Impact Factor
ISSN: 1837-9664
J Cancer 2021; 12(8):2173-2180. doi:10.7150/jca.50299 This issue Cite
Research Paper
Genome-wide Expression Analysis Identifies the Association between SEC14L2 and Castration-resistant Prostate Cancer Survival
1. Department of Urology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China.
2. Department of Urology, Shanghai General Hospital, Shanghai Jiao Tong University, Shanghai, China.
3. Program for Personalized Cancer Care, NorthShore University HealthSystem (Teaching affiliation of University of Chicago), Evanston, IL 60201, USA.
* These authors contributed equally to this study.
Received 2020-7-4; Accepted 2021-2-11; Published 2021-2-22
Abstract
Background: Mechanism of castration-resistant prostate cancer (CRPC) is still unclear. Our objective is to investigate the association between genes expression and CRPC through the genome-wide approach and functional researches.
Methods: Differentially expressed genes (DEGs) between PCa and CRPC tissues were identified using expression profile obtained from Gene Expression Omnibus database (GEO). Survival analysis was performed using online database Gene Expression Profiling Interactive Analysis (GEPIA). Oncomine database was further used to explore the relationship between DEGs expression levels with clinical parameters. After in silico study, SEC14L2-knockdown CRPC cells and normal prostatic epithelial cells were used for in vitro study to verify its biological functions.
Results: A total of 3 consistently changed DEGs (SEC14L2, DMD, SEL1L) were identified correlating with CRPC after cross validation in three independent datasets. Low expression of SEC14L2 was associated with poorer disease-free survival and higher Gleason score than normal/high expression of SEC14L2. SEC14L2 knockdown promoted cell proliferation, migration, invasion as well as cell cycle progression in CRPC cells (all P<0.05) while no significant effects were observed in normal prostatic epithelial cells.
Conclusions: Low expression of SEC14L2 was significantly associated with CRPC, and correlated with PCa aggressiveness and poorer prognosis. SEC14L2 might be a potential biomarker or drug target for CRPC.
Keywords: Expression, Castration-resistant prostate cancer, prognosis.
Introduction
Prostate cancer (PCa) has become one of the most common cancers in the world [1]. To date, androgen-deprivation therapy (ADT) has remained the most essential and first-line treatment for men with advanced PCa since it was introduced in 1970s [2, 3]. Despite its efficacy in treating PCa, most patients ultimately progress to castration-resistant prostate cancer (CRPC) within years. They eventually become insensitive to ADT treatment [4]. As a more lethal stage of the disease, CRPC is critically challenging for clinical treatment due to the lack of effective therapies and poor prognosis [4].
Given that the effectiveness of ADT relies on the critical role of androgen receptor (AR) in the progression of PCa, enormous efforts have been devoted to discover other hub genes. So far, scientists have reported that TMPRSS2:ERG fusion gene and deletion of PTEN are associated with poorer prognosis [5, 6]. SRD5α2, CYP17, BRAC1 and BRAC2 mutations are found to be related with elevated risk for prostate cancer [5]. Splicing variation of AR, overexpression of growth factor receptors and elevated level of YAP, STAT-3 are associated with the development of CRPC [5, 7]. Although numerous biomarkers have been reported, most of them are not actionable targets.
In this study, via bioinformatic methods based on gene expression databases, we aimed to investigate additional CRPC related molecules and to verify their biological functions in CRPC cell line. Our purpose is to find potential biomarkers and actionable targets in CRPC.
Materials and Methods
Microarray Gene Expression Data Analysis
Genome-wide expression microarray data comparing PCa with CRPC were obtained from Gene Expression Omnibus database (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The inclusion criteria includes: (a) accessible microarray data from both PCa and CRPC samples in the dataset; (b) the platform and array used in the study were well accepted, and passed the quality control processes during the experimental period; (c) no treatment effect (either no treatment, or pre-treatment data could be obtained). Finally, expression profiles of CRPC and PCa samples from GSE28403, GSE74367 and GSE101607 were used for further analysis [8-10].
