Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(18):5293-5308. doi:10.7150/jca.42816 This issue Cite
Review
The new horizon of liquid biopsy in sarcoma: the potential utility of circulating tumor nucleic acids
1. Department of bone and soft tissue tumor, Tianjin Medical University Cancer Institute and Hospital, Tianjin, 300060, China
2. National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin's Medical University Cancer Institute and Hospital, Tianjin, 300060, China.
3. Department of Orthopedics, Affiliated Hospital of Chengde Medical College, Chengde, Hebei, 067000, China
Received 2019-12-7; Accepted 2020-6-11; Published 2020-7-9
Abstract
The diagnosis, treatment and prognosis of sarcoma are mainly dependent on tissue biopsy, which is limited in its ability to provide a panoramic view into the dynamics of tumor progression. In addition, effective biomarkers to monitor the progression and therapeutic response of sarcoma are lacking. Liquid biopsy, a recent technological breakthrough, has gained great attention in the last few decades. Nucleic acids (such as DNA, mRNAs, microRNAs, and long non-coding RNAs) that are released from tumors circulate in the blood of cancer patients and can be evaluated through liquid biopsy. Circulating tumor nucleic acids reflect the intertumoral and intratumoral heterogeneity, and thus liquid biopsy provides a noninvasive strategy to examine these molecules compared with traditional tissue biopsy. Over the past decade, a great deal of information on the potential utilization of circulating tumor nucleic acids in sarcoma screening, prognosis and therapy efficacy monitoring has emerged. Several specific gene mutations in sarcoma can be detected in peripheral blood samples from patients and can be found in circulating tumor DNA to monitor sarcoma. In addition, circulating tumor non-coding RNA may also be a promising biomarker in sarcoma. In this review, we discuss the clinical application of circulating tumor nucleic acids as blood-borne biomarkers in sarcoma.
Keywords: sarcoma, liquid biopsy, circulating tumor nucleic acids, bloodborne biomarkers
Introduction
Sarcomas are a heterogeneous group of malignant solid tumors that originate from mesenchymal tissues and encompass more than 70 histological subtypes [1, 2]. Sarcomas account for nearly 1% of all adult malignancies and approximately 15%-21% of paediatric malignant tumors [2, 3]. Although the 5-year survival rate of malignant bone tumors, such as osteosarcoma (OS), Ewing sarcoma (ES) and chondrosarcoma (CS), has increased to 50%-70% with the advent of effective chemotherapeutic agents, the survival rate has reached a plateau over the past decades [1]. The same trend is observed in soft tissue sarcomas [1, 4]. The mortality of bone and soft tissue sarcomas in the USA in 2019 is estimated to be as high as 41% and 47%, respectively [3]. Based on the data of the “Surveillance of rare cancer in Europe” (RARECARE) project, the five-year relative survival was 58% for soft tissue sarcomas (STSs) and 62% for bony sarcomas in European regions between 2000 and 2002 [5]. However, there is a paucity of overall sarcoma data from Asia. The National Central Cancer Registry of China estimated that there were 28,000 new bone sarcoma diagnoses and 20,700 deaths (73.9%) from bone sarcoma in China in 2015 [6]. Approximately 39,900 new STS cases occurred nationwide in China in 2014, but the data on mortality were lacking [7]. By analysing data from the Bone and Soft Tissue Tumor (BSTT) registry of Japan from 2006 to 2013, the cumulative disease-specific survival values of bone sarcoma at 2 and 5 years were 80.3% and 70.3%, respectively [8]. Existing evidence has shown that racial and ethnic disparities do exist with respect to occurrence and mortality in sarcoma. Studies have revealed that sarcomas are more prevalent in Caucasian descent (58.1%) than Hispanic (21.4%) or African Americans (13.0%), and Hispanic or African descent Americans had significantly poorer overall survival at the 5-years follow-up point than Caucasian descent [9-11]. The poor prognosis of sarcoma is partly attributed to a lack of effective biomarkers for early diagnosis, screening, prognosis and tumor monitoring.
