Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(20):5941-5952. doi:10.7150/jca.46703 This issue Cite
Research Paper
Elevated double-strand break repair protein RAD50 predicts poor prognosis in hepatitis B virus-related hepatocellular carcinoma: A study based on Chinese high-risk cohorts
1. Department of Neurosurgery, Affiliated Hospital of Youjiang Medical University for Nationalities, Guangxi, 533000, China.
2. Clinical College of Youjiang Medical University for Nationalities, Baise, Guangxi, 533000, China.
3. Department of Urology, Fudan University Shanghai Cancer Center, Shanghai 200032, China.
4. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 20032, China.
5. Department of Gastrointestinal Surgery, Nantong University Affiliated Hospital, Nantong, Jiangsu, 226001, China.
6. Gastroenterology Department, Second affiliated Hospital of Nantong University, Nantong, Jiangsu, 226001, China.
7. Department of Pathogenic Biology, Medical College, Nantong University, Nantong, Jiangsu, 226001, China.
*These authors contributed equally to this work. (Missing from manuscript?)
Received 2020-4-6; Accepted 2020-7-17; Published 2020-8-14
Abstract
Objective: Increasing evidence indicates that RAD50, which is involved in the repair process of DNA double-strand break (DSB), is also involved in cancer outcomes. However, its role in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) remains unclear. This study was designed to investigate the expression of RAD50 and its prognostic value in HBV-related HCC patients.
Methods: 107 and 100 patients with HBV-related HCC from the Affiliated Hospital of Youjiang Medical University of Nationalities (AHYMUN) and the Affiliated Hospital of Nantong University (AHNU), respectively, were enrolled in the study. The distribution of the categorical clinical-pathological data and the levels of RAD50 expression were compared with a χ2 test. Immunohistochemistry (IHC) staining of RAD50 was performed. A partial likelihood test based on univariate and multivariate Cox regression analysis was developed to address the influence of independent factors on disease-free survival (DFS) and overall survival (OS). The Oncomine online database was used to analyse and validate the differential expression of RAD50. The Kaplan-Meier method and a log-rank test were performed to assess the influence of RAD50 on survival at different levels.
Results: RAD50 was highly expressed in HCC tissues compared to normal tissues and was significantly correlated with OS in the Cancer Genome Atlas (TCGA) cohort. The validation analysis indicated that significantly increased levels of RAD50 were expressed in HCC tissues in the two independent cohorts. In addition, HCC patients with elevated RAD50 expression levels showed poor OS and DFS in the AHYMUN cohort and decreased OS and DFS in the AHNTU cohort.
Conclusion: In conclusion, our study reveals that elevated RAD50 expression is significantly correlated with cancer progression and poor survival in HBV-related HCC patients. These data suggest that RAD50 may act as an oncogene and may serve as a promising target for the therapy of HBV-related HCC patients.
Keywords: RAD50, hepatocellular carcinoma, DNA repair, multiple cohorts, prognosis
Introduction
In China, the incidence and mortality of primary hepatocellular carcinoma (HCC) is the second highest of all malignant cancers [1]. In addition, in China, because the incidence of hepatitis B virus (HBV) infection is high, HBV accounts for at least 80% of HCC cases. In the Guangxi Zhuang Autonomous Region (Guangxi province) [2] and Qidong in Nantong Prefecture (Jiangsu province) [3], HBV infection is very common, leading to a high incidence of HCC [4, 5]. Although multiple markers, such as alpha fetoprotein (AFP), have been widely used in clinical practice, the accuracy and sensitivity of diagnosis and targeted therapy need to be improved. Therefore, to reduce the cost of treatment for patients with HBV-related HCC and to improve their quality of life, it is important to understand the underlying molecular mechanisms and apply them to the development of targeted treatments.
The DNA repair component protein Rad50 (RAD50) is a component of the MRE11-RAD50-NBS1 (MRN) complex, which is a heterotrimer composed of meiotic recombination 11 (MRE11), RAD50, and Nijmegen fracture syndrome 1 (NBS1), participating in the DNA damage response (DDR). Recent studies have found that DDR may be related to the occurrence and development of cancers. Defective mitosis-linked DNA damage response and chromosomal instability facilitate the progression of liver cancer [6]. In addition, the mutation of RAD50 has an adverse effect on the prognosis of prostate cancer patients [7]. Increased RAD50 expression has been found in multiple tumours, including breast cancer, ovarian cancer, lung cancer and rectal cancer [8-11]. For example, RAD50 increases in colorectal cancer patients have been shown to be positively related to tumour development and prognosis [12]. It is speculated that the high expression of RAD50 leads to cell instability, which increases the tendency toward DNA damage accumulation and malignant transformation. Following HBV infection, RAD50 expression in hepatocytes has been shown to be increased [13].
To investigate the expression and prognostic value of RAD50 in HBV-related HCC, we recruited 207 HBV-related HCC patients from two centres. The prognostic role of RAD50 in HCC patients was validated and functionally annotated in silico. We speculate that RAD50 is associated with poor prognosis and may serve as a potential therapeutic target for HBV-related HCC.
Materials and Methods
Patients and Variables
In this study, a total of 107 HCC patients were recruited from the Affiliated Hospital of Youjiang Medical University of Nationalities (AHYMUM) (Youjiang, Guangxi, China) between April 2008 and October 2018, and 100 HCC patients were also recruited from the Affiliated Hospital of Nantong University (AHNTU) (Nantong, Jiangsu, China) for analyses between July 2013 and May 2017. The inclusion criteria consisted of the following: (1) HCC diagnosed by pathology (surgery or biopsy), typical dynamic imaging studies, and AFP serology according to the American Association for the Study of Liver Diseases (AASLD) guidelines [17]; (2) chronic hepatitis B virus infection [hepatitis B surface antigen (+)]; (3) no previous oncological treatment or liver resection; (4) no tumour invasion in the branches or trunk of the portal vein or hepatic veins according to MRI; (5) no other diffuse liver disease, including primary sclerosing cholangitis or primary biliary cirrhosis. The clinical and pathological parameters in the two cohorts, including age at surgery, gender, tumour size, American Joint Committee on Cancer (AJCC) stage, primary tumour lesion, the degree of pathological nuclear differential expression, Okuda score, albumin level, bilirubin level, presence of liver cirrhosis, and the degrees of microvascular invasion and capsular invasion are summarized in Table 1. Tissue samples, including HCC and adjacent normal tissues, were collected during surgery and were available from the AHYMUM and AHNTU tissue banks. All study designs and test procedures were performed in accordance with the Helsinki Declaration II. Ethical approval and consent to participate in the current study were obtained from the ethics committee.