Bioconductor packages in R Software (Version 3.5.0) were used to analyze the array data [11]. Differentially expressed genes (DEGs) analyses were performed after normalization based on different platform. Any DEG with P value <0.05 and absolute log2FC≥1 was obtained from each dataset for cross validation among the datasets.
Validation of DEGs in Clinical Databases
Gene Expression Profiling Interactive Analysis (GEPIA) was used to obtained expression data from The Cancer Genome Atlas (TCGA) and perform survival analysis [12]. Oncomine database was used to verify the expression levels of DEGs in PCa among different Gleason scores [13].
Functional Experiments
Cell line and culture
DU145 (castration-resistant prostate cancer cell), PC3 (castration-resistant prostate cancer cell) and RWPE-1 (normal prostatic epithelial cell) were purchased from the Cell Bank of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured as protocol in RPMI-1640 medium (Gibco, Grand Island, NY, USA) with 10% FBS at 37℃ in the environment with 5% CO2. RWPE-1 cells were grown in Keratinocyte Serum Free Medium (Gibco, Grand Island, NY, USA) at 37℃ in the environment with 5% CO2.
Lentivirus vectors for SEC14L2 small hairpin RNA (shRNA) construction and transfection
Three SEC14L2 shRNA lentivirus vectors and the control vector with green fluorescent protein (GFP) were purchased from GeneChem Co.,Ltd (Shanghai, China). DU145, PC3 and RWPE-1 cells in the logarithmic growth phase were harvested and seeded in 6-well plates. After cells grew at about 20% confluence, lentiviruses were used for transfection according to the manufacturer's protocols. Fluorescence images were taken 72h after the transfection and RT-qPCR was used to validate the interference efficiency.
RT-qPCR
Trizol agent (Pufei, Shanghai, China) was used to extract total RNA from DU145, PC3 and RWPE-1 cells and mRNA reverse transcription was then implemented using M-MLV Reverse Transcriptase kit (Progema, Madison, USA). RNA quality and quantification were assessed spectrophotometrically with a 260/280 ratio of >1.8. Primers for SEC14L2 and GAPDH were synthesized by GeneChem Co.,Ltd (Shanghai, China) and sequences were shown as follow: SEC14L2 sense 5'-CGTCAATGTTGGCTACTCT-3', antisense 5'-ACAATCCTGGGTTCAAATC -3', GAPDH 5'- TGACTTCAACAGCGACACCCA -3', antisense 5'-CACCCTGTTGCTGTAGCCAAA-3'. RT-qPCR using SYBR Master Mixture (TAKARA, Dalian, China) was performed to detect mRNA expression. The reaction system (12μl) contained 6μl SYBR premix ex taq, 0.5μl forward primer, 0.5μl reverse primer, 1μl DNA template and 4μl RNase-Free water. The relative mRNA level of SEC14L2 were standardized to GAPDH based on 2-ΔΔCt method.
CCK-8 assay
DU145, PC3 and RWPE-1 cells were seeded in 96-well plates at a density of about 2500 cells/well and the supernatant was replaced with complete medium containing 10% CCK-8 agent 2h before detection. After incubation at 37℃ for 2h, the absorbance was measured by microplate reader at 450nm. The proliferation rates were tested at 1, 2, 3, 4d after transfection.
Clone formation assay
DU145, PC3 and RWPE-1 cells transfected with si-SEC14L2 or control vector were seeded in 6-well plates at a concentration of 500 cells/well. Clones were fixed with paraformaldehyde and stained with crystal violet after 14 days.
Flowcytometry assay
Cells were harvested when reaching about 80% confluency and washed twice with ice-cold PBS. Cells were then incubated with 1000μl PBS containing 25μl propidium iodide (2mg/ml, Sigma-Aldrich), 10μl RNase A (100μg /ml, Thermo Fisher) and 40μl Triton X-100 (Sigma-Aldrich) for 30 minutes at room temperature and analyzed with BD Accuri C6 plus flow cytometer (BD Biosciences).
