24]. Unlike previous reports, we selected ESCC cell lines with relatively low HMGB1 expression for overexpression in the present study. We verified the changes in HMGB1 protein localization in KYSE30 and KYSE510 cells before and after irradiation by immunofluorescence laser confocal technique. Because radiotherapy is an important treatment for ESCC, patients who had only received radiotherapy were included in this study for survival analysis to clarify the correlation between HMGB1 expression levels and survival of ESCC patients after radiotherapy alone.
HMGB1 protein overexpression regulated cell cycle distribution of ESCC cells. A. HMGB1 alleviated the G0/G1 arrest of KYSE30 and KYSE510 after irradiation; B. There was a correlation between HMGB1 and γ-H2AX proteins after 6 Gy X-ray irradiation. Western blotting was used to detect the expression levels of γ-H2AX in KYSE30 and KYSE510 cells exposed or not to irradiation. Each experiment was replicated three times. The comparison with NC group is indicated by asterisks (*P<0.05), and the comparison with the corresponding non-irradiated group is indicated by triangles (▲P<0.05, P<0.01 and ▲▲▲P<0.001). OEH, overexpression of HMGB1; NC, negative control.
We included 39 stage I-III ESCC patients who received radiotherapy in the present study. HMGB1 expression levels were associated with the 10-year survival rates of these patients, but no correlation was found in the 1-, 3-, and 5-year survival data.
This study explored the relationship between HMGB1 and ESCC radiosensitivity from a new perspective of HMGB1 overexpression. We observed that the overexpression of HMGB1 could enhance the migration, proliferation, and radioresistance of ESCC cells before and after radiation treatment in vitro, and HMGB1 may play an important role in the occurrence and development of EC. Amornsupak et al. believed that tumor cytoplasmic levels of HMGB1 could be used as an independent prognostic indicator for metastasis or recurrence in breast cancer patients [35]. Li et al. reported that overexpression of HMGB1 can facilitate cervical cell migration by IHC analysis of cervical cancer tissues from 239 patients [36].
Shi et al. provided evidence supporting a close correlation between HMGB1 overexpression and gallbladder cancer progression and poor prognosis [37]. Moreover, Dong et al. suggested that HMGB1 was associated with the 5-year overall survival of gastric cancer patients after analyzing the survival data of 100 patients with stage IIIA gastric cancer [38].
γ-H2AX plays a critical role in forming chromatin-remodeling complexes, and it also participates in the DNA damage and repair work associated with radiotherapy [39]. γ-H2AX is closely related to DNA repair in tumor cells after radiotherapy. It is involved in radiotherapy-related DNA damage and repair in many cancers, including colorectal cancer [40], hepatocellular carcinoma [41], and others. Moreover, according to Kawashima et al., after colorectal cancer cells were irradiated, their γ-H2AX expression levels increased, and cell viability also decreased time-dependent [40]. We found that ESCC cells overexpressing HMGB1 had higher γ-H2AX levels than the NC groups with 6 Gy irradiation.
Moreover, it has been confirmed that radiotherapy could induce a G0/G1 cell cycle arrest and pro-apoptotic effect in EC [42, 43], nasopharyngeal carcinoma [44], meningiomas [45], and breast cancer [46]. Our results corroborate these findings, which revealed that KYSE30 and KYSE510 cells showed cell cycle arrest at the G0/G1 phase after radiotherapy, and this cell cycle arrest was weakened after overexpressed HMGB1. The limitation of the article is that validation of normal esophageal cells and animal experiments were not performed. Follow-up external validation and in-depth studies are still necessary.
Conclusion
In this study, we demonstrated that positive HMGB1 expression in ESCC tissues was negatively correlated with 10-year survival rates of patients. Overexpressing HMGB1 promoted the migration, proliferation, and radioresistance of ESCC cells by reducing G0/G1 phase arrest. In conclusion, this study provides a theoretical basis that HMGB1 may be a potential target for overcoming ESCC radioresistance.