Immunohistochemistry (IHC)
Immunostaining of RAD50 was performed using a mouse monoclonal anti-RAD50 antibody (1:1000, #Ab89, Abcam, USA). Positive or negative staining of a specific protein on one formalin-fixed paraffin-embedded (FFPE) slide was independently assessed by two experienced pathologists and determined according to the following descriptions. The staining intensity level was graded from 0 to 3. Samples with no staining or weak, median or strong staining were graded at the level of 0, 1, 2 or 3, respectively. The staining extent ranged from 0 to 4 based on the percentage of coverage of the immunoreactive tumour cells (0%, 1-25%, 26-50%, 51-75%, or 76-100%, respectively). The overall IHC score, which was graded on a scale from 0 to 12, was generated by multiplying the staining intensity and extent scores. Negative staining was indicated by a grade from 0 to 3 and positive staining was indicated by a grade from 4 to 12 for each sample. All samples were classified into the tumour or normal group to confirm the differential expression of RAD50.
Statistical analysis
To determine the association of different RAD50 mRNA expression levels with clinicopathological characteristics, a χ2-test was performed to compare the distribution of the categorical data between groups. A scatter plot was utilized to represent the differential expression of RAD50 in normal and HCC tissues. The primary end point was overall survival (OS) for patients who survived for a specific period of time, which was determined according to the length of time from the date of surgery to the date of death or date of last follow-up. Disease-free survival (DFS), as the secondary end point, was defined as the length of time from the initiation of curative treatment when no disease could be detected until the date of progression, the initiation of second-line treatment or the date of death, which ever occurred first. The follow-up duration was estimated using the Kaplan-Meier method with a 95% confidence interval (CI) and a log-rank test with separate curves. The hazard ratios were derived from Cox proportional hazard regression models based on a high-versus-low comparison to identify the independent predictors. Univariate and multivariate Cox regression models were independently analysed to evaluate the influence of the confounding covariates, including age at surgery, gender, tumour size, AJCC stage, primary tumour site, the degree of pathological nuclear differential expression, Okuda score, albumin level, bilirubin level, presence of liver cirrhosis, presence of microvascular invasion, presence of capsular invasion, and RAD50 expression, on survival. Statistical analyses were performed with SPSS software (version 23.0, SPSS Inc., Chicago, IL, USA). All hypothetical tests were two-sided, and P-values less than 0.05 were considered significant for all tests.
Determination of differential RAD50 expression based on Oncomine datasets
RAD50 expression profiles in hepatocellular tumour and normal tissue were analysed and displayed using the Oncomine online database (http://www.oncomine.com). A dataset including the known gene expression patterns in human HCC were included to validate the mRNA expression of RAD50 in the two groups. An unpaired t test was used to analyse the significant differences.
Data processing of Gene Set Enrichment Analysis (GSEA)
The Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) (version 10.0) online database was used to predict the protein-protein interaction (PPI) network for RAD50. Any interaction with a combined score > 0.4 was considered statistically significant. The significant genes were included in functional and pathway enrichment analyses in a bubble chart using DAVID bioinformatics resources, which indicated the most significant signalling pathways according to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. (GO) enrichment analysis, including biological processes (BP), cellular components (CC), and molecular functions (MF). Datasets from TCGA database were analysed with the GSEA method using the category version 2.10.1 package in R. For each separate analysis, the Student's t-test statistical score was obtained for each of the consistent pathways, and the mean differential expression of each gene was calculated. A permutation test was performed 1000 times to identify the pathways with significant changes. The adjusted P values (adj. P), which were obtained by default using the Benjamini and Hochberg (BH) false discovery rate (FDR) method, were utilized to correct for the occurrence of false positive results. Significantly related genes were defined by an adj. P less than 0.01 and an FDR less than 0.25.
In this study, the staining intensity level was graded from 0 to 3 (0-25%, 26-50%, 51-75%, or 76-100%). Samples with no staining or weak, median or strong staining were graded at the level of 0, 1, 2 or 3, respectively. The staining extent ranged from 0 to 4 based on the percentage of coverage of the immunoreactive tumour cells (0%, 1-25%, 26-50%, 51-75%, or 76-100%, respectively). The overall IHC score, which was graded on a scale from 0 to 12, was generated by multiplying the staining intensity and extent scores. Negative staining was indicated by a grade from 0 to 3 and positive staining was indicated by a grade from 4 to 12 for each sample. The different intensity of RAD50 expression ranging from 0-25%, 26-50%, 51-75%, and 76-100% was shown in Fig. S1.
Results
In this study, the research was conducted in three steps. In the first step, the differential expression of RAD50 in normal and liver tissues was analysed; in the second step, RAD50 expression in HCC patients was assessed as an indicator of progression and prognosis; in the third step, the differential expression of RAD50 was validated in the Oncomine datasets, and GSEA was used to identify the relationships between significant genes and the involved signalling pathways.
Prognostic role of RAD50 in HCC based on TCGA
As shown in Table 1, in the AHYMUM cohort, increased RAD50 expression was significantly correlated with decreased age (p = 0.021), advanced AJCC stage (p = 0.001), the presence of multiple primary tumour lesions (p = 0.001), the presence of micro vascular invasion (p = 0.028), the presence of capsular invasion (p = 0.015), higher HBV DNA level (p = 0.004), increased Okuda score (p = 0.009) and increased Child-Pugh Stage (p = 0.025). In the AHNTU cohort, increased RAD50 expression was significantly associated with larger tumour size (p < 0.001), advanced AJCC stage (p = 0.003), the presence of multiple primary tumour lesions (p = 0.010), the degree of pathological nuclear differential expression (p = 0.005), the presence of microvascular invasion (p = 0.001), the presence of capsular invasion (p = 0.003), and higher Okuda score (p = 0.008). A chi-square test showed that the baseline data were balanced in terms of the distribution of the categorical data, including gender, albumin level, bilirubin level and the presence of liver cirrhosis (p > 0.05).