Transwell migration and invasion assay
For the migration assays, the 24-well Transwell chamber (Corning, USA) was used and 100μl complete medium containing 1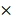 105 cells were added to the upper chambers while 600μl RPMI-1640 medium with 30% FBS was added to the lower chambers. After culture for 24 hours, cells remaining at the inner face of the chamber were removed and those on the outer membrane were fixed with 4% paraformaldehyde, labelled with crystal violet and counted with inverted fluorescence microscope.
105 cells were added to the upper chambers while 600μl RPMI-1640 medium with 30% FBS was added to the lower chambers. After culture for 24 hours, cells remaining at the inner face of the chamber were removed and those on the outer membrane were fixed with 4% paraformaldehyde, labelled with crystal violet and counted with inverted fluorescence microscope.
For the invasion assays, the sample procedure was performed except the inner face of the upper chamber was coated by Matrigel.
Statistical analysis
R software (Version 3.5.0) were used to perform DEGs analyses. Heatmap was used to describe the differential expression level. Cross validation among the datasets were performed and presented using vennDiagram analysis. SPSS 19.0.0 software (IBM SPSS, USA) was used to analyze all the experimental data. In the functional study, two-tail Student's T test was implemented and P-value<0.05 was considered as statistical significance. All experiments were triplicated for validation.
Results
Three GEO datasets (GSE28403, GSE74367, GSE101607) were investigated to explore differential expressed mRNAs between CRPC and PCa samples. A total of 94 CRPC (45 in GSE74367, 40 in GSE101607 and 9 in GSE28403 using Affymetrix Genechip platform) and 23 PCa samples (11 in GSE74367, 8 in GSE101607 and 4 in GSE28403 using Affymetrix Genechip platform) were obtained for further analysis. Details of the datasets were shown in supplementary Table 1.
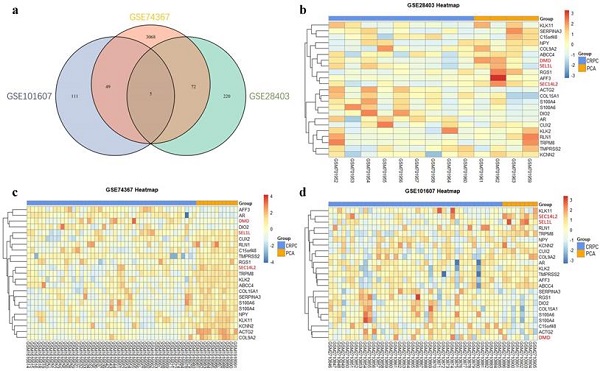
Identification of differentially expressed genes. (a) Venn plot showing 5 common DEGs among GSE28403, GSE74367 and GSE101607. (b, c and d) Volcano plot showing DEGs in CRPC compared with PCa samples. The gradual color ranging from blue to red represents the process from downregulation to upregulation. The consistently changed 3 DEGs (DMD, SEC14L2 and SEL1L) were highlighted in red.
