Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(17):5331-5337. doi:10.7150/jca.59193 This issue Cite
Research Paper
Prognostic Value of KRAS Exon 3 and Exon 4 Mutations in Colorectal Cancer Patients
1. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai, China.
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China.
3. Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, China.
4. Department of Biostatistics and Bioinformatics, Emory University, Atlanta, GA.
*These authors contributed equally to this study.
Received 2021-2-8; Accepted 2021-5-23; Published 2021-7-3
Abstract
Background: The clinical significance of KRAS exon 3/4 mutations in colorectal cancer (CRC) remains unclear. We aimed to assess the prognostic value of KRAS exons 3 and 4 mutations to determine the necessity for their testing.
Methods: KRAS mutations in exon 2/3/4 were evaluated in 1816 stage I-IV patients with colorectal adenocarcinoma.
Results: The mutation rates of KRAS and KRAS exons 2, 3, and 4 were 49.0%, 43.0%, 1.9%, and 4.1%, respectively. Univariate survival analysis showed that patients with exon 3 mutation had worse overall survival (OS) compared to those with KRAS exon 2 mutation or wild-type KRAS (P = 0.044, and P = 0.001). Meanwhile, there was no difference in survival between patients with wild-type KRAS and with exon 4 mutation (P = 0.128). In multivariate analysis, KRAS mutations in exon 3 and 2 were both independent factors for worse OS (Exon 3, P = 0.032, HR = 1.861, 95% CI: 1.021-3.391; Exon 2, P = 0.049, HR = 1.298, 95% CI: 1.002-1.682). Among the patients with KRAS exon 2 mutations, those that had mutations in codon 13 had significantly worse prognosis than those with wild-type KRAS (P = 0.001) or KRAS codon 12 mutations (P = 0.003).
Conclusions: In KRAS-mutated CRC, exon 3 mutations predict the worst prognosis, while exon 4 mutations predict the best prognosis. Among KRAS exon 2 mutated patients, codon 13 mutations predict worse prognosis than codon 12 mutations. Mutations of different KRAS exons should be analyzed separately.
Keywords: KRAS exon 3, KRAS exon 4, KRAS mutations, colorectal cancer, clinicopathologic features, prognosis
Introduction
Colorectal cancer (CRC) is the third and second most common malignancy in men and women worldwide, respectively, and the fourth leading cause of cancer-related mortality [1, 2]. KRAS is one of the first genes to be identified as an oncogene in CRC. Detection of KRAS mutations has emerged as an important assessment method for patients with CRC due to its clinical value in predicting prognosis and resistance to targeted therapies [3]. KRAS mutations are often found in exons 2, 3, and 4, with KRAS exon 2 mutations being the most common, accounting for 81-96% of all KRAS mutations. The remaining 4-19% of mutations are located in KRAS exons 3 and 4 [4-6]. Despite the lower frequency of mutations in KRAS exons 3 and 4, they should not be neglected given the high prevalence of CRC.
Currently, mutations in KRAS exon 2 are routinely tested for metastatic CRC in most clinical institutions, and the identification of such mutations has been wildly reported to be associated with poor prognosis and resistance to anti-epidermal growth factor receptor (EGFR) therapy [3, 7, 8]. In contrast, mutations in exons 3 and exon 4 of KRAS are not widely tested due to their low mutation rate. Thus, the prognostic value of mutations in KRAS exon 3/4 remains unclear, and patients harboring KRAS mutations in exon 3 or exon 4 are usually combined into one group to meet the number requirements for analysis [5, 9, 10]. Further, no consensus has been reached on the clinicopathologic features and prognosis of patients with KRAS mutations in exon 3 or exon 4 [11-13].
Patient inclusion flowchart.
Specific mutations in the KRAS gene are closely related to the precise treatment of colorectal cancer. We have previously analyzed the clinicopathologic features and prognostic value of KRAS, NRAS, and BRAF mutations in our cohort [14]. The present study aimed to assess the prognostic value of KRAS mutations in exons 3 and 4 to elucidate the necessity for their testing as well as to identify the clinicopathologic characteristics of patients harboring these mutations.