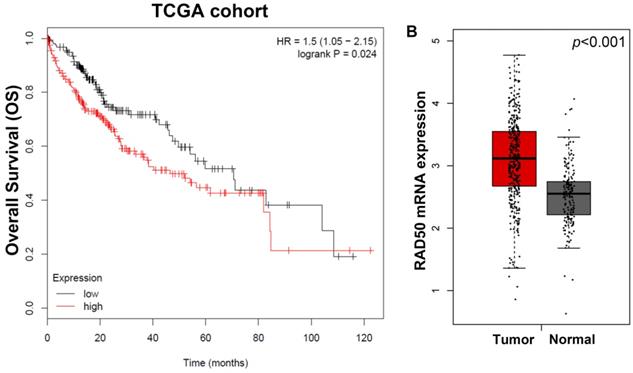
As shown in Fig. 1A, the Kaplan-Meier survival analyses of different RAD50 expression groups based on OS in 207 patients with HCC from the TCGA database, patients with high RAD50 expression have significantly lower OS than patients with low expression. Moreover, in the TCGA cohorts, the expression of RAD50 in tumour tissues is significantly higher than that in normal tissues (Fig. 1B).
External validation of differential expressed RAD50 in multiply cohorts
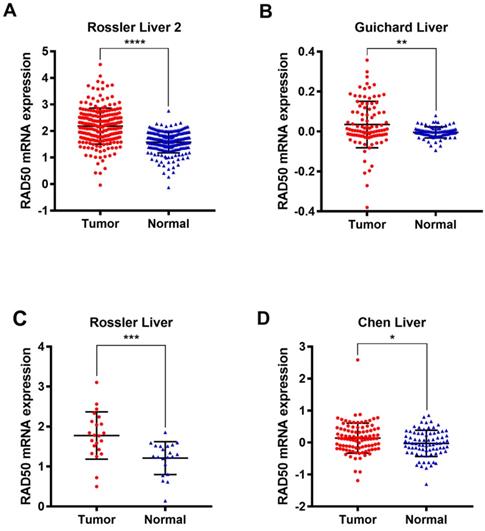
Four datasets (Rossler Liver 2 [14], Guichard Liver [15], Rossler Liver [14], and Chen Liver [16]) from the Oncomine database were analysed to validate the differential expression of RAD50 in HCC tumour and normal tissues (Fig. 2).
Clinicopathological characteristics in relation to RAD50 expression status in two cohorts were shown
| Characteristics N (%) | AHYMUM cohort (N=107) | RAD50 expression | χ2 | P | AHNTU cohort (N=100) | RAD50 expression | χ2 | P | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Low IHC score (N=53) | High IHC score (N=54) | Low IHC score (N=52) | High IHC score (N=48) | |||||||
| Age | 5.287 | 0.021 | 0.038 | 0.844 | ||||||
| <60 years | 85 (79.4) | 40 (75.5) | 45 (83.3) | 72 (72.0) | 37 (71.2) | 35 (72.9) | ||||
| ≥60 years | 22 (20.6) | 13 (24.5) | 9 (16.7) | 28 (28.0) | 15 (28.2) | 13 (27.1) | ||||
| Gender | 0.698 | 0.403 | 1.512 | 0.219 | ||||||
| Male | 90 (84.1) | 43 (81.1) | 47 (87.0) | 82 (82.0) | 45 (86.5) | 37 (77.1) | ||||
| Female | 17 (15.9) | 10 (18.9) | 7 (13.0) | 18 (18.0) | 7 (13.5) | 11 (22.9) | ||||
| Tumor size | 3.447 | 0.063 | 13.117 | <0.001 | ||||||
| <5 cm | 59 (55.1) | 34 (64.2) | 25 (46.3) | 48 (48.0) | 34 (65.4) | 14 (29.2) | ||||
| ≥5 cm | 48 (44.9) | 19 (35.8) | 29 (53.7) | 52 (52.0) | 18 (34.6) | 34 (70.8) | ||||
| AJCC stage | 10.363 | 0.001 | 9.324 | 0.003 | ||||||
| I - II | 94 (87.9) | 52 (98.1) | 42 (77.8) | 66 (66.0) | 42 (80.8) | 14 (29.2) | ||||
| III - IV | 13 (12.1) | 1 (1.9) | 12 (22.2) | 34 (34.0) | 10 (19.2) | 24 (70.8) | ||||
| Primary lesion | 10.194 | 0.001 | 6.732 | 0.010 | ||||||
| Single | 83 (77.8) | 48 (90.6) | 35 (64.8) | 79 (79.0) | 46 (88.5) | 33 (68.8) | ||||
| ≥2 | 24 (22.4) | 5 (9.4) | 19 (35.2) | 21 (21.0) | 6 (11.5) | 15 (31.2) | ||||
| Albumin level | 0.291 | 0.590 | 0.496 | 0.503 | ||||||
| <35 g/L | 37 (34.6) | 17 (32.1) | 20 (37.0) | 51 (51.0) | 23 (44.2) | 28 (58.3) | ||||
| ≥35 g/L | 70 (65.4) | 36 (67.9) | 34 (63.0) | 49 (49.0) | 29 (55.8) | 20 (41.7) | ||||
| Bilirubin level | 0.641 | 0.423 | 1.803 | 0.179 | ||||||
| <20.4 umol/L | 77 (72.0) | 40 (75.5) | 37 (68.5) | 65 (65.0) | 37 (71.2) | 28 (58.3) | ||||
| ≥20.4 umol /L | 30 (28.0) | 13 (24.5) | 17 (31.5) | 35 (35.0) | 15 (28.8) | 20 (41.7) | ||||
| Liver cirrhosis | 2.905 | 0.088 | 0.550 | 0.458 | ||||||
| Absent | 6 (5.6) | 5 (9.4) | 1 (1.9) | 6 (6.0) | 4 (7.7) | 2 (4.2) | ||||
| Present | 101 (94.4) | 48 (90.6) | 53 (98.1) | 94 (94.0) | 48 (92.3) | 46 (95.8) | ||||
| Differential degree | 0.849 | 0.357 | 7.771 | 0.005 | ||||||
| Low | 30 (28.0) | 17 (32.1) | 13 (24.1) | 62 (62.0) | 39 (75.0) | 23 (47.9) | ||||
| High | 77 (72.0) | 36 (67.9) | 41 (75.9) | 38 (38.0) | 13 (25.0) | 25 (52.1) | ||||
| Microvascular invasion | 4.821 | 0.028 | 10.359 | 0.001 | ||||||
| Absent | 96 (89.7) | 51 (96.2) | 45 (83.3) | 62 (62.0) | 40 (88.9) | 22 (45.8) | ||||
| Present | 11 (10.3) | 2 (3.8) | 9 (16.7) | 38 (38.0) | 12 (11.1) | 26 (54.2) | ||||
| Capsular invasion | 5.922 | 0.015 | 9.324 | 0.003 | ||||||
| Absent | 87 (81.3) | 48 (90.6) | 39 (72.2) | 67 (67.0) | 42 (80.8) | 25 (52.1) | ||||
| Present | 20 (18.7) | 5 (9.4) | 15 (27.8) | 33 (33.0) | 10 (19.2) | 23 (47.9) | ||||
| Okuda score | 6.805 | 0.009 | 7.015 | 0.008 | ||||||
| I | 51 (47.7) | 32 (60.4) | 19 (35.2) | 69 (69.0) | 42 (80.8) | 27 (56.3) | ||||
| II | 56 (52.3) | 21 (39.6) | 35 (64.8) | 31 (31.0) | 10 (19.2) | 21 (43.8) | ||||
A. Kaplan-Meier survival analyses of different RAD50 expression groups based on OS in 207 patients with hepatocellular carcinoma from the TCGA database; B. Differential transcriptomic expression of RAD50 in tumour and normal tissues in the TCGA cohorts.