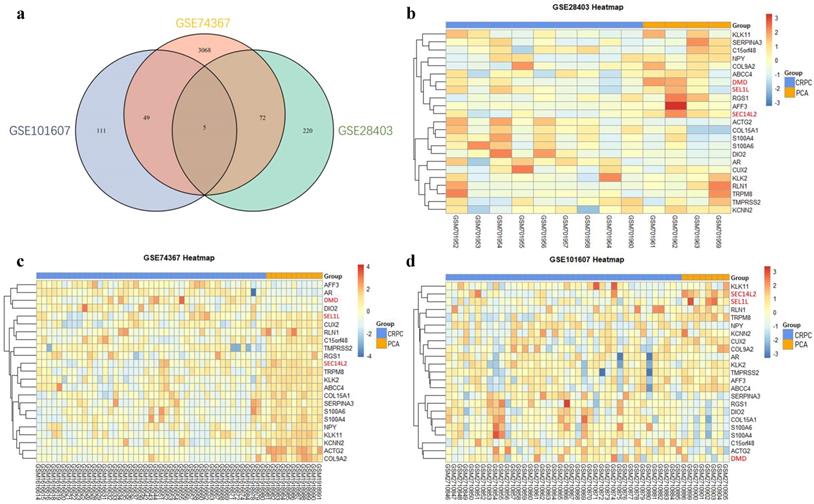
The correlation between DEGs expression and clinical parameters. (a) SEC14L2 downregulation was associated with shorter DFS survival but had no effect on OS. (b and c) Neither DMD nor SEL1L had siginificant effect on PCa survival. (d) The Oncomine dataset demonstrated SEC14L2 expression declined with the increase of Gleason score. SEC14L2 expression in Gleason score 9 samples was siginificantly lower than Gleason score 7. *p<0.05 were considered as statistically significant.
After the normalization, 297 DEGs were identified in GSE28403, 3194 DEGs were identified in GSE74367 and 165 DEGs were identified in GSE101607, using the criteria of P value <0.05 and absolute log2FC ≥1. A total of 5 shared DEGs were further sorted using Venndiagram, of which DMD, SEC14L2 and SEL1L were consistently downregulated (Fig. 1a and Supplementary Table 2). The heatmap plot was used to present DEGs respectively (Fig. 1b, c and d).
To further explore the relationship between co-differentially expressed DEGs with the prognosis of PCa, GEPIA database was used to perform survival analysis. The results showed that high expression of SEC14L2 was associated with longer disease free survival of PCa (HR=0.6, p<0.05, Fig. 2a), while the other two genes had no significant association with disease prognosis (p>0.05, Fig. 2b, c). Additionally, Oncomine database was further applied to reveal the association between SEC14L2 expression and clinicopathological features (Fig. 2d). Consistent with survival analysis, the Gleason score upgraded along with the decline of SEC14L2 expression (Gleason score 7 vs. Gleason score 9, p<0.05) [14].
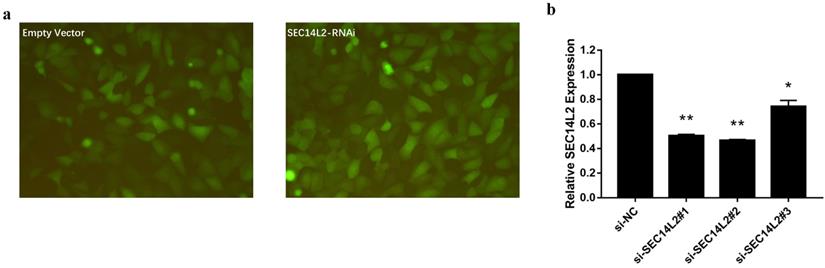
We then knocked down (KD) SEC14L2 by introducing shRNAs targeted against SEC14L2 into DU145 (a CRPC cell line) via lentivirus vector system. GFP was set as a reporter gene. Fluorescence microscope images of the exposed DU145 cells were used to evaluate the transfection efficiency. The results showed that more than 80% cells expressed GFP, indicating a high transfection efficiency of lentiviral system (Fig. 3a). RT-qPCR was further implemented to verify the silencing effect of shRNAs and the results confirmed that SEC14L2 was successfully downregulated (p<0.05). In addition, si-SEC14L2#1 and si-SEC14L2#2 showed similar interference effect but both were more potent than si-SEC14L2#3. Therefore, si-SEC14L2#2 was chosen for further experiments (Fig. 3b). The shRNA was then introduced to PC3 and RWPE-1 cells and decreased expressions of SEC14L2 were verified by RT-qPCR (Fig. S2).