Methods
Patients
This was a retrospective study of patients who underwent radical surgery and were pathologically diagnosed with CRC between July 2010 and June 2018. Of the 18604 patients whose tumor tissues were screened for mutations, 1949 patients were identified to have mutations in KRAS, NRAS, or BRAF. We excluded three patients due to a second primary malignant tumor (two with hepatocellular carcinoma and one with lung adenocarcinoma). Further, considering the effect of BRAF and NRAS mutations on prognosis, we also excluded 128 patients with infrequent mutations in BRAF (V600E or non-V600E) or NRAS (exon 2/3/4) and 2 patients that harbored KRAS mutations in both exons 2 and 3 or in exons 2 and 4. Finally, 1816 patients were included in the analysis (Figure 1). This study was approved by the Research Ethics Committee of the Fudan University Shanghai Cancer Center in China, and all patients provided written informed consent.
Mutation analysis
Mutation analysis was performed in formaldehyde fixed-paraffin embedded tissues after confirmation by two pathologists in hematoxylin and eosin-stained slides. Genomic DNA was extracted using the QIAamp DNA Mini Kit following the manufacturer's protocol (Qiagen, Valencia, CA, USA). DNA content was quantified using NanoDrop ND-1000 (Nanodrop, Wilmington, DE, USA). Sequencing was performed in 1311 patients. KRAS exon 2, 3, and 4; NRAS exon 2, 3, and 4; and BRAF exon 15 were amplified using the Real-time PCR master mix (TOYOBA, Osaka, Japan) and bidirectionally sequenced via ABI 3730XL and BigDye Terminator v. 3.1 Cycle Sequencing kit (Applied Biosystems, Carlsbad, CA, USA). The positive samples were further confirmed by three independent experiments. For the other 505 patients, amplification refractory mutation system (ARMS) analysis was conducted for mutations in KRAS exon 2 (codon 12/13), 3 (codon 59/61), and 4 (codon 117/146) using the AmoyDx KRAS/NRAS/BRAF Mutations Detection Kit (Amoy Diagnostics, Xiamen, China). All experiments were conducted as per the manufacturer's recommendations.
Statistical Analysis
Statistical analysis was performed using SPSS software version 25.0 (IBM Corporation, Armonk, NY, USA). A two-sided P-value < 0.05 was considered statistically significant. Chi-square tests and Fisher's exact tests were used to compare the categorical variables. For continuous variables, the Kolmogorov-Smirnov test was performed to verify the normal distribution assumptions. The exploratory comparison of normally distributed and non-normally distributed independent groups was performed using t-tests and Mann-Whitney U tests, respectively, for comparison between two groups and using analysis of variance (ANOVA) for comparison between more than two groups. Logistic regression was used for multivariate analysis. Overall survival (OS) was defined as the time between the first surgery and death from any cause. For survival analysis, curves were plotted using the Kaplan-Meier method and analyzed using the log-rank test. Univariate and multivariate analyses to identify prognostic biomarkers were performed using Cox proportional hazard models.
Results
Mutational landscape and clinicopathologic characteristics of the patients
The total KRAS mutation rate was 49.0% (889/1816), while the KRAS exon 2, 3, and 4 mutations rates were 43.0% (780/1816), 1.9% (34/1816), and 4.1% (75/1816), respectively. The mutation analysis results are shown in Table 1. The patients' clinicopathologic characteristics and the results of univariate analysis are shown in Table 2. Meanwhile, the results of multivariate analysis are shown in Table 3. In both univariate and multivariate analyses, a high rate of KRAS exon 2 mutation was associated with the following factors: female sex, advanced age, right-colon tumor, high carcinoembryonic antigen (CEA) levels, stage IV tumor, tumor type histology, and extranodal tumor deposit. The KRAS exon 3 mutation rate was higher in female patients and in patients with extranodal tumor deposit. Factors associated with a high KRAS exon 4 mutation rate included advanced age, tumor type histology, and mucinous carcinoma.