Analysis of expression in hepatocellular tumour vs. normal tissues based on data from the Oncomine database. A. Roessler, et al., A unique metastasis gene signature enables prediction of tumour relapse in early-stage hepatocellular carcinoma patients, Cancer Res, 2010. B. Guichard C, et al., Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma, Nature Genetics, 2012. C. Roessler, et al., A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients, Cancer Res, 2010. D. Chen X, et al., Gene expression patterns in human liver cancers, MolBiol Cell, 2002.
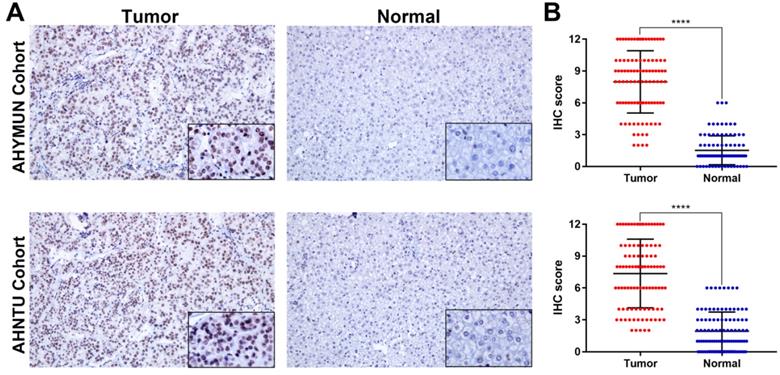
To analyse the RAD50 expression, IHC was performed to reveal the staining distribution in tumour and normal tissues (Fig. 3A). The scatter plot of the IHC scores revealed that RAD50 expression was significantly elevated in HCC tissues in the AHYMUM (p < 0.001) and AHNTU (p < 0.001) cohorts (Fig. 3B).
IHC staining and scatter plot analysis of hepatocellular carcinoma and normal tissues of patients in the AHYMUN and AHNTU cohorts. A. IHC staining of hepatocellular carcinoma and normal tissues from patients in the AHYMUN and AHNTU cohorts; B. Scatter plot showing the IHC scores derived from normal and tumour tissues (p < 0.001) from different cohorts.
Multivariate Cox regression analysis of DFS in AHYMUM and AHNTU cohorts (DFS: disease-free survival) was shown
| Covariates | AHYMUM | AHNTU | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Tumor size (ref. <5cm) | - | - | - | 0.839 | 0.412-1.707 | 0.627 |
| AJCC stage (ref. II - IV) | 0.660 | 0.267-1.631 | 0.269 | 1.796 | 0.912-3.538 | 0.090 |
| Primary lesion (ref. Single) | 1.156 | 0.706-2.349 | 0.155 | 2.563 | 1.286-5.110 | 0.007 |
| Differential degree (ref. Low) | 2.052 | 0.964-4.371 | 0.062 | 1.404 | 0.633-3.110 | 0.404 |
| Microvascular invasion (ref. Absent) | 1.020 | 0.374-2.780 | 0.969 | 1.317 | 0.626-2.769 | 0.468 |
| Capsular invasion (ref. Absent) | 1.281 | 0.672-2.441 | 1.281 | 0.899 | 0.418-1.933 | 0.784 |
| Okuda score (ref. I) | 1.914 | 1.014-3.611 | 0.045 | 3.084 | 1.632-5.828 | 0.001 |
| RAD50 expression (ref. low) | 4.612 | 2.336-9.106 | <0.001 | 2.660 | 1.331-5.317 | 0.006 |
Cox regression analyses
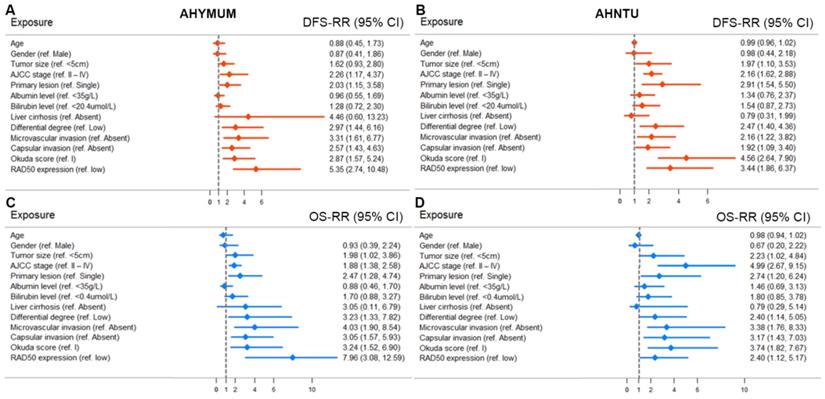
The univariate Cox regression analysis, as shown in the forest plots, revealed that age, gender, albumin level, bilirubin level and liver cirrhosis were not significantly associated with DFS in the patients form both AHYMUN (Fig. 4A) and AHNTU (Fig. 4B) cohorts. Meanwhile, tumour size was not an independent predictor of DFS in the AHYMUN cohort. In the multivariate models, the Okuda score (ref. I) was significantly associated with DFS in HCC patients, indicating the good representativeness of the population in the AHYMUN (HR: 1.914, p = 0.045) and AHNTU (HR: 3.084, p = 0.001) cohorts. More importantly, the subgroup analysis of RAD50 expression showed that RAD50 amplification was significantly correlated with DFS in the AHYMUN (HR: 4.612, p < 0.001) and AHNTU (HR: 2.660, p = 0.006) cohorts. Additionally, in the AHNTU cohort, a number of primary lesions was significantly correlated with poor DFS (HR: 2.563; p = 0.007) according to the multivariate model used for the Cox regression analyses (Table 2).
