3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(8):2230-2242. doi:10.7150/jca.52156 This issue Cite
Research Paper
Identification of Prognosis-related Hub RNA Binding Proteins Function through Regulating Metabolic Processes in Tongue Cancer
1. Department of Geriatrics, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, Hefei National Laboratory for Physical Sciences at the Microscale, University of Science and Technology of China, Hefei, 230026, China.
2. College of Liren, Yanshan University, Qinhuangdao, 066004, China.
3. Department of Neurobiology and Biophysics, School of Life Sciences, University of Science and Technology of China, Hefei, 230026, China.
Abstract
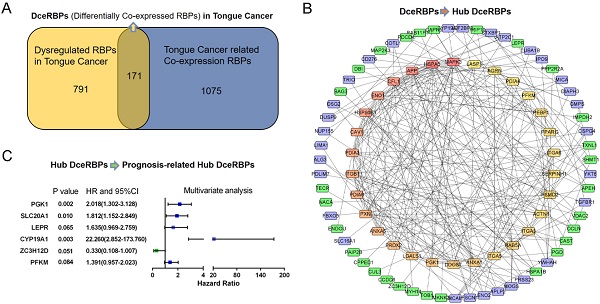
RNA binding proteins (RBPs) are dysregulated and associated with the occurrence and development in various malignant tumors. However, the role of RBPs in tongue cancer are largely unclear. Here, by integrating the differential gene expression analysis and the Weighted Gene Co-expression Network Analysis (WGCNA) of TCGA-retrieved RNA-seq data, we identified a total of 171 differential co-expression RBPs. Then, in a protein-protein interaction (PPI) network containing 134 nodes (RBPs) and 315 network edges (RBP-RBP interacting networks), the top 30 hub RBPs were identified using the CytoHubba plugin of Cytoscape. Furthermore, we investigated the expression and prognostic value of these RBPs and their highly correlated networks. Among them, six RBPs (PGK1, SLC20A1, LEPR, CYP19A1, ZC3H12D, and PFKM) were shown to be the prognosis-related hub RBPs (prhRBPs). Based on these hub RBPs, we constructed a prognostic model and found that the patients in the high-risk group had dramatically poor overall survival compared to those in low-risk group. In addition, we validated the prognostic model in GSE41613, another tongue cancer patient cohort from GEO datasets. The time-dependent receiver operating characteristic (ROC) analysis of the prognostic model further confirmed the predictive capability of the risk model for tongue cancer. As suggested in functional annotation analysis, we found an intensive enrichment of these prhRBPs in metabolic pathways, including AMPK, HIF-1 signaling pathway, Glycolysis, and steroid hormone biosynthesis. Together, our study revealed the underlying role of RBP in tongue cancer biology and potentially unveiled novel targets for cancer therapy.
Keywords: RNA binding protein, tongue cancer, metabolic process, risk score, prognostic model.

 Global reach, higher impact
Global reach, higher impact