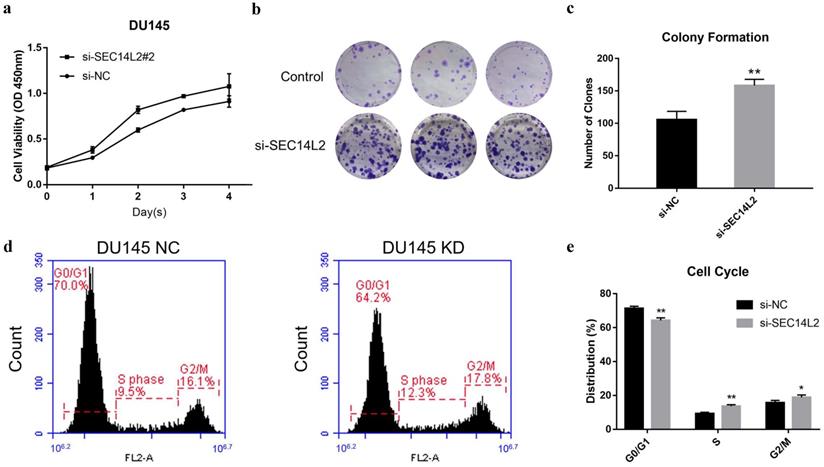
CCK8 assay and clone formation assay was performed to evaluate the impact of SEC14L2 expression on the proliferation capacity of DU145 cells. The results of CCK8 assay revealed that the cell growth rate in SCE14L2 KD group was significantly promoted comparing with control group (p<0.05, Fig. 4a). Similarly, more clones were observed in the SEC14L2 KD group (NC vs KD 105±7 vs 158±5, p<0.05, Fig. 4b, c). Downregulation of SEC14L2 also promoted clonogenic growth of PC3 cells (NC vs KD 121±5 vs 153±6, p<0.05, Fig. S3e, f) but not RWPE-1 cells (NC vs KD 74±2 vs 83±3, p<0.05, Fig. S3b, c). Flow cytometry was then utilized for cell cycle analysis. We observed a larger proportion of SEC14L2 KD cells in the S and G2 phase, as known as a mitotic active stage. Meanwhile, more cells in in the G1 phase were found in the control group (p<0.05, Fig. 4d, e). The similar G1-to-S transition was also identified in PC3 cells (p<0.05, Fig. S4c, d). However, SEC14L2 downregulation had no significant effect on the cell cycle of RWPE-1 cells (Fig. S4a, b). These results suggested that SEC14L2 KD may enhance mitosis in CRPC cells.
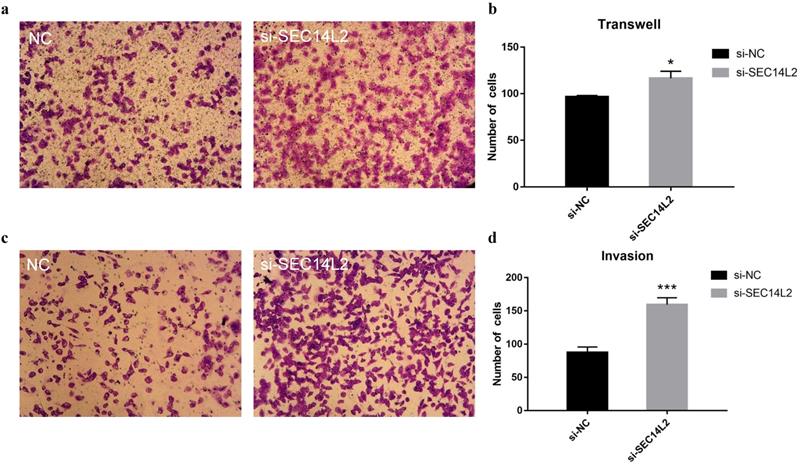
To further investigate the role of SEC14L2 in cancer progression, cell migration assays and cell invasion assays were performed. Compared with control group, more DU145 cells were observed on the lower chamber after 24 hours in the SEC14L2 KD group (NC vs KD 96±1.56 vs 117±7.56, p<0.05), indicating a strengthened motor capacity (Fig. 5a, b). The similar phenomenon was also observed in PC3 cells (NC vs KD 94±2.03 vs 121±5.78, p<0.05, Fig. S5a, b). Afterwards, the upper chambers were covered by Matrigel so as to mimic the basement membrane and the results confirmed the enhanced invasion capacity of SEC14L2 downregulated DU145 cells (NC vs KD 88±7.93 vs 159±10.47, p<0.05, Fig. 5c, d). Likewise, more PC3 cells in the SEC14L2 KD group invaded through Matrigel (NC vs KD 80±3.18 vs 141±6.74, p<0.05, Fig. S5c, d). Above phenomena proved that interference of SEC14L2 expression markedly promote CRPC cells migration and invasion.