At present, the diagnosis, treatment and monitoring of tumors mainly depend on tissue biopsy and imaging examination. Tissue biopsy currently remains the gold standard for diagnosis and therapy [12, 13]. However, tissue biopsy is associated with several limitations (Table 1). For example, tissue biopsy is invasive and may result in some complications [14, 15]. Furthermore, tissue biopsy only provides a snapshot of the genetic alterations in a single part of the tumor and does not reflect the dynamic changes of the tumor [16, 17]. Tumor progression and response to chemotherapy or surgery also rely on radiological imaging investigation, but the imaging methods have limited sensitivity and specificity [17]. Distinguishing tissues with residual tumor from tissues affected by inflammatory changes after surgery can be difficult in some cases [18]. In addition, the potential side-effects from radiological examination is a health hazard that need to be considered. Therefore, identifying a more effective and non-invasive method that can be used to monitor tumor dynamics is urgently needed.
Liquid biopsy has gained great attention in the last few decades as an alternative to monitor malignant neoplasms and was listed as one of the top ten technological breakthroughs in 2015 [19]. Compared with tissue biopsy, liquid biopsy has several advantages, including a non-invasive procedure, real-time nature and the potential to provide a panoramic molecular portrait of the tumor (Table 1). Although liquid biopsy mainly refers to the monitoring of circulating tumor nucleic acids (ctNAs), it also includes the detection of circulating tumor cells (CTCs) and tumor-derived exosomes [20-22] (Figure 1). A comparison of the three approaches to liquid biopsy is listed in Table 2.
Comparison of tissue biopsy and liquid biopsy in sarcoma
| Advantages | Limitations | |
|---|---|---|
| Tissue biopsy | 1. The gold standard of diagnosis, classification and staging of sarcoma [12, 13] 2. Guides treatment in sarcoma [12, 13] 3. Well-developed technical process and widely used in the clinic [12, 13] | 1. Invasive procedure [14, 15] 2. Difficult to detect intratumoral and intertumoral heterogeneity with a single sample from a single site [16, 17] 3. Possible clinical complications associated with tissue biopsy [14, 15] 4. Difficult to sample certain parts of the body [12, 13] 5. Difficult to sample continuously during the follow-up period [12, 13] |
| Liquid biopsy | 1. Non-invasive procedure [19, 22] 2. Easily repeated sampling during the follow-up period [19, 22] 3. Body fluid samples can reflect a panoramic view of the tumor in real time, including the primary and metastatic sites [19, 22] 4. Dynamic evaluation of therapeutic effects [19, 22] 5. Formation of personalized therapy based on tumor heterogeneity [19, 22] 6. Easy sampling and not limited by the anatomic location of the tumor [19, 22] | 1. Difficult to find sensitive and specific biomarkers due to the high heterogeneity of sarcoma [13, 16, 195] 2. Significance of biomarkers and clinicopathological features in sarcomas have not been fully established [16, 195] 3. Differences between samples and sampling processes lead to poor consistency in results between different studies [16, 195] |
Comparison of three types of liquid biopsy molecules (CTCs, ctNAs and exosomes)
| Type | Source | Enrichmental or analytical methods | Strength | Limitation |
|---|---|---|---|---|
| CTCs | Peripheral blood | Physical property-based methods: tumor cell size [29, 197]; biological property-based methods: mesenchymal cell markers, specific chromosomal translocations, specific gene mutation [24, 197] | Non-invasive [29, 197]; DNAs, RNAs and proteins can be analyzed and can be used as diagnostic or prognostic biomarkers [29, 197]; can be used to evaluate the response to therapy and for early detection of residual and recurrent lesions [29, 197] | Impact of heterogeneity on various CTC enrichment methods [24]; low concentration of CTCs [24]; absence of specific markers expressed by sarcoma tumor cells and highly heterogeneous phenotype [24] |
| ctDNA and ctRNA | Serum or plasma | PCR(e.g., conventional RT-qPCR; dPCR; ddPCR); NGS; RNA-seq [16, 58] | Non-invasive [16, 58]; Can be used to analyze the tumor molecular heterogeneity; for precision therapeutic decision making based on specific gene mutation; for monitoring tumor burden and prognosis [16, 58]; for early detection of emerging chemotherapy resistance [16, 58] | Sample type and preparation protocols are different in different studies [16, 195]; the relation between ctNAs and clinicopathological features is unclear [16, 195]; sequencing data require professional bioinformatics analysis [16] |
| Exosomes | Serum, plasma or other body fluids | Ultracentrifugation; density gradient centrifugation; membrane filtration; size exclusion chromatography; microfludic technologies [16, 58] | Non-invasive; abundant in multiple body fluids; exosomes contain DNAs, RNAs and proteins that have diagnostic and prognostic value [16, 58] | The purity of tumor-derived exosomes is different for different isolation methods; identification of the exosomes is difficult due to the lack of specific markers [16, 58] |
Abbreviations: CTCs: circulating tumor cells; ctNAs: circulating tumor nucleic acids, including circulating tumor DNA(ctDNA), specific gene transcripts, non-coding RNA; NGS: next generation sequence; RT-PCR: real time polymerase chain reaction; dPCR: digital polymerase chain reaction; ddPCR: droplet digital polymerase chain reaction.