Abbreviations
HMGB1: high mobility group box 1; ESCC: esophageal squamous cell carcinoma; γ-H2A: phosphorylated histone H2AX; EC: esophageal cancer; DSBs: double-strand breaks; CSC: cancer stem cell; EMT: epithelial-mesenchymal transition; IHC: immunohistochemistry; FBS: fetal bovine serum; PBS: phosphate buffer saline; NC: negative control; RT-qPCR: real-time quantitative polymerase chain reaction; PVDF: polyvinylidene fluoride; TBST: TBS containing 0.1% Tween 20; SF: survival curves; KM: Kaplan-Meier; GTV: gross tumor volume; SERs: sensitizer enhancement ratios.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81872456), Natural Science Foundation of Hebei Province (no. H2020206583), and a grant from the Education Department of Hebei Province (no. CXZZBS2021078).
Availability of data and materials
The datasets that were used and/or analyzed in this study are available from the corresponding author upon reasonable request.
Disclosure
This article does not contain any studies with animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Author contributions
SZ conceived and designed the study. JD conducted the experiments as well as data collection and drafted the manuscript. XZ, XD and NZ designed the experimental approach and contributed to data interpretation. WS, MM, and YW helped with the investigation and formal analyses of data. SZ approved the final version of the manuscript. JD and NZ checked all the raw data for authenticity. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kuntz S, Krieghoff-Henning E, Kather JN. et al. Gastrointestinal cancer classification and prognostication from histology using deep learning: Systematic review. Eur J Cancer. 2021;155:200-215
2. Nejat Pish-Kenari F, Qujeq D, Maghsoudi H. Some of the effective factors in the pathogenesis of gastro-oesophageal reflux disease. J Cell Mol Med. 2018;22(12):6401-6404
3. Burke AM, Yeh C, Kim S. et al. A Prospective Study of Early Radiation Associated Cardiac Toxicity Following Neoadjuvant Chemoradiation for Distal Esophageal Cancer. Front Oncol. 2020;10:1169
4. Zhu Z, Song J, Gu J. et al. FMS-Related Tyrosine Kinase 3 Ligand Promotes Radioresistance in Esophageal Squamous Cell Carcinoma. Front Pharmacol. 2021;12:659735
5. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424
6. Lee JJ, Park IH, Rhee WJ. et al. HMGB1 modulates the balance between senescence and apoptosis in response to genotoxic stress. FASEB J. 2019;33(10):10942-10953
7. Tan G, Huang C, Chen J. et al. HMGB1 released from GSDME-mediated pyroptotic epithelial cells participates in the tumorigenesis of colitis-associated colorectal cancer through the ERK1/2 pathway. J Hematol Oncol. 2020;13(1):149
8. Jiao X, Zhang S, Jiao J. et al. Promoter methylation of SEPT9 as a potential biomarker for early detection of cervical cancer and its overexpression predicts radioresistance. Clin Epigenetics. 2019;11(1):120
9. Wang S, Zhang Y. HMGB1 in inflammation and cancer. J Hematol Oncol. 2020;13(1):116
10. Zhang Z, Wang M, Zhou L. et al. Increased HMGB1 and cleaved caspase-3 stimulate the proliferation of tumor cells and are correlated with the poor prognosis in colorectal cancer. J Exp Clin Cancer Res. 2015;34(1):51
11. Liao Y, Liu S, Fu S. et al. HMGB1 in Radiotherapy: A Two Headed Signal Regulating Tumor Radiosensitivity and Immunity. Onco Targets Ther. 2020;13:6859-6871
12. Bertheloot D, Latz E. HMGB1, IL-1α, IL-33 and S100 proteins: dual-function alarmins. Cell Mol Immunol. 2017;14(1):43-64