Mutations detected in the 1816 BRAF/NRAS wild-type patients
| Mutations | Codons | Amino acid alteration | Number of patients |
|---|---|---|---|
| Wild type | 927 | ||
| KRAS exon 2 mutation | Codon 12 | p.G12D | 174 |
| p.G12V | 121 | ||
| p.G12S | 36 | ||
| p.G12C | 26 | ||
| p.G12A | 15 | ||
| p.G12R | 8 | ||
| Other amino acid alteration | 7 | ||
| Unknown amino acid alteration | 153 | ||
| Codon 13 | P.G13D | 184 | |
| p.G13R | 3 | ||
| p.G13C | 1 | ||
| p.G13V | 1 | ||
| Unknown codon | 51 | ||
| KRAS exon 3 mutation | Codon 61 | p.Q61H | 8 |
| p.Q61L | 7 | ||
| p.Q61R | 5 | ||
| p.Q61K | 2 | ||
| Codon 59 | p.A59T | 2 | |
| Unknown codon | 10 | ||
| KRAS exon 4 mutation | Codon 146 | p.A146T | 44 |
| p.A146V | 4 | ||
| p.A146N | 1 | ||
| Codon 117 | p.K117D | 5 | |
| p.K117N | 3 | ||
| p.K117R | 1 | ||
| Codon 131 | p.Q131fs | 1 | |
| Unknown codon | 16 |
Univariate analysis of the clinicopathologic features
| Variables | Wild type N = 927 (%) | KRAS | KRAS exon 2 | KRAS exon 3 | KRAS exon 4 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mutant N = 889 (%) | P-value | Mutant N = 780 (%) | P-value | Mutant N = 34 (%) | P-value | MutantN = 75 (%) | P-value | ||
| Sex | 0.001 | 0.003 | 0.020 | 0.379 | |||||
| Male | 591 (63.8) | 501 (56.4) | 442 (56.7) | 15 (44.1) | 44 (58.7) | ||||
| Female | 336 (36.2) | 388 (43.6) | 338 (43.3) | 19 (55.9) | 31 (41.3) | ||||
| Age | 60.0 (22-89) | 62.0 (20-91) | 0.003 | 62.0 (20-91) | 0.011 | 60.0 (37-90) | 0.749 | 64.0 (34-89) | 0.017 |
| Tumor site | 0.001 | 0.001 | 0.140 | 0.004 | |||||
| Rectum | 430 (46.4) | 429 (48.3) | 373 (47.8) | 12 (35.3) | 44 (58.7) | ||||
| Left | 321 (34.6) | 186 (20.9) | 164 (21.0) | 11 (32.4) | 11 (14.7) | ||||
| Right | 176 (19.0) | 274 (30.8) | 243 (31.2) | 11 (32.4) | 20 (26.6) | ||||
| Tumor size | 4.0 (0.5-17.0) | 4.2 (0.5-22.0) | 0.040 | 4.1 (0.5-22.0) | 0.089 | 3.9 (1.5-7.5) | 0.933 | 4.5 (1.2-11.0) | 0.014 |
| CEA level | 0.001 | 0.001 | 0.113 | 0.175 | |||||
| ≤5 μg/L | 520 (56.1) | 411 (46.2) | 361 (46.3) | 14 (41.2) | (48.0) | ||||
| >5 μg/L | 407 (43.9) | 478 (53.8) | 419 (53.7) | 20 (58.8) | 39 (52.0) | ||||
| T stage | 0.676 | 0.514 | 0.148 | 0.665 | |||||
| T1 | 31 (3.3) | 25 (2.8) | 21 (2.7) | 4 (5.3) | |||||
| T2 | 126 (13.6) | 107 (12.0) | 92 (11.8) | 3 (8.8) | 12 (16.0) | ||||
| T3 | 512 (55.2) | 506 (56.9) | 453 (58.1) | 16 (47.1) | 37 (49.4) | ||||
| T4 | 258 (27.9) | 251 (28.2) | 214 (27.4) | 15 (44.1) | 22 (29.3) | ||||
| N stage | 0.124 | 0.111 | 0.114 | 0.542 | |||||
| N0 | 399 (43.0) | 351 (39.5) | 306 (39.2) | 10 (29.4) | 35 (46.7) | ||||
| N1-2 | 528 (57.0) | 538 (60.5) | 474 (60.8) | 24 (70.6) | 40 (53.3) | ||||
| TNM stage | 0.002 | 0.001 | 0.057 | 0.588 | |||||
| I | 102 (11.0) | 74 (8.3) | 60 (7.7) | 2 (5.9) | 12 (16.0) | ||||