The biological processes and detection methods of circulating nucleic acids and a schematic of liquid biopsy in sarcoma. Liquid biopsy includes the isolation of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA) and circulating tumor RNA (ctRNA) and exosomes from the blood. Clinical application of liquid biopsy in sarcoma involve diagnosis in the early stage, monitoring of sarcoma progression, (e.g., local recurrence and distant metastasis), prediction of prognosis and detection of response to therapy. The biological processes of circulating nucleic acids are shown in the magnified view in step 5, and ctDNA, ctRNAs, and exosomes can be discharged into the circulation by apoptotic or necrotic CTCs. Some of the circulating nucleic acids are degraded by the liver and kidney, and the remaining circulating nucleic acids can be detected and amplified by PCR and NGS, which can be used to monitor sarcomas. Bloodborne CTCs, ctDNA, ctRNAs and exosomes can be analysed to diagnose sarcoma, predict prognosis and evaluate response to therapy. This process is called “liquid biopsy”. Based on these steps, we can monitor the tumor. Abbreviations: CTCs: circulating tumor cells; ctDNA: circulating tumor DNA; ctRNA: circulating RNA; RBC: red blood cell
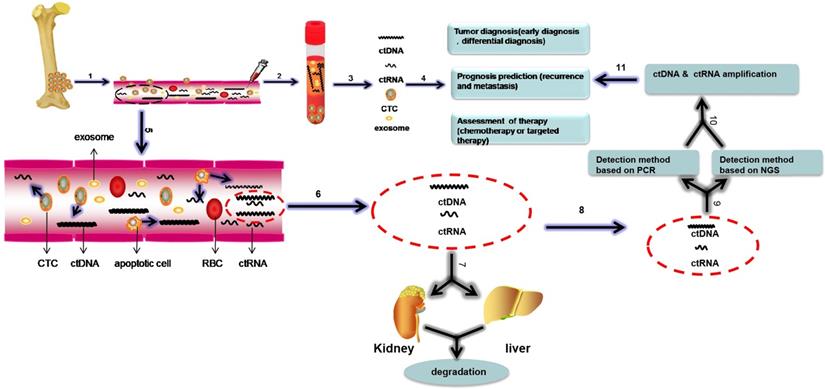
Because of the rarity of sarcoma and its high heterogeneity, the literature on CTCs and tumor-derived exosomes in sarcoma is limited [23, 24]. The production of ctNAs is a more frequent event in the course of cancer progression. Studies have estimated that a tumor that weighs 100g is equivalent to 3×1010 tumor cells, and 3.3% of tumor DNA will be released into blood circulation per day [25]. ctNAs include circulating tumor DNA (ctDNA) as well as circulating tumor RNAs (ctRNAs), such as mRNAs, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). The dynamic change of these molecules can reflect the pathological process of tumor progression and indicate the presence of a benign or malignant lesion [26]. ctNAs can also reflect the profile of the tumor genotype in real-time and with a non-invasive approach compared with tissue biopsy which reflects a single point in time of a single site of the tumor [19].
In this review, we will discuss recent advances in the identification of ctNAs in sarcoma and their potential clinical applications. We will also review the current limitations of ctNAs and the prospects for translation into clinical practice.