13. Zhang L, Shi H, Chen H. et al. Dedifferentiation process driven by radiotherapy-induced HMGB1/TLR2/YAP/HIF-1α signaling enhances pancreatic cancer stemness. Cell Death Dis. 2019;10(10):724
14. Chen X, Zhang L, Jiang Y. et al. Radiotherapy-induced cell death activates paracrine HMGB1-TLR2 signaling and accelerates pancreatic carcinoma metastasis. J Exp Clin Cancer Res. 2018;37(1):77
15. Bai L, Zhang J, Gao D. et al. Downregulation of high mobility group box 1 enhances the radiosensitivity of non-small cell lung cancer by acting as a crucial target of microRNA-107. Exp Ther Med. 2021;22(1):679
16. Shrivastava S, Mansure JJ, Almajed W. et al. The Role of HMGB1 in Radioresistance of Bladder Cancer. Mol Cancer Ther. 2016;15(3):471-479
17. Ayoub M, Shinde-Jadhav S, Mansure JJ. et al. The immune mediated role of extracellular HMGB1 in a heterotopic model of bladder cancer radioresistance. Sci Rep. 2019;9(1):6348
18. Matsubara D, Konishi H, Arita T. et al. Involvement of Intracellular and Extracellular High-Mobility Group Box-1 in the Progression of Esophageal Squamous Cell Carcinoma. Ann Surg Oncol. 2020;27(9):3233-3244
19. Takahata R, Ono S, Tsujimoto H. et al. Preoperative chemoradiation therapy for esophageal cancer is a risk factor for the elevation of high mobility group box-1, leading to an increase in postoperative severe pulmonary complications. Dis Esophagus. 2016;29(1):70-78
20. Chuangui C, Peng T, Zhentao Y. The expression of high mobility group box 1 is associated with lymph node metastasis and poor prognosis in esophageal squamous cell carcinoma. Pathol Oncol Res. 2012;18(4):1021-1027
21. Ma H, Zheng S, Zhang X. et al. High mobility group box 1 promotes radioresistance in esophageal squamous cell carcinoma cell lines by modulating autophagy. Cell Death Dis. 2019;10(2):136
22. Di X, He G, Chen H. et al. High-mobility group box 1 protein modulated proliferation and radioresistance in esophageal squamous cell carcinoma. J Gastroenterol Hepatol. 2019;34(4):728-735
23. Zhao Y, Yi J, Tao L. et al. Wnt signaling induces radioresistance through upregulating HMGB1 in esophageal squamous cell carcinoma. Cell Death Dis. 2018;9(4):433
24. Zhang X, Yang X, Zhu S. et al. Radiosensitization of esophageal carcinoma cells by knockdown of HMGB1 expression. Oncol Rep. 2019;41(3):1960-1970
25. Cheng DD, Li J, Li SJ. et al. CNOT1 cooperates with LMNA to aggravate osteosarcoma tumorigenesis through the Hedgehog signaling pathway. Mol Oncol. 2017;11(4):388-404
26. Soumaoro LT, Uetake H, Higuchi T. et al. Cyclooxygenase-2 expression: a significant prognostic indicator for patients with colorectal cancer. Clin Cancer Res. 2004;10(24):8465-8471
27. Liu Y, Liu X, Jin X. et al. The dependence of radiation enhancement effect on the concentration of gold nanoparticles exposed to low- and high-LET radiations. Phys Med. 2015;31(3):210-218
28. Gustafsson NMS, Färnegårdh K, Bonagas N. et al. Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination. Nat Commun. 2018;9(1):3872
29. Lou MM, Tang XQ, Wang GM. et al. Long noncoding RNA BS-DRL1 modulates the DNA damage response and genome stability by interacting with HMGB1 in neurons. Nat Commun. 2021;12(1):4075
30. Bolidong D, Domoto T, Uehara M. et al. Potential therapeutic effect of targeting glycogen synthase kinase 3β in esophageal squamous cell carcinoma. Sci Rep. 2020;10(1):11807