In Fig. 4C and 4D, several parameters, including age, gender, albumin level, bilirubin level and the presence of liver cirrhosis, were not significantly associated with OS in the two cohorts. As shown in Table 3, the multivariate Cox analysis indicated that the Okuda score (ref. I) was associated with OS in both the AHYMUN (HR: 2.329, p = 0.031) and AHNTU (HR: 2.819, p = 0.030) cohorts. RAD50 expression (ref. low) was associated with OS in AHYMUM patients (HR: 6.807; p < 0.001) but was not markedly correlated with OS in HCC patients from the AHNTU (HR: 1.735; p = 0.195) cohort. The other factors, including tumour size (ref. < 5 cm), the presence of multiple primary lesions (ref. single), the degree of pathological nuclear differential expression (ref. low), the presence of microvascular invasion (ref. absent) and the presence of capsular invasion (ref. absent), were not identified as prognostic indicators of OS (p > 0.05).
Forest plots were used to visualize the univariate Cox regression analysis of DFS and OS in the AHYMUM and AHNTU cohorts.
Multivariate Cox regression analysis of OS in AHYMUM and AHNTU cohorts (OS: overall survival) was shown
| Covariates | AHYMUM | AHNTU | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Tumor size (ref. <5cm) | 1.199 | 0.463-3.104 | 0.708 | 0.905 | 0.358-2.283 | 0.832 |
| AJCC stage (ref. II - IV) | 0.763 | 0.256-2.277 | 0.628 | 2.826 | 1.150-6.947 | 0.024 |
| Primary lesion (ref. Single) | 1.151 | 0.514-0.576 | 0.733 | 2.151 | 0.898-5.154 | 0.086 |
| Differential degree (ref. Low) | 2.019 | 0.792-5.147 | 0.141 | 0.925 | 0.377-2.269 | 0.865 |
| Microvascular invasion (ref. Absent) | 1.488 | 0.719-3.081 | 0.284 | 2.023 | 0.783-5.228 | 0.146 |
| Capsular invasion (ref. Absent) | 0.984 | 0.326-2.973 | 0.978 | 1.363 | 0.518-3.585 | 0.530 |
| Okuda score (ref. I) | 2.329 | 1.079-5.027 | 0.031 | 2.819 | 1.105-7.194 | 0.030 |
| RAD50 expression (ref. low) | 6.807 | 2.598-17.839 | <0.001 | 1.735 | 0.753-3.996 | 0.195 |
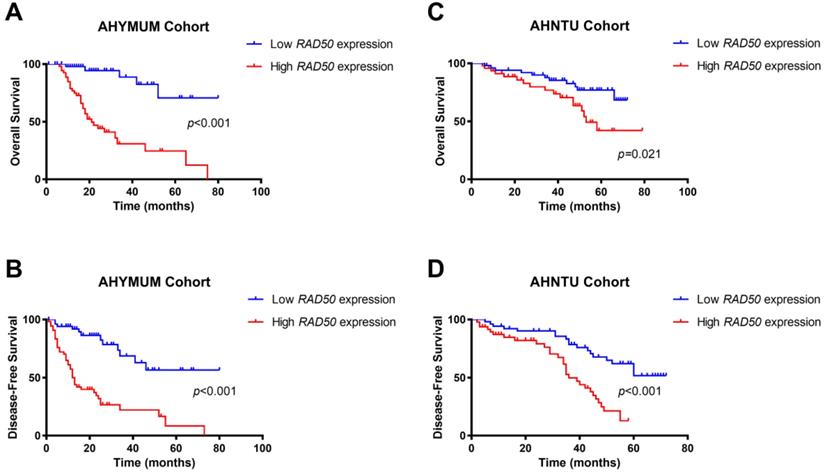
The survival curves suggested that HCC patients in the AHYMUN cohort with elevated RAD50 expression levels showed poor OS (p < 0.001) and poor DFS (p < 0.001) (Fig. 5A and 5B). In addition, in the AHNTU cohort, increased RAD50 expression was significantly associated with decreased OS (p = 0.021) and DFS (p < 0.001) (Fig. 5C and 5D).