| II | 235 (25.4) | 206 (23.2) | 180 (23.1) | 7 (20.6) | 19 (25.4) | ||||
| III | 310 (33.4) | 280 (31.5) | 251 (32.2) | 9 (26.5) | 20 (26.6) | ||||
| IV | 280 (30.2) | 329 (37.0) | 289 (37.0) | 16 (47.0) | 24 (32.0) | ||||
| Histological | 0.001 | 0.001 | 0.596 | 0.003 | |||||
| Ulcer type | 662 (71.4) | 565 (63.6) | 495 (63.5) | 27 (79.4) | 43 (57.3) | ||||
| Tumor type | 225 (24.3) | 294 (33.1) | 257 (32.9) | 6 (17.6) | 31 (41.3) | ||||
| Invasive type | 40 (4.3) | 30 (3.3) | 28 (3.6) | 1 (3.0) | 1 (1.4) | ||||
| Pathology | 0.002 | 0.010 | 0.460 | 0.001 | |||||
| Adenocarcinoma | 851 (91.8) | 776 (87.3) | 687 (88.1) | 30 (88.2) | 59 (78.7) | ||||
| Mucinous | 76 (8.2) | 113 (12.7) | 93 (11.9) | 4 (11.8) | 16 (21.3) | ||||
| Differentiation | 0.023 | 0.092 | 0.157 | 0.011 | |||||
| G3-G4 | 252 (27.2) | 285 (32.1) | 241 (30.9) | 13 (38.2) | 31 (41.3) | ||||
| G1-G2 | 675 (72.8) | 604 (67.9) | 539 (69.1) | 21 (61.8) | 44 (58.7) | ||||
| Lymphovascular invasion | 0.811 | 0.990 | 0.447 | 0.095 | |||||
| Negative | 604 (65.2) | 584 (65.7) | 508 (65.1) | 20 (58.8) | 56 (74.7) | ||||
| Positive | 323 (34.8) | 305 (34.3) | 272 (34.9) | 14 (41.2) | 19 (25.3) | ||||
| Perineural invasion | 0.164 | 0.097 | 0.358 | 0.237 | |||||
| Negative | 618 (66.7) | 565 (63.6) | 490 (62.8) | 20 (58.8) | 55 (73.3) | ||||
| Positive | 309 (33.3) | 324 (36.4) | 290 (37.2) | 14 (41.2) | 20 (26.6) | ||||
| Extranodal tumor deposit | 0.011 | 0.013 | 0.024 | 0.878 | |||||
| Negative | 755 (81.4) | 681 (76.6) | 597 (76.5) | 22 (64.7) | 62 (82.7) | ||||
| Positive | 172 (18.6) | 208 (23.4) | 183 (23.5) | 12 (35.3) | 13 (17.3) | ||||
Multivariate analysis of the clinicopathologic features
| Variables | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| KRAS | KRAS exon 2 | KRAS exon 3 | KRAS exon 4 | |||||
| P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | |
| Sex (Male) | 0.002 | 0.728 (0.598-0.887) | 0.006 | 0.752 (0.614-0.922) | 0.035 | 0.473 (0.236-0.947) | ||
| Age | 0.001 | 1.016 (1.008-1.025) | 0.002 | 1.014 (1.005-1.022) | 0.002 | 1.035 (1.013-1.058) | ||
| Tumor site | ||||||||
| Rectum | Ref | 1 | Ref | 1 | Ref | 1 | ||
| Left | 0.001 | 0.535 (0.424-0.675) | 0.001 | 0.545 (0.428-0.694) | 0.001 | 0.328 (0.165-0.651) | ||
| Right | 0.008 | 1.388 (1.088-1.773) | 0.002 | 1.475 (1.151-1.892) | 0.750 | 0.909 (0.505-1.637) | ||
| CEA level (>5 μg/L) | 0.001 | 1.390 (1.140-1.695) | 0.002 | 1.376 (1.121-1.689) | ||||
| TNM stage | ||||||||
| I | Ref | 1 | Ref | 1 | ||||
| II | 0.160 | 1.310 (0.899-1.908) | 0.068 | 1.448 (0.973-2.155) | ||||
| III | 0.244 | 1.248 (0.860-1.810) | 0.066 | 1.443 (0.975-2.134) | ||||
| IV | 0.008 | 1.678 (1.143-2.464) | 0.002 | 1.868 (1.246-2.801) | ||||
| Histological | ||||||||