31. Qin G, Lian J, Yue D. et al. Musashi1, a potential prognostic marker in esophageal squamous cell carcinoma. Oncol Rep. 2017;38(3):1724-1732
32. Zhou YC, Chen LL, Xu HB. et al. Aging-related prognosis analysis of definitive radiotherapy for very elderly esophageal cancer. Cancer Med. 2018;7(5):1837-1844
33. Choi HS, Kim JH, Jang SJ. et al. Synergistic Tumoricidal Effects of Alpha-Lipoic Acid and Radiotherapy on Human Breast Cancer Cells via HMGB1. Cancer Res Treat. 2021;53(3):685-694
34. Jiang H, Hu X, Zhang H. et al. Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat Oncol. 2017;12(1):65
35. Amornsupak K, Jamjuntra P, Warnnissorn M. et al. High ASMA+ Fibroblasts and Low Cytoplasmic HMGB1+ Breast Cancer Cells Predict Poor Prognosis. Clin Breast Cancer. 2017;17(6):441-452
36. Li P, Xu M, Cai H. et al. The effect of HMGB1 on the clinicopathological and prognostic features of cervical cancer. Biosci Rep. 2019;39(5):BSR20181016
37. Shi Z, Huang Q, Chen J. et al. Correlation of HMGB1 expression to progression and poor prognosis of adenocarcinoma and squamous cell/adenosquamous carcinoma of gallbladder. Am J Transl Res. 2015;7(10):2015-2025
38. Dong J, Li J, Liu S. et al. Prognostic potential of an immune score based on the density of CD8+ T cells, CD20+ B cells, and CD33+/p-STAT1+ double-positive cells and HMGB1 expression within cancer nests in stage IIIA gastric cancer patients. Chin J Cancer Res. 2016;28(5):543-552
39. Pouliliou S, Koukourakis MI. Gamma histone 2AX (γ-H2AX) as a predictive tool in radiation oncology. Biomarkers. 2014;19(3):167-180
40. Kawashima S, Kawaguchi N, Taniguchi K. et al. γ-H2AX as a potential indicator of radiosensitivity in colorectal cancer cells. Oncol Lett. 2020;20(3):2331-2337
41. Sun J, Zhu Z, Li W. et al. UBE2T-regulated H2AX monoubiquitination induces hepatocellular carcinoma radioresistance by facilitating CHK1 activation. J Exp Clin Cancer Res. 2020;39(1):222
42. Jin J, Guo Y, Dong X. et al. Methylation-associated silencing of miR-193b improves the radiotherapy sensitivity of esophageal cancer cells by targeting cyclin D1 in areas with zinc deficiency. Radiother Oncol. 2020;150:104-113
43. Zou N, Zhang X, Li S. et al. Elevated HNF1A expression promotes radiation-resistance via driving PI3K/AKT signaling pathway in esophageal squamous cell carcinoma cells. J Cancer. 2021;12(16):5013-5024
44. Zhu H, Ding W, Wu J. et al. TRAF2 Knockdown in Nasopharyngeal Carcinoma Induced Cell Cycle Arrest and Enhanced the Sensitivity to Radiotherapy. Biomed Res Int. 2020;2020:1641340
45. Zhang Q, Song LR, Huo XL. et al. MicroRNA-221/222 Inhibits the Radiation-Induced Invasiveness and Promotes the Radiosensitivity of Malignant Meningioma Cells. Front Oncol. 2020;10:1441
46. Ding X, Yang Q, Kong X. et al. Moran. Radiosensitization effect of Huaier on breast cancer cells. Oncol Rep. 2016;35(5):2843-2850
Author contact
![]() Corresponding author: Professor Shuchai Zhu, Department of Radiation Oncology, The Fourth Hospital of Hebei Medical University, 12 Jiankang Road, Shijiazhuang, Hebei 050011, P.R. China; Tel: +86-311-8609-5673; Fax: +86-311-8609-5819; E-mail: sczhu1965com
Corresponding author: Professor Shuchai Zhu, Department of Radiation Oncology, The Fourth Hospital of Hebei Medical University, 12 Jiankang Road, Shijiazhuang, Hebei 050011, P.R. China; Tel: +86-311-8609-5673; Fax: +86-311-8609-5819; E-mail: sczhu1965com

 Global reach, higher impact
Global reach, higher impact