Expression validation and GSEA
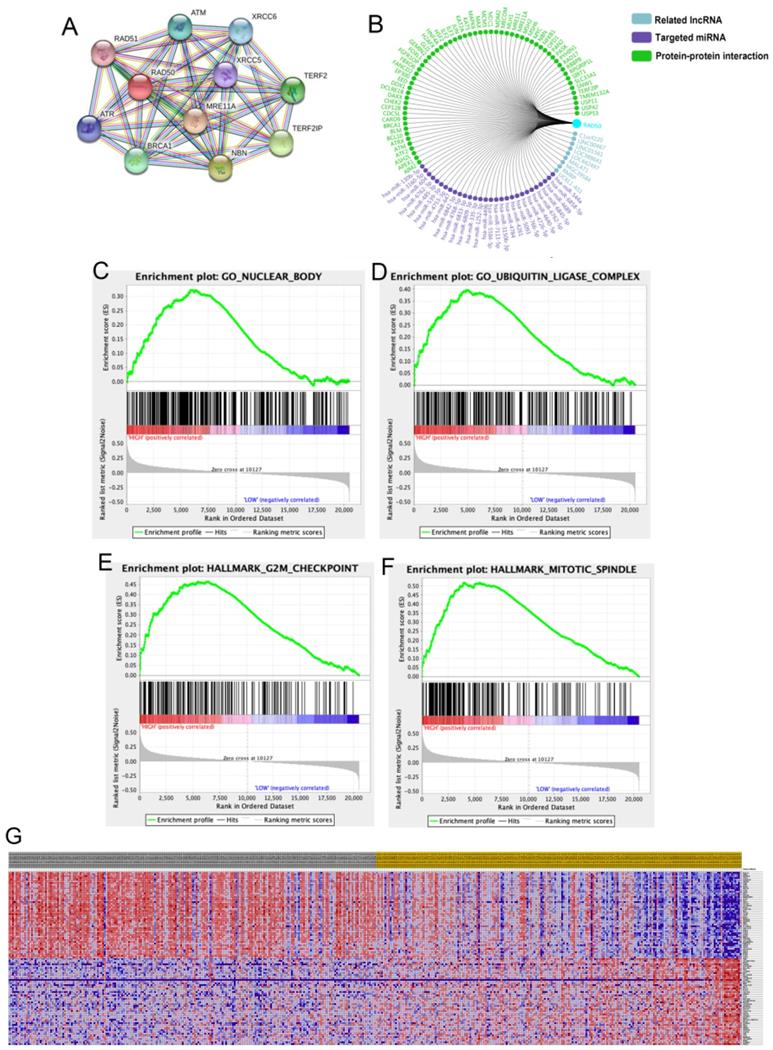
A total of 11 significant genes, including RAD51, ATM, XRCC6, RAD50, XRCC5, TERF2, ATR, MRE11A, BRAC1, NBN and TERF2IP, were included in the molecular model shown in Fig. 6A. The significant and potentially related Inc RNAs, targeted miRNAs and protein-protein interaction nodes are shown in Fig. 6B. Significant differences were found among the four validation datasets (p < 0.05). A total of 100 significant genes were obtained from GSEA, and the genes with positive correlations were plotted. In addition, RAD50 was found to be involved in pathways from most functional annotation categories, including the nuclear body, ubiquitin ligase complex, G2M checkpoint and mitotic spindle signalling pathways. The details are shown in Fig. 6.
Functional enrichment analysis
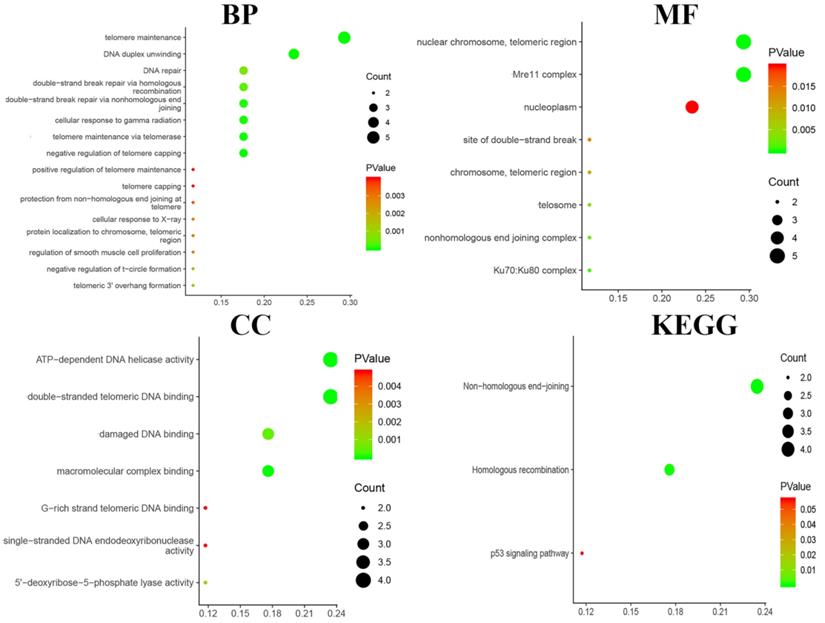
Functional and pathway enrichment analyses, including analyses of biological processes, cellular components, molecular functions and KEGG pathways, were performed using DAVID and are represented in the bubble chart in Fig. 7. The results of GO analyses showed that changes in BPs were significantly enriched in telomere maintenance, DNA duplex unwinding and DNA repair; changes in MFs were mostly enriched in nuclear chromosome and telomeric region; changes in CCs were mostly enriched in ATP-dependent DNA helicase activity and damaged DNA binding. Changes in KEGG pathway enrichment analyses were existed in non-homologous end-joining and homologous recombination.
Kaplan-Meier survival analyses of different RAD50 expression groups based on OS and DFS in 107 patients with HBV-related HCC from the AHYMUM cohort (A & B) and in 100 patients from the AHNTU cohort (C & D).
Discussion
Herein, we demonstrated that RAD50 was positively correlated with poor prognosis in HCC patients in the TCGA cohort. In the two validated cohorts, increased RAD50 expression was observed in cancer tissues compared to that in para-cancer tissues, which was shown be associated with decreased OS and DFS. Four validation cohorts from the Oncomine database also showed elevated RAD50 expression in HCC tissues compared with that in adjacent normal tissues. Additionally, the functional annotation bioinformatics analysis identified significant related hub genes and signalling pathways.
According to the Int Act molecular interaction database, human RAD50 potentially interacts with the transcription factor c-Jun/activator protein 1 (AP-1) (website: https://www.ebi.ac.uk/intact/interactors/id:Q92878*). C-Jun induces HBV-related liver tumour genes in mice via upregulating the transcription of its target gene osteo pontin (OPN) [17]. OPN is overexpressed in several human carcinomas and contributes to inflammation, tumour progression, and metastasis [18, 19]. Therefore, we speculated that the interaction between RAD50 and c-Jun promoted the transcription activity of c-Jun, upregulated OPN expression, facilitated the development of HBV-related HCC, and contributed to poor prognosis.