Verification of SEC14L2 knockdown in DU145 cells. (a) Fluorescence microscope images of DU145 cells 72h after transfection with or without SEC14L2 RNAi. (b) Relative SEC14L2 mRNA expression level normalized to GAPDH in different shRNA groups or empty vector. * p<0.05, ** p<0.01 were considered as statistically significant.
SEC14L2 knockdown promoted DU145 cell proliferation. (a) CCK8 assays revealed that downregulation of SEC14L2 promoted the growth rate of DU145. (b, c) Downregulation of SEC14L2 increased the number of clones in DU145 cells. (d, e) Downregulation of SEC14L2 fueled the G1-to-S phase transition. *p<0.05, **p<0.01 were considered as statistically significant.
SEC14L2 knockdown promoted DU145 mobility. (a) Transwell migration images of SEC14L2 silenced group and control group (b) SEC14L2 KD enhanced cell mobility (c) Transwell invasion images of SEC14L2 silenced group and control group (d) SEC14L2 KD enhanced cell invasion capacity. *p<0.05, ***p<0.001 were considered as statistically significant.
Discussion
After a period of ADT treatment, disease will always develop to a more malignant and lethal stage, as known as CRPC [4]. With the high throughput sequencing and bioinformatics advancement being widely applied, researchers are able to investigate additional biomarkers or actionable targets via open access databases. In the present study, we identified 3 DEGs (DMD, SEC14L2 and SEL1L) that are associated with CRPC by analyzing three independent series from GEO database. Cross validation via venn diagram, survival analysis and clinicopathological correlation study further confirmed that only SEC14L2 was associated with CRPC aggressiveness and survival. Further functional experiments proved that SEC14L2 knockdown promoted cell proliferation, migration, invasion as well as cell cycle progression in CRPC cells but not in normal prostatic epithelial cells. Taken together, these results implicated the potential tumor suppressive role of SEC14L2 in CRPC.
As one of the six members of SEC14 family, SEC14L2 encodes lipid binding proteins such as α-tocopherol transfer protein, which facilitates the uptake of Vitamin E [15]. Though SEC14L2 expression is relatively low in many human tissues, it is highly expressed in liver, brain, small intestine and prostate [16]. Previous studies demonstrated that low expression of TAP (protein encoded by SEC14L2) was associated with higher Ki-67 expression in prostate cancer tissue. Lower expression of TAP was also found to be related with elevated PSA level, larger tumor size, higher clinical stage and poorer survival [17]. Our study further confirmed these findings. In additional, we found that low expression of SEC14L2 was significantly associated with disease aggressiveness and poorer survival.
Vitamin E, which acts as the substrate of TAP has been reported as a protective factor in prostate cancer, especially in smokers or those in advanced stage [18-22]. Whereas, other studies proclaim that Vitamin E supplement at a dosage of 400 IU/day may not protect males from prostate cancer [23, 24] or even increase the prostate cancer risk [25]. Some researchers believe that limited understanding of metabolic and physiologic mechanism behind Vitamin E shall account for the controversial role of Vitamin E in the prostate cancer [26, 27]. In vitro experiments confirm that SEC14L2 suppresses PCa cells growth by facilitating the uptake of Vitamin E and α-Tocopheryl succinate, an analogue of Vitamin E with proapoptotic effect [28, 29]. Therefore, given the inhibitory effect SEC14L2 posed on CRPC cell lines and the α-tocopherol transfer protein it encoded, we could safely speculate that SEC14L2 coupled with Vitamin E or its analogues might be conducive to the development of next generation therapy. Vitamin E may also be a supplement for localized PCa patients which will protect them from developing CRPC. Further study based on animal model and population level should be conducted for this hypothesis.