| Ulcer type | Ref | 1 | Ref | 1 | Ref | 1 | ||
| Tumor type | 0.001 | 1.632 (1.307-2.037) | 0.001 | 1.699 (1.352-2.136) | 0.006 | 2.009 (1.219-3.311) | ||
| Invasive type | 0.270 | 0.751 (0.452-1.248) | 0.407 | 0.803 (0.479-1.348) | 0.263 | 0.314 (0.041-2.385) | ||
| Pathology (Mucinous) | 0.038 | 1.310 (1.015-1.692) | 0.001 | 3.800 (1.974-7.313) | ||||
| Extranodal tumor deposit | 0.038 | 1.310 (1.015-1.692) | 0.034 | 1.329 (1.022-1.729) | 0.029 | 2.249 (1.087-4.653) | ||
Survival analysis
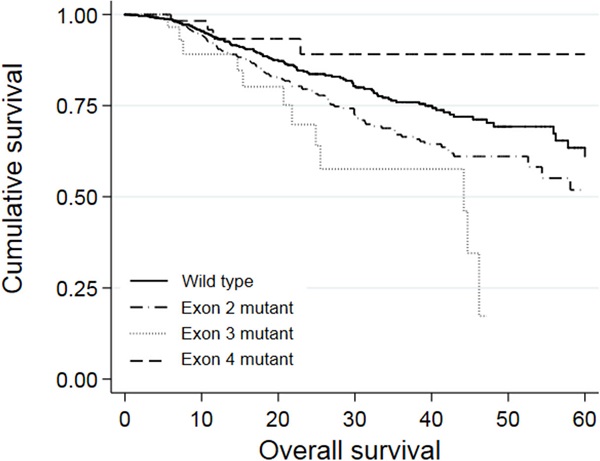
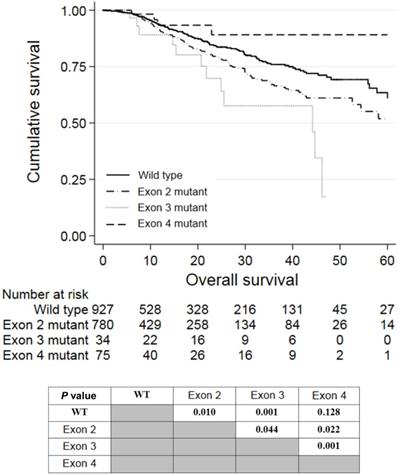
Univariate analysis revealed that a right-colon tumor, larger tumor size, higher CEA levels, neoadjuvant treatment, adjuvant treatment, stage III and stage IV tumor, palliative resection, mucinous carcinoma, poor differentiation, perineural invasion, extranodal tumor deposit, and KRAS mutations in exon 2 or 3 were associated with shorter OS. Among these factors, larger tumor size (P = 0.001, HR = 1.073, 95% CI: 1.028-1.120), stage IV tumor (P = 0.027, HR = 3.203, 95% CI: 1.140-9.005), palliative resection (P = 0.001, HR = 2.371, 95% CI: 1.678-3.350), poor differentiation (P = 0.002, HR = 1.520, 95% CI: 1.167-1.978), perineural invasion (P = 0.016, HR = 1.385, 95% CI: 1.062-1.805), extranodal tumor deposit (P = 0.001, HR = 1.573, 95% CI: 1.198-2.066), and KRAS mutations in exon 2 (P = 0.049, HR = 1.298, 95% CI: 1.002-1.682) or 3 (P = 0.032, HR = 1.861, 95% CI: 1.021-3.391) were independently associated with worse OS in multivariate analyses. The survival curves of patients with wild-type KRAS and KRAS mutations in different exons are shown in Figure 2. Patients who harbored KRAS exon 2 mutations had poorer prognosis than those with wild-type KRAS (P = 0.010) and KRAS exon 4 mutations (P = 0.022). Meanwhile, patients with KRAS exon 3 mutations had an even worse prognosis than those with exon 2 mutations (P = 0.044). However, patients with KRAS exon 4 mutations survived slightly longer than wild-type patients, but the difference was not significant (P = 0.128).
Kaplan-Meier analysis of patients with KRAS exon 2, 3, or 4 mutations.
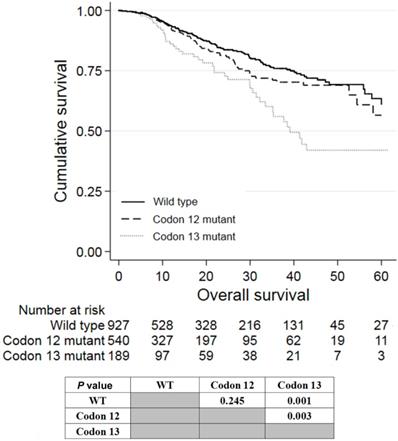
Of the 780 patients with KRAS exon 2 mutations, 540 and 189 patients had mutations in codon 12 and 13, respectively. Patients with mutations in codon 13 had worse prognosis than wild-type patients (P = 0.001; Figure 3) and patients with mutations in codon 12 (P = 0.003). Patients with KRAS codon 12 mutations tended to have worse prognosis than wild-type patients, although it was not statistically significant (P = 0.245). The survival of patients having codon 59/61 and codon 117/131/146 mutations was not analyzed because of the small number of patients with these mutations.