Our study suggested that increased RAD50 expression in HBV-related HCC is a marker of poor prognosis. The researchers have designed a chemical forward genetic screen to identify a small-molecule inhibitor of the MRN complex, named Z-5-(4-hydroxybenzylidene)-2-imino-1, 3-thiazolidin-4-one (mirin) [20]. In 2009, other researchers have confirmed the structure of mirin [21]. However, there have been no reports about the applications of mirin in cellular, animal and clinical levels. We questioned whether mirin could be used in an HBV-related HCC animal model to determine the therapeutic effect via the inhibition of the MRN complex.
The predictive power of RAD50 may be as strong as that of the classical factors commonly used in clinical practice. Serum tumour markers are the most commonly used markers for the monitoring and early diagnosis of HBV-related HCC because they are non-invasive, objective, and reproducible. The most common serological marker is AFP. Disappointingly, in a retrospective case-control study, the sensitivity of AFP test for the most effective cut-off value (10-20 ng/ml) in the diagnosis of HCC was approximately 60%, and its specificity was 80% [22, 23]. These values are much lower than those required for the treatment and prediction of prognosis of HCC patients. However, RAD50 was shown to be a significant and independent biomarker [24]. The data indicate that RAD50 may be used to shed further light on the mechanisms underlying HCC progression, to facilitate the discovery of new therapies, and to open up new avenues for the personalized treatment of HCC patients.
RAD50 related protein analysis. A-B. A total of 11 significant genes, including RAD51, ATM, XRCC6, RAD50, XRCC5, TERF2, ATR, MRE11A, BRAC1, NBN and TERF2IP, were identified in the molecular model. C-G. Datasets from the TCGA database were analysed with the GSEA method.
Functional and pathway enrichment analyses were performed using DAVID to obtain a bubble chart. Biological processes, cellular components, molecular functions and the KEGG analysis of the DEGs indicated the most significant signalling pathways.
In the study, the analysis of the data from the two cohorts supported our hypothesis and clearly demonstrated the high expression of RAD50 in tumour tissues from HCC patients, which results in increases in the HCC recurrence rate and poor overall survival. Furthermore, GSEA indicated that RAD50 is involved in some of the most important pathways, including the mitotic spindle, ultraviolet (UV) response and transforming growth factor beta (TGF-β) signalling pathways that are enriched in HCC specimens. In addition to participating in the DSB repair pathway, RAD50 also interacts with WD repeat and coiled coil containing (MMAP), which is expressed by the mitotic-specific MRN complex to maintain optimal genomic stability during mitosis [25]. The question whether RAD50 contributes to HBV-related HCC progression via interacting with MMAP needs further investigation.
The relationship between RAD50 and HCC has rarely been investigated [26]. However, it is worth noting that RAD50 is a repair factor that has been identified in the DSB reaction and is highly expressed in many cancers. With this in mind, to further explain the role of RAD50 in the invasion and metastasis of HCC, we used GSEA to analyse data from public databases to identify important genes and pathways that may be involved in the relevance of RAD50 to the initiation of carcinogenesis.
The limitations of this study are described as follows. First, the retrospective nature of the data set is obvious. Although data was obtained from AHYMUN and AHNTU cohorts, the sample size is relatively small, and the patients in the two Chinese cohorts exhibit economic and geographical differences, which may lead differences in therapeutic strategies and overall prognoses. Second, while our research involved a thorough functional annotation of RAD50 in HBV-related HCC, we failed to confirm the underlying mechanisms. In the future, we will conduct multi-centre research in Europe and the United States to further explore the role of RAD50 in the development and prognosis of HCC. RAD50 shows as an indicator of poor prognosis in HBV-related HCC, suggesting a potential target for the therapy of HBV-related HCC.
Conclusion
Our study reveals that elevated RAD50 expression is significantly correlated with cancer progression and poor survival in HBV-related HCC patients. These data suggest that RAD50, which is a potential therapeutic target, may act as an oncoprotein and could serve as a promising prognostic marker in HCC patients. In this regard, randomized clinical trials and further investigation are required to identify the true value of RAD50 in terms of its clinical application in HCC patients.
Abbreviations
AHNTU: Affiliated Hospital of Nantong University; AHYMUM: Affiliated Hospital of YouJiang Medical University for Nationalities; BP: biological process; CC: cellular component; CI: confidence interval; DD: DNA damage response; DFS: disease-free survival; GO: gene ontology; HBV: hepatitis B virus; HCC: Hepatocellular carcinoma; HR: hazard ratio; IHC: immunohistochemistry; KEGG: Kyoto Encyclopedia of Genes and Genomes; MF: molecular function; OS: overall survival; TCGA: the Cancer Genome Atlas.
Supplementary Material
Supplementary figure S1.
Acknowledgements
We especially appreciate the Affiliated Hospital of Youjiang Medical University of Nationalities and the Affiliated Hospital of Nantong University for the clinical sample collection.
Authors' contributions
The work presented here was carried out with the collaboration of all authors. DW Ye and HL Zhang defined the research topic and discussed the analyses and their interpretation, and presentation. HN Huang and XJ Liu developed the algorithm, provided pathological samples, provided clinicopathological data and performed the statistical analysis. WH Xu, WR Liu and YY Chen drafted the manuscript, recorded the clinical data, analysed the IHC data and interpreted the results. LG Gu and LQ He collected clinical and pathological records and helped to analyse the IHC data. HK Wang and YY Qu participated in reviewing all clinical records and performed the associated data collection. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The ethics approval and consent to participate in the current study were obtained from the ethics committees of AHYMUM and AHNTU.
Availability of data and materials
The datasets and/or analysis results used during the current study are available from the corresponding author upon reasonable request.