Some limitations should be noted in our study. The sample size was relatively small. However, based on the 3 independent studies, we were able to find 3 differentially expressed genes (DMD, SEC14L2 and SEL1L) that were significantly associated with CRPC in each of the dataset. In the further investigation, we would also like to confirmed our findings via in vivo experiments and population-based studies.
Conclusion
Low expression of SEC14L2 was significantly associated with CRPC, and correlated with PCa aggressiveness and poorer prognosis. SEC14L2 might be a potential biomarker or drug target for CRPC.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was in supported by grants from National Natural Science Foundation of China (Grant No. 81772741 and No. 81972645), Shanghai Rising-Star Program (Grant No. 18QA1402800), the “Chen Guang” project from Shanghai Municipal Education Commission and Shanghai Education Development Foundation (Grant No. 17CG09), Shanghai Jiao Tong University School of Medicine Gaofeng-Clinical Medicine Grant Support (Grant No. 20181701), Shanghai Municipal Human Resources and Social Security Bureau (Grant No. 2018052), and Shanghai Jiao Tong University SMC-Chenxing Scholar Project to Rong Na. All the funders had no role in study design, data collection, data analysis, interpretation, and writing of the report.
Authors' contributions
RN conceived and designed the study. SL, JH, JY, TC and DX contributed materials and collected the data. SL, DH, NZ, GJ, YG and RN analyzed the data. SL, DH, and RN wrote the manuscript. SL and DH contributed equally to this study. All authors have read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M. et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol. 2017;71:618-29
2. Cornford P, Bellmunt J, Bolla M, Briers E, De Santis M, Gross T. et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part II: Treatment of Relapsing, Metastatic, and Castration-Resistant Prostate Cancer. Eur Urol. 2017;71:630-42
3. Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22:232-40
4. Loriot Y, Supiot S, Beauval J-B, Schlürmann F, Pasticier G, Sargos P. et al. Management of non-metastatic castrate-resistant prostate cancer: A systematic review. Cancer Treat Rev. 2018;70:223-31
5. Gandhi J, Afridi A, Vatsia S, Joshi G, Joshi G, Kaplan SA. et al. The molecular biology of prostate cancer: current understanding and clinical implications. Prostate cancer and prostatic diseases. 2018;21:22-36
6. Boström PJ, Bjartell AS, Catto JWF, Eggener SE, Lilja H, Loeb S. et al. Genomic Predictors of Outcome in Prostate Cancer. Eur Urol. 2015;68:1033-44
7. Salem O, Hansen CG. The Hippo Pathway in Prostate Cancer. Cells. 2019 8
8. Roudier MP, Winters BR, Coleman I, Lam H-M, Zhang X, Coleman R. et al. Characterizing the molecular features of ERG-positive tumors in primary and castration resistant prostate cancer. Prostate. 2016;76:810-22
9. Vainio P, Wolf M, Edgren H, He T, Kohonen P, Mpindi J-P. et al. Integrative genomic, transcriptomic, and RNAi analysis indicates a potential oncogenic role for FAM110B in castration-resistant prostate cancer. Prostate. 2012;72:789-802
10. Ylitalo EB, Thysell E, Jernberg E, Lundholm M, Crnalic S, Egevad L. et al. Subgroups of Castration-resistant Prostate Cancer Bone Metastases Defined Through an Inverse Relationship Between Androgen Receptor Activity and Immune Response. Eur Urol. 2017;71:776-87
11. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40
12. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017 45
13. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D. et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1-6
14. Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L. et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790-9
15. Wright ME, Peters U, Gunter MJ, Moore SC, Lawson KA, Yeager M. et al. Association of variants in two vitamin e transport genes with circulating vitamin e concentrations and prostate cancer risk. Cancer Res. 2009;69:1429-38
16. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397-406
17. Wen XQ, Li XJ, Su ZL, Liu Y, Zhou XF, Cai YB. et al. Reduced expression of alpha-tocopherol-associated protein is associated with tumor cell proliferation and the increased risk of prostate cancer recurrence. Asian journal of andrology. 2007;9:206-12
18. Bidoli E, Talamini R, Zucchetto A, Bosetti C, Negri E, Lenardon O. et al. Dietary vitamins E and C and prostate cancer risk. Acta Oncol. 2009;48:890-4
19. Heinonen OP, Albanes D, Virtamo J, Taylor PR, Huttunen JK, Hartman AM. et al. Prostate cancer and supplementation with alpha-tocopherol and beta-carotene: incidence and mortality in a controlled trial. J Natl Cancer Inst. 1998;90:440-6
20. Helzlsouer KJ, Huang HY, Alberg AJ, Hoffman S, Burke A, Norkus EP. et al. Association between alpha-tocopherol, gamma-tocopherol, selenium, and subsequent prostate cancer. Journal of the National Cancer Institute. 2000;92:2018-23
21. Chan JM, Stampfer MJ, Ma J, Rimm EB, Willett WC, Giovannucci EL. Supplemental vitamin E intake and prostate cancer risk in a large cohort of men in the United States. Cancer Epidemiol Biomarkers Prev. 1999;8:893-9
22. Kirsh VA, Hayes RB, Mayne ST, Chatterjee N, Subar AF, Dixon LB. et al. Supplemental and dietary vitamin E, beta-carotene, and vitamin C intakes and prostate cancer risk. J Natl Cancer Inst. 2006;98:245-54
23. Wang L, Sesso HD, Glynn RJ, Christen WG, Bubes V, Manson JE. et al. Vitamin E and C supplementation and risk of cancer in men: posttrial follow-up in the Physicians' Health Study II randomized trial. The American journal of clinical nutrition. 2014;100:915-23
24. Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG. et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). Jama. 2009;301:39-51
25. Klein EA, Thompson IM Jr, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ. et al. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). Jama. 2011;306:1549-56
26. Ledesma MC, Jung-Hynes B, Schmit TL, Kumar R, Mukhtar H, Ahmad N. Selenium and vitamin E for prostate cancer: post-SELECT (Selenium and Vitamin E Cancer Prevention Trial) status. Molecular medicine (Cambridge, Mass). 2011;17:134-43
27. Chan JM, Darke AK, Penney KL, Tangen CM, Goodman PJ, Lee G-SM. et al. Selenium- or Vitamin E-Related Gene Variants, Interaction with Supplementation, and Risk of High-Grade Prostate Cancer in SELECT. Cancer Epidemiol Biomarkers Prev. 2016;25:1050-8
28. Ni J, Pang S-T, Yeh S. Differential retention of alpha-vitamin E is correlated with its transporter gene expression and growth inhibition efficacy in prostate cancer cells. Prostate. 2007;67:463-71
29. Ni J, Wen X, Yao J, Chang H-C, Yin Y, Zhang M. et al. Tocopherol-associated protein suppresses prostate cancer cell growth by inhibition of the phosphoinositide 3-kinase pathway. Cancer Res. 2005;65:9807-16
Author contact
![]() Corresponding author: Rong Na, M.D., Ph.D., Department of Urology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Address: 197 Ruijin 2nd Road, Shanghai, China 200025. Email: narong.hscom
Corresponding author: Rong Na, M.D., Ph.D., Department of Urology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. Address: 197 Ruijin 2nd Road, Shanghai, China 200025. Email: narong.hscom

 Global reach, higher impact
Global reach, higher impact