Biology of ctNAs
Current studies have suggested that cell-free DNA (cfDNA) that circulates in the blood is derived from primary tumors, CTCs, distant micrometastatic lesions, normal haemocytes, and normal stromal cells [27]. Existing evidence has shown that a primary tumor releases 106 CTCs per gram of tumor every day, the majority of which die, as only approximately 0.01% of CTCs survive [28, 29]. ctDNA is the DNA released from tumor cells in cancer patients [21]. It is released into the bloodstream during the necrotic and apoptotic processes of tumor cells and metabolized in the liver and kidney (Figure 1). It comprises 0.1% to 90% of cfDNA [30, 31]. The half-life of ctDNA varies in different types of tumors and different individuals, ranging from 15 minutes to several hours [21, 30]. In addition to ctDNA, other types of ctNAs, including circulating tumor transcripts and non-coding RNAs such as miRNAs and lncRNAs, are also discharged from dying tumor cells into the circulation [26, 32] (Figure 1). Increasing evidence has indicated that circulating tumor transcripts and non-coding RNAs are highly stable [33, 34]. They are protected from degradation by their packaging into exosomes, microvesicles or other vesicle types [33-36] or in an EV-independent fashion, and they are detected in complexes with the protein Argonaute 2 (AGO2) [37, 38] or high-density lipoproteins (HDLs) [39] in the extracellular space and body fluids. Therefore, circulating transcripts and non-coding RNAs are stable and can survive in the bloodstream for a long time. However, the half-life of circulating tumor RNAs is not established [40]. Although ctNAs reflect the tumor dynamics and tumor burden, the enrichment and detection of such low concentrations of ctNAs in circulation remain challenging.
Detection Methods for ctNAs
Currently, the two most common methods used to detect ctDNA include polymerase chain reaction (PCR)-based methods and next generation sequencing (NGS)-based methods (Figure 1). Digital PCR (dPCR) is the latest technology and can quantify nucleic acid molecules in tumor samples. This technique allows the quantification of a number of DNA molecules at once. Droplet-digital PCR (ddPCR) can be applied to distinguish and quantify alterations with allele frequencies as low as 0.01% or less in cfDNA. Although short amplicons, less than 100 bp, can be amplified from formalin-fixed and paraffin-embedded (FFPE) samples by PCR analyses and fragmentation of DNA and crosslinking do not markedly affect the final results, it is obvious that fresh tumor sample are better [41-43]. In addition, PCR is restricted by the requirement of prior knowledge of specific gene mutations or copy number variations to be analysed [22].
NGS can be used to detect tumor-specific gene mutations at frequencies as low as 0.05% in the whole genome or exome [44, 45]. NGS can independently capture the complete panel of mutated genes in a cancer and is not based on prior knowledge of target genes. A hybrid approach incorporating droplet-digital PCR and NGS in liquid biopsy has been proposed to upgrade the sensitivity and specificity of the detection method [46].
Quantitative reverse-transcription PCR (RT-qPCR) is a fast, sensitive and low-throughput method, while microarray and deep sequencing are high-throughput assays that can be used to detect the whole transcriptome and non-coding RNAs in various types of samples including body fluids [47-49]. Therefore, circulating tumor RNAs, including mRNAs, miRNAs, and lncRNAs, can be detected by RT-qPCR, microarray analysis and RNA-deep sequencing technology [49].
Clinical applications of different ctNAs in sarcomas
The analysis of ctNAs allows for the detection of genetic and epigenetic variations in tumors over time. For sarcomas, genetic variations including mutational changes, translocations and fusions, have been identified from ctDNA in liquid biopsy [50, 51]. Correspondingly, epigenetics variations in sarcomas mainly encompass three types: DNA methylation, histone modification and non-coding RNA modifications [52, 53]. To date, only circulating non-coding RNAs , mainly microRNAs, have been studied in sarcoma liquid biopsy samples [38, 40, 54, 55]. Previous studies have demonstrated that the levels of ctNAs in sarcomas are associated with tumorigenesis, progression and resistance to therapy [40, 46, 50, 54, 56-60]. Therefore, ctNAs show the potential to serve as non-invasive biomarkers in monitoring sarcoma progression and therapeutic response.