Funding
This work is supported by grants from the National Natural Science Foundation of China (No. 81202004, 81802525, 81472377) and the Shanghai Natural Science Foundation of China (No. 16ZR1406400). This study was partly supported by the major project (No. MS22018009) and the general project (No. JC2018087) of Nantong city.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Chen JG, Zhang SW. Liver cancer epidemic in China: past, present and future. Seminars in cancer biology. 2011;21:59-69
2. Zheng D, Deng W, Huang T, Li X, Li Z. [Relationship between hepatitis B virus genotype, BCP/Pre-C region mutations and risk of hepatocellular carcinoma in Guangxi Zhuang Autonomous Region]. Zhonghua Liu Xing Bing Xue Za Zhi. 2015;36:725-9
3. Chen T, Qian G, Fan C, Sun Y, Wang J, Lu P. et al. Qidong hepatitis B virus infection cohort: a 25-year prospective study in high risk area of primary liver cancer. Hepatoma Res. 2018 4
4. Bei C, Tan C, Zhu X, Wang Z, Tan S. Association Between Polymorphisms in CMTM Family Genes and Hepatocellular Carcinoma in Guangxi of China. DNA Cell Biol. 2018;37:691-6
5. Fan C, Li M, Gan Y, Chen T, Sun Y, Lu J. et al. A simple AGED score for risk classification of primary liver cancer: development and validation with long-term prospective HBsAg-positive cohorts in Qidong, China. Gut. 2019;68:948-9
6. Tahmasebi-Birgani M, Ansari H, Carloni V. Defective mitosis-linked DNA damage response and chromosomal instability in liver cancer. Biochim Biophys Acta Rev Cancer. 2019;1872:60-5
7. Xu WH, Wang J, Sheng HY, Qu YY, Wang HK, Zhu Y. et al. Prognostic implication and functional annotations of Rad50 expression in patients with prostate cancer. J Cell Biochem. 2020;121:3124-34
8. Tommiska J, Seal S, Renwick A, Barfoot R, Baskcomb L, Jayatilake H. et al. Evaluation of RAD50 in familial breast cancer predisposition. Int J Cancer. 2006;118:2911-6
9. Zhang M, Liu G, Xue F, Edwards R, Sood AK, Zhang W. et al. Copy number deletion of RAD50 as predictive marker of BRCAness and PARP inhibitor response in BRCA wild type ovarian cancer. Gynecol Oncol. 2016;141:57-64
10. Adams DL, Adams DK, He J, Kalhor N, Zhang M, Xu T. et al. Sequential Tracking of PD-L1 Expression and RAD50 Induction in Circulating Tumor and Stromal Cells of Lung Cancer Patients Undergoing Radiotherapy. Clin Cancer Res. 2017;23:5948-58
11. Ho V, Chung L, Singh A, Lea V, Abubakar A, Lim SH. et al. Overexpression of the MRE11-RAD50-NBS1 (MRN) complex in rectal cancer correlates with poor response to neoadjuvant radiotherapy and prognosis. BMC Cancer. 2018;18:869
12. Gao J, Zhang H, Arbman G, Sun XF. RAD50/MRE11/NBS1 proteins in relation to tumour development and prognosis in patients with microsatellite stable colorectal cancer. Histol Histopathol. 2008;23:1495-502
13. Ryu HM, Park SG, Yea SS, Jang WH, Yang YI, Jung G. Gene expression analysis of primary normal human hepatocytes infected with human hepatitis B virus. World J Gastroenterol. 2006;12:4986-95
14. Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer research. 2010;70:10202-12
15. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB. et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nature genetics. 2012;44:694-8
16. Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J. et al. Gene expression patterns in human liver cancers. Molecular biology of the cell. 2002;13:1929-39
17. Trierweiler C, Hockenjos B, Zatloukal K, Thimme R, Blum HE, Wagner EF. et al. The transcription factor c-JUN/AP-1 promotes HBV-related liver tumorigenesis in mice. Cell Death Differ. 2016;23:576-82
18. Weber CE, Kothari AN, Wai PY, Li NY, Driver J, Zapf MA. et al. Osteopontin mediates an MZF1-TGF-beta1-dependent transformation of mesenchymal stem cells into cancer-associated fibroblasts in breast cancer. Oncogene. 2015;34:4821-33
19. Qin X, Yan M, Wang X, Xu Q, Wang X, Zhu X. et al. Cancer-associated Fibroblast-derived IL-6 Promotes Head and Neck Cancer Progression via the Osteopontin-NF-kappa B Signaling Pathway. Theranostics. 2018;8:921-40
20. Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML. et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119-25
21. Garner KM, Pletnev AA, Eastman A. Corrected structure of mirin, a small-molecule inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2009;5:129-30 author reply 30
22. Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR. et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110-8
23. Lok AS, Sterling RK, Everhart JE, Wright EC, Hoefs JC, Di Bisceglie AM. et al. Des-gamma-carboxy prothrombin and alpha-fetoprotein as biomarkers for the early detection of hepatocellular carcinoma. Gastroenterology. 2010;138:493-502
24. Chen C, Wang Y, Mei JF, Li SS, Xu HX, Xiong HP. et al. Targeting RAD50 increases sensitivity to radiotherapy in colorectal cancer cells. Neoplasma. 2018;65:75-80
25. Xu R, Xu Y, Huo W, Lv Z, Yuan J, Ning S. et al. Mitosis-specific MRN complex promotes a mitotic signaling cascade to regulate spindle dynamics and chromosome segregation. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E10079-e88
26. Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071-8
Author contact
![]() Corresponding authors: Hailiang Zhang, Department of Urology, Fudan University Shanghai Cancer Center, China. E-mail: zhanghl918com; Tel.: 86-21-64175590-2805; Xiaojuan Liu, Department of Pathogenic Biology, Medical College, Nantong University, Nantong, Jiangsu, China. E-mail: lxjedu.cn; Tel.: 86-13773672280; Haineng Huang, Department of Neurosurgery, Affiliated Hospital of Youjiang Medical University for Nationalities, Guangxi, 533000, China; Clinical College of Youjiang Medical University for Nationalities, Baise, Guangxi, 533000, China. E-mail: bshuanghncom; Tel.: 86-0776-2824233.
Corresponding authors: Hailiang Zhang, Department of Urology, Fudan University Shanghai Cancer Center, China. E-mail: zhanghl918com; Tel.: 86-21-64175590-2805; Xiaojuan Liu, Department of Pathogenic Biology, Medical College, Nantong University, Nantong, Jiangsu, China. E-mail: lxjedu.cn; Tel.: 86-13773672280; Haineng Huang, Department of Neurosurgery, Affiliated Hospital of Youjiang Medical University for Nationalities, Guangxi, 533000, China; Clinical College of Youjiang Medical University for Nationalities, Baise, Guangxi, 533000, China. E-mail: bshuanghncom; Tel.: 86-0776-2824233.

 Global reach, higher impact
Global reach, higher impact