Discussion
In this study, we retrospectively analyzed the prognostic value of mutations in KRAS exons 2, 3, and 4. We found that patients with KRAS exon 3 mutations had the worst prognosis, while patients with exon 4 mutations had the best prognosis. Further analysis of codon 12 and 13 sequences in KRAS exon 2 mutation showed that patients harboring mutations in codon 13 had worse prognosis than those with mutations in codon 12. To our best knowledge, this is the first study to (1) report significant differences in prognosis between KRAS exon 3 and exon 4 mutations and (2) report worse prognosis of patients with KRAS codon 13 mutations compared to those with mutations in codon 12. Further, our cohort of 1816 patients is the largest cohort size in which KRAS exon 3/4 mutations have been tested.
Univariate and multivariate analyses of the prognostic variables for OS
| Prognostic variables | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| P-value | HR (95% CI) | P-value | HR (95% CI) | |
| Sex (Male) | 0.097 | 0.810 (0.632-1.039) | ||
| Age | 0.150 | 0.993 (0.982-1.003) | ||
| Tumor site | ||||
| Rectum | Ref | |||
| Left | 0.233 | 1.199 (0.890-1.617) | ||
| Right | 0.001 | 1.629 (1.206-2.200) | ||
| Tumor size | 0.001 | 1.133 (1.083-1.186) | 0.001 | 1.073 (1.028-1.120) |
| Elevated CEA level | 0.001 | 1.862 (1.439-2.408) | ||
| Neoadjuvant treatment | 0.001 | 2.062 (1.575-2.700) | ||
| Adjuvant treatment | 0.001 | 1.972 (1.401-2.775) | ||
| TNM stage | ||||
| I | Ref | Ref | ||
| II | 0.262 | 1.881 (0.624-5.668) | 0.541 | 1.412 (0.467-4.270) |
| III | 0.012 | 3.705 (1.330-10.322) | 0.114 | 2.300 (0.818-6.469) |
| IV | 0.001 | 10.058(3.735-27.084) | 0.027 | 3.203(1.140-9.005) |
| Palliative resectiona | 0.001 | 4.310 (3.347-5.552) | 0.001 | 2.371(1.678-3.350) |
| Histology | ||||
| Ulcer type | Ref | |||
| Tumor type | 0.260 | 0.849 (0.639-1.129) | ||
| Invasive type | 0.325 | 1.285 (0.780-2.116) | ||
| Pathology (mucinous) | 0.001 | 1.741 (1.261-2.404) | ||
| Differentiation (G3-G4) | 0.001 | 2.271 (1.773-2.920) | 0.002 | 1.520 (1.167-1.978) |
| Lymphovascular invasion | 0.001 | 2.100 (1.639-2.691) | ||
| Perineural Invasion | 0.001 | 2.042 (1.591-2.622) | 0.016 | 1.385 (1.062-1.805) |
| Extranodal tumor deposit | 0.001 | 2.718 (2.116-3.493) | 0.001 | 1.573 (1.198-2.066) |
| KRAS exon 2 mutant | 0.010 | 1.399 (1.084-1.806) | 0.049 | 1.298 (1.002-1.682) |
| KRAS exon 3 mutant | 0.001 | 2.518 (1.389-4.567) | 0.032 | 1.861 (1.021-3.391) |
| KRAS exon 4 mutant | 0.128 | 0.569 (0.205-1.580) | 0.141 | 0.472 (0.174-1.282) |
a) Cases were considered as palliative excision when primary and metastatic lesions were not both radically resected.
Despite evidence that mutations in RAS genes, particularly KRAS, play an essential role in predicting resistance to anti-EGFR therapy and worse prognosis in CRC patients, only a few studies have investigated the clinical value of mutations in exons 3 and 4. Information on the clinical relevance of KRAS mutations is largely based on KRAS exon 2 testing. Although KRAS exon 3/4 mutations are associated with clinicopathologic features or patient survival, the necessity of KRAS exon 3/4 testing has not been determined to date. Thus, we conducted this single-center retrospective study to explore the impact of KRAS exons 3 and 4 mutations on patient survival.