A ctDNA detection assay was recently approved by the US Food and Drug Administration for use in lung cancer to identify the molecular composition of tumors [22]. Tumor-derived mutated genes and fusion gene existed in many sarcoma subtypes [61, 62], and researches focusing on the potential of these mutated gene as non-invasive marker is ongoing [18, 50, 57, 59, 63-65].
miRNAs are a set of endogenous regulatory single-strand non-coding RNAs found in eukaryotes that are approximately 19-25 nucleotides in length [66]. Recent studies have shown that miRNAs are involved in cell proliferation, differentiation and apoptosis as well as development and disease [67-73]. Accumulating research has demonstrated that miRNAs are expressed in cell lines and tissue samples from bone and soft tissue sarcoma [58, 66, 73, 74], and merging evidence has indicated that circulating miRNAs can be detected in body fluids of cancer patients [40, 75, 76]. Because of their stability in plasma or serum, miRNAs are considered promising biomarkers for early cancer diagnosis and prognosis prediction [40, 75, 76].
Below, we will discuss the current literature regarding the association of ctDNAs and ctRNAs, including mRNAs, miRNAs, and lncRNAs, and their clinical utility in various subtypes of sarcomas. All of the ctNAs identified thus far and their roles in sarcoma are summarized in Table 3.
Osteosarcoma
Osteosarcoma (OS) is the most common malignant bone tumor and mainly affects the long bones of children and adolescents. In genetics, OS is associated with complex genomic alterations, including point mutations, deletions, amplifications and structural variants of various genes [77]. The application of ctDNA in OS patients is limited. At the time of writing, only two studies evaluating liquid biopsy in OS have been performed [59, 64]. In one study, the authors examined whether common somatic mutations could be detected in ctDNA from OS patient plasma [64]. Mutations in TP53, ATRX, DLG2 and MET in ctDNA were identified by targeted NGS in primary tissue samples and plasma samples, and these mutations were associated with the clinical course [64]. In another study, copy number gains of chromosome arm 8q were detected in peripheral blood in OS patients [57]. The changing level of ctDNA was correlated with tumor burden [59]. An understanding of the clinical utility of ctDNA detection in this highly heterogeneous tumor is in the early stages, and further research is required.
Multiple circulating miRNAs have been detected that play oncogenic or antitumor-suppressor roles in OS. miR-542-3p was initially shown to enhance U2OS cell proliferation and migration [78]. In a subsequent study, higher serum levels of miR-542-3p were detected in OS patients than in healthy individuals with sufficient sensitivity and specificity, and elevated miR-542-3p was associated with poor progression-free survival and overall survival [79]. Other miRNAs, such as miR-221, miR-191, miR-421, and miR-124, have also been found in the circulation of OS patients and play oncogenic roles [80-83]. Furthermore, miR-221, miR-191 and miR-421 levels were significantly upregulated in osteosarcoma tissues and serum compared with the respective levels in normal controls, however, miR-124 was remarkably decreased [80-83]. These miRNAs may serve as diagnostic indicators or prognostic biomarkers in the future.
Summary of circulating tumor nucleic acids in bone and soft tissue sarcomas.