It has been well demonstrated that patients harboring KRAS exon 2 mutations do not benefit from anti-EGFR therapy. Lièvre et al. [3] found that KRAS exon 2 mutations were associated with resistance to cetuximab and poor prognosis. Amado et al. [8] also reported that KRAS exon 2 mutations influence response to panitumumab. Recent clinical trials also showed that patients with KRAS exons 3 and 4 mutations lack response to anti-EGFR therapy. However, patients with KRAS mutations in exons 3 or 4 were usually analyzed together as one group due to the small cohort size. In the PRIME study, mutations in the exon 3 of KRAS were detected in 24 out of 641 metastatic CRC patients, while 36 patients were detected with mutations in exon 4. These patients, along with 48 patients with NRAS mutations, were further analyzed as one group [5]. Similar situation was also seen in the CRYSTAL study, where 63 patients harboring mutations in KRAS exons 3/4 or NRAS exon 2/3/4 were categorized into one group [10]. These findings formed the basis for international guidelines to recommend that all patients with metastatic CRC should be tested for KRAS exon 2/3/4 mutations and that patients detected with mutations in any of these exons should not be treated with anti-EGFR therapy. However, we believe that randomized, controlled trials with larger patient cohorts are needed to determine whether these findings are also representative for patients harboring mutations in exons 3 or 4.
Kaplan-Meier analysis of patients with KRAS mutations in codon 12 or 13 of exon 2.
The KRAS exon 2/3/4 mutation rates in the current study cohort were consistent with those in two previous studies conducted in relatively large populations [5, 15]. In the study by Vaughn et al. [15], KRAS exon 2 mutations were detected in 42.4% (900/2121) of their patients, while mutations in exon 3 and 4 were detected in only 3.7% (19/513) and 3.3% (17/513) of 513 exon 2 wild-type patients, respectively. However, in their study, no mutations in codons 59 or 117 were detected, which might explain why their results differ from ours. Similarly, Douillard et al. [5] reported mutation rates of KRAS mutations in exons 2, 3, and 4 of 40.1% (440/1096), 3.7% (24/641), and 5.6% (36/641), respectively. KRAS codon 61 mutations were also detected in that study, but mutations in codon 59 were not. In an Eastern population, Guo et al. [16] evaluated 353 stage I-IV Chinese CRC patients, and the KRAS mutation rates in exons 2, 3, and 4 were 42.2% (149/353), 2.3% (8/353), and 8.2% (29/353), respectively. The differences in mutation rates might be due to the differences in cohort size.
With respect to clinicopathologic features, our study showed that KRAS mutations in exon 2/3 were associated with a high rate of positive extranodal tumor deposit, and extranodal tumor deposit was known as a predictor of poor prognosis [17, 18]. This might be related to the poor prognosis of patients harboring KRAS exon 2/3 mutations. No relevant findings from other studies have been reported yet.
The prognostic value of KRAS exon 3/4 remained unclear before the current study [19-21]. Similar survival trends of patients with mutations in KRAS exon 2/3/4 were reported by Frankel et al. [22] in a cohort of 165 stage IV CRC patients, although the small cohort size did not allow for robust statistical analyses. In most studies, patients with mutations in KRAS exon 3/4 were combined for analysis [11-13], which was not reasonable according to our results because mutations in these exons resulted in very different prognosis.
The poor prognosis of patients with KRAS exon 2 mutations has been widely reported. However, the prognostic value of mutations in codons 12/13 is controversial. In a study of 1075 stage I-IV CRC patients by Imamura et al. [23], survival analysis showed that mutations in codon 12, but not codon 13, was associated with a worse prognosis than wild-type KRAS. Similar results were found by Margonis et al. [24] in a study of 512 stage IV CRC patients. Passot et al. [25] evaluated 524 stage IV CRC patients and reported that patients with mutations in codon 12 or 13 had worse prognosis than KRAS wild-type patients. However, there was no significant difference in the prognosis of patients with codon 12 and codon 13 mutations, consistent with the findings of some other studies [13, 26]. The varying results in these studies could be caused by the differences in cohort size, data analysis methods, or race.
This study was subject to limitations because of its single-center and retrospective design. Further, the influence of specific amino acid alterations on prognosis was unclear due to the small sample size of the subgroups. Further studies with a larger sample size and longer follow-up are needed to validate our findings. We will focus on response analysis of chemotherapies (irinotecan based or oxaliplatin based) and targeted therapies (cetuximab or bevacizumab) when a sufficient number of cases are accumulated.
In summary, this large-scale retrospective study demonstrated that KRAS mutations in exon 3 predict the worst prognosis, while those in exon 4 predict the best prognosis. KRAS mutations in codon 13 predict worse prognosis compared to mutations in codon 12. Thus, further studies on treatment efficacy should evaluate patients with KRAS exon 3 mutations separately from those with KRAS exon 4 mutations.
Abbreviations
CRC: colorectal cancer; OS: overall survival; EGFR: epidermal growth factor receptor; ARMS: the amplification refractory mutation system; CEA: carcinoembryonic antigen; OR: odds ratio; CI: confidence interval; HR: hazard ratio.