| Sarcoma subtype | Circulating nucleic acid | Molecular target | Analysis method | Sample | Diagnostic value | Prognostic assessment | Response to therapy | Reference |
|---|---|---|---|---|---|---|---|---|
| OS | ctDNA | TP53,ATRX,DLG2,andMET mutations | tNGS | Plasma | √ | [64] | ||
| Chrosome 8q | ddPCR+NGS | Plasma | √ | [59] | ||||
| OS | ctRNA | miR-542-3p | RT-qPCR | Serum | √ | √ | [79] | |
| miR-221 | RT-qPCR | Serum | √ | √ | [80] | |||
| miR-191 | RT-qPCR | Serum | √ | √ | [81] | |||
| miR-124 | RT-qPCR | Serum | √ | √ | [83] | |||
| miR-421 | RT-qPCR | Serum | √ | √ | √ | [82] | ||
| miR-25-3p | RT-qPCR | Serum | √ | √ | √ | [84] | ||
| miR-21 | RT-qPCR | Serum | √ | √ | [85] | |||
| miR-491 | RT-qPCR | Serum | √ | [87] | ||||
| miR-101 | RT-qPCR | Serum | √ | [89] | ||||
| miR-497 | RT-qPCR | Serum | √ | [90] | ||||
| OS | ctRNA | miR-195 | RT-qPCR | Serum | √ | √ | [88] | |
| miR-375 | RT-qPCR | Serum | √ | √ | [91] | |||
| miR133b and miR-206 | RT-qPCR | Serum | √ | √ | √ | [92] | ||
| miR199a-3p, miR-21 and miR-143 | RT-qPCR | Serum | √ | √ | [93] | |||
| miR-215-5p andmiR-642a-5p | RT-qPCR | Serum | √ | [94] | ||||
| miR-21, miR-221 and miR-106a | RT-qPCR | Serum | √ | [95] | ||||
| miR-106a-5p, miR16-5p, miR-20a-5p, miR-425-5p, miR451a, miR-25-3P and miR139-5p | RT-qPCR | Serum | √ | √ | [96] | |||
| OS | ctRNA | lncRNA TUG1 | RT-qPCR | Whole blood | √ | [111] | ||
| LncRNA UCA1 | RT-qPCR | Plasma | √ | [112] | ||||
| ES | ctDNA | STAG2 and TP53 | NGS | Plasma | √ | [59, 118] | ||
| EWS/FLI and EWS/ERG fusion sequences | dPCR | Plasma | √ | √ | [18, 59, 63] | |||
| mtDNA | RT-qPCR | Serum | √ | √ | [126] | |||
| ES | ctRNA | EWS/FLI1 and EWS/ERG transcripts | RT-qPCR | Plasma | √ | √ | [119-122] | |
| miR-125b | RT-qPCR | Serum | √ | [127] | ||||
| CS | ctDNA | mutant IDH1/IDH2 | dPCR | Plasma | √ | √ | [133] | |
| LPS | ctDNA | TERT C228T break point t(12:16) | qPCR | Plasma | √ | [60] | ||
| TP53 | NGS | Plasma | √ | [147] | ||||
| ctRNA | miR-3613-3p | RT-qPCR | Whole blood | √ | [149] | |||
| SS | ctDNA | SYT-SSX fusion sequence | Nested PCR | Peripheral blood | √ | [150-152] | ||
| ctRNA | miR-92b-3p | RT-qPCR | Serum | √ | √ | √ | [155] | |
| miR-99a-5p, miR-146b-5p, miR-148b-3p, miR-195-5p, miR-223-3p, miR-500b-3p and miR-505-3p | RT-qPCR | Serum | √ | √ | [56] | |||
| RMS | ctDNA | PAX3-FOXO1 fusion | ddPCR | Plasma | √ | √ | [159, 160] | |
| miR-1, miR-133a, miR-133b and miR-206 | RT-qPCR | Serum | √ | [162] | ||||
| LMS | ctDNA | TP53, RB1 and PTEN | ULP-WGS | Plasma | √ | [166] | ||
| ctDNA | TP53, PIK3CA and HRAS | tNGS | Plasma | √ | [50] | |||
| ctRNA | miR-199b-5p, miR-320a, miR-199a-3p, miR-126, andmiR-22 | RT-qPCR | Plasma | √ | √ | [170] | ||
| GIST | ctDNA | KIT or PDGFRA | L-PCR or dPCR | Plasma | √ | √ | √ | [174-180] |
Abbreviations: tNGS: targeted next generation sequencing; cNGS: custom next generation sequencing; L-NGS: ligation-next generation sequencing; ULP-WGS: ultra-low passage whole-genome sequencing; ddPCR: droplet-digital polymerase chain reaction; dPCR: digital polymerase chain reaction; RT-qPCR: real time quantitative polymerase chain reaction; L-PCR: ligation-polymerase chain reaction
Owing to the application of neoadjuvant chemotherapy, the 5-year survival rate of OS patients after limb salvage surgery has reached 60%-70%, but the prognosis of patients with chemotherapy resistance is poor because of the lack of predictive biomarkers. Recently, Fujiwara et al. found that circulating miR-25-3p levels were inversely correlated with chemotherapy response in OS patients [74, 84]. Simultaneously, they found that miR-25-3p induces chemoresistance via Dickkopf WNT Signaling pathway inhibitor 3 (DKK3) [74, 84]. Moreover, elevated serum levels of miR-21 were also linked with chemo-resistance in OS patients and indicated a poor progression-free survival and overall survival [85]. These miRNAs may be potential therapeutic targets for OS. Besides above, Allen-Rhoades et al reported that miR-214 is a prognostic marker in metastatic OS, and its level is negatively correlated with prognosis [86]. This indicates that some miRNAs are worth studying as prognostic markers in OS patients.