Acknowledgements
The authors would thank all the patients included and the pathologists of the Department of Pathology of Fudan University Shanghai Cancer Center participating in this study. This work was supported by the Shanghai Committee of Science and Technology [grant numbers 20DZ1100101, 18140903702, and 19511121202].
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA-CANCER J CLIN. 2015;65:87-108
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-86
3. Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E. et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374-9
4. De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594-603
5. Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M. et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-34
6. Schirripa M, Cremolini C, Loupakis F, Morvillo M, Bergamo F, Zoratto F. et al. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer. 2015;136:83-90
7. Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC. et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757-65
8. Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ. et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626-34
9. Sorich MJ, Wiese MD, Rowland A, Kichenadasse G, McKinnon RA, Karapetis CS. et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13-21
10. Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezínek I. et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol. 2015;33:692-700
11. Janakiraman M, Vakiani E, Zeng Z, Pratilas CA, Taylor BS, Chitale D. et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901-11
12. Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D. et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101:715-21
13. Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X. et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135
14. Guo TA, Wu YC, Tan C, Jin YT, Sheng WQ, Cai SJ. et al. Clinicopathologic features and prognostic value of KRAS, NRAS and BRAF mutations and DNA mismatch repair status: A single-center retrospective study of 1,834 Chinese patients with Stage I-IV colorectal cancer. Int J Cancer. 2019;145:1625-34
15. Vaughn CP, ZoBell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes, Chromosomes Cancer. 2011;50:307-12
16. Guo F, Gong H, Zhao H, Chen J, Zhang Y, Zhang L. et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep. 2018;8:6076
17. Tong L, Gao P, Wang Z, Song YX, Xu YY, Sun Z. et al. Is the Seventh Edition of the UICC/AJCC TNM Staging System reasonable for patients with tumor deposits in colorectal cancer? Ann Surg. 2012;255:208-13
18. Nagtegaal ID, Tot T, Jayne DG, McShane P, Nihlberg A, Marshall HC. et al. Lymph nodes, tumor deposits, and TNM: are we getting better? J Clin Oncol. 2011;29:2487-92
19. Karagkounis G, Torbenson MS, Daniel HD, Azad NS, Diaz LA, Donehower RC. et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer-Am Cancer Soc. 2013;119:4137-44
20. Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM. et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931-7
21. Taieb J, Le Malicot K, Shi Q, Penault-Llorca F, Bouché O, Tabernero J. et al. Prognostic value of BRAF and KRAS mutations in MSI and MSS stage III colon cancer. J NATL CANCER I. 2016;109(5):djw272
22. Frankel TL, Vakiani E, Nathan H DeMatteo RP, Kingham TP, Allen PJ. et al. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer-Am Cancer Soc. 2017;123:568-75
23. Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M. et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18:4753-63
24. Margonis GA, Kim Y, Sasaki K, Samaha M, Amini N, Pawlik TM. et al. Codon 13 KRAS mutation predicts patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Cancer-Am Cancer Soc. 2016;122:2698-707
25. Passot G, Denbo JW, Yamashita S, Kopetz SE, Chun YS, Maru D. et al. Is hepatectomy justified for patients with RAS mutant colorectal liver metastases? An analysis of 524 patients undergoing curative liver resection. Surgery. 2017;161:332-40
26. Yokota T, Ura T, Shibata N Takahari D, Shitara K, Nomura M. et al. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Brit J Cancer. 2011;104:856-62
Author contact
![]() Corresponding authors: Xiaoli Zhu, Fang-Qi Liu, Ye Xu, Department of Pathology, Fudan University Shanghai Cancer Center (XLZ), Department of Colorectal Surgery, Fudan University Shanghai Cancer Center (FQL and YX). No. 270 Dong-An Road, Shanghai 200032, China. Tel: +86-021-64175590, Fax: +86-021-64035387, E-mail: shhzxl22com (XLZ); liufq021com (FQL); yexuedu.cn (YX).
Corresponding authors: Xiaoli Zhu, Fang-Qi Liu, Ye Xu, Department of Pathology, Fudan University Shanghai Cancer Center (XLZ), Department of Colorectal Surgery, Fudan University Shanghai Cancer Center (FQL and YX). No. 270 Dong-An Road, Shanghai 200032, China. Tel: +86-021-64175590, Fax: +86-021-64035387, E-mail: shhzxl22com (XLZ); liufq021com (FQL); yexuedu.cn (YX).

 Global reach, higher impact
Global reach, higher impact