In addition to oncogenic miRNAs, some studies have shown that other miRNAs act as tumor suppressors in OS. miR-491 binds to the 3′ untranslated region of CRYAB to mediate apoptosis in OS cells [87].The decreased level of miR-491 in OS patient serum was associated with lung metastasis, chemoresistance and a poor overall survival rate [87]. miR-491 was an independent prognostic factor in OS patients and a potential biomarker in liquid biopsy. The levels of other miRNAs, such as miR-124, miR-101, miR-497 and miR-195, could be detected in peripheral blood samples and were downregulated in OS patients compared with the respective levels in healthy individuals [83, 88-91]. Furthermore, these miRNAs are also related to higher pathological grade, metastasis, local recurrence and chemo-resistance [83, 88-91].
Some studies have shown that different combinations of miRNAs are associated with prognosis in OS. Decreased miR-133b or miR-206 alone in OS patient serum was associated with high tumor grade, metastasis, and recurrence [92]. Moreover, a reduced level of miR-133b was linked with chemo-resistance and poor disease-free survival and overall survival in OS [92]. However, a decrease in both miR-133b and miR-206 was correlated with a worse outcome than a decrease in miR-133b or miR-206 alone [92]. Combinations of two or more circulating miRNAs have been used to predict the outcome in OS patients in other studies [93-96]. Asano et al. recently analysed miRNA levels in the serum of 1,002 patients with bone and soft tissue tumors and formed an index that when combined with seven serum microRNAs can effectively distinguish patients with sarcoma from those with benign tumors, with a high sensitivity of 90% and a specificity of 95% [55]. These results indicate that the use of combinations of miRNAs is a promising strategy for early and precise diagnosis and prediction of the prognosis of sarcoma in liquid biopsy samples.
LncRNAs are a group of non-protein-coding molecules of more than 200 nucleotides in length [97]. LncRNAs are also involved in the development, proliferation, differentiation and apoptosis of cancer cells [98-100]. Multiple studies have shown that lncRNAs play an important role in the regulation of progression and metastasis in many types of cancer, such as hepatocellular carcinoma [101], colorectal cancer [102-104] and breast cancer [105, 106].
The role of lncRNAs in sarcoma,especially in OS, has been the subject of investigation. Many lncRNAs participate in proliferation, differentiation, invasiveness, and even chemo-resistance in OS, and the levels of lncRNAs have been associated with clinicopathological features in OS patients [107-110].Therefore, circulating lncRNAs are attractive as potential noninvasive biomarkers. However, at the time of writing, only two studies have been reported. In 2016,Ma et al. showed that the plasma lncRNATUG1 was overexpressed in OS patients and that its level correlated with poor prognosis and disease status [111]. The level of the circulating lncRNA UCA1 showed significant differences between OS patients and healthy individuals [112]. Furthermore, compared with that in healthy individuals, the level of UCA1 expression in OS patients was higher, and the overall survival and disease-free survival were poorer [112]. These studies suggest that the lncRNAs TUG1 and UCA1 could be potential non-invasive biomarkers for the diagnosis and prognosis of OS patients.
Ewing sarcoma
Ewing sarcoma (ES) is a highly aggressive cancer that is predominantly founded in children and adolescents [113]. ES is characterized by chromosomal translocation of the EWS gene to locations carrying different ETS-related genes [113, 114].Previous studies have indicated that ES has few recurrent somatic event besides EWSR1-ETS fusions [113, 115, 116]. Only a few studies have indicated that mutations in STAG2 and Tp53 can be detected by NGS and are associated with a worse prognosis in ES [115-117]. Therefore, reports on somatic events in liquid biopsy samples in ES are insufficient. Recently, two reports indicated that STAG2 and Tp53 mutations were found in ES patient peripheral blood samples [59, 118]. Furthermore, they found that changes in the ctDNA level over time corresponded to response to therapy [59, 118].
Genetically, the most common gene fusions in ES cases are the EWS-FLI1 (90%-95%) and EWS-ERG (5%-10%) gene fusions [113]. The well-characterized genetic characteristics in ES provide the possibility to use liquid biopsy to monitor these gene fusion events. Hayashi et al. [63] found that circulating gene fusion levels paralleled with tumor burden and tumor metastasis in an ES xenotransplant mouse model, which prompted the authors to study the correlation between circulating gene fusion levels and tumor dynamics in ES patients. Subsequently, they found that the level of the EWS-FLI1 fusion gene in the circulation decreased after chemotherapy or surgery and then started to rise when the tumor relapsed before clinical or radiographic evidence was confirmed [63]. This result suggests that fusion genes can be used as non-invasive biomarkers for the prognosis and evaluation of therapeutic effects in ES patients. Although only three ES patients were analysed in the study, these findings suggest promising applications for liquid biopsy in ES. Accumulating evidence has shown that the level of the EWSR1 fusion gene in the circulation decreases rapidly after initial chemotherapy in most patients and increases with tumor recurrence [18, 118]. These results indicate that the levels of circulating EWSR1-FLI1and EWSR1-ERG in ES patients can reflect the tumor burden and relapse status [18, 63, 118]. These studies further support the therapeutic application of the liquid biopsy strategies. Shulman and colleagues showed that ctDNA levels were inversely associated with disease-free survival and overall survival in a larger cohort of cancer patients, including 94 ES cases, indicating the potential of ctDNA as a non-invasive prognostic biomarker in clinical practice [59].
As early as 1997, a study showed that EWS-FLI1 transcripts could be detected in peripheral blood and bone marrow samples from ES patients by RT-PCR [119]. Although the exact physiological and clinical significance of circulating EWS-FLI1 transcripts was not fully understood at the time, the results provided an important foundation for subsequent study. Alava et al. later demonstrated that changes in circulating EWS-FLI1/ERG transcript levels correlated with the disease status of ES patients [120]. The circulating EWS-FLI1/ERG transcripts became undetectable after therapy and were then detectable before the disease progressed [120]. Avigadet al. reported nearly identical results and showed that circulating mutated fusion gene transcripts in ES patients could predict prognosis [121]. These studies can be regarded as early explorations into liquid biopsy in ES patients, and the results indicate that circulating tumor-specific fusion gene transcripts in peripheral blood can reflect the therapeutic effect in tumors and predict tumor progression. A recent study by Allegretti demonstrated that the number of plasma EWS-FLI1 fusion gene transcripts paralleled the positron emission tomography volumetric parameters in ES patients, which provided preliminary evidence for using circulating gene transcripts to monitor the therapeutic efficacy of treatments in ES and evaluate the presence of residual lesions [122].
While the majority of reports have focused on mutated nuclear DNA transcripts as circulating biomarkers in ES, studies have shown that mutated mitochondrial DNA (mtDNA), an extra-chromosomal genome that replicates independently of nuclear DNA [123, 124], can be detected in many types of body fluids in patients with various cancers [123-125]. Yu and collaborators demonstrated that the mtDNA level in the serum of ES patients was approximately two-fold lower than that of healthy adults, indicating the potential for mtDNA as a diagnostic marker to identify ES patients [126]. In addition, the concentration of mtDNA in the serum was related to tumor metastasis in ES patients.
Compared with that in OS, research on circulating miRNAs in ES is rare. At the time of writing, only one study has been published, in which the authors examined circulating miR-125b in a group of Chinese ES patients [127]. The results demonstrated that the circulating miR-125b level was decreased in the serum from ES patients compared with healthy individuals and was linked to poor response to chemotherapy. Previous research showed that miR-125b plays a tumor suppressor role in ES cells by targeting the PI3K-AKT-mTOR signalling pathway [128], and the chemoresistance function induced by miR-125b occurs via suppression of the apoptotic mediators p53 and BAK [129]. These results suggest that circulating miR-125b may be a non-invasive biomarker in ES patients and requires further validation in a larger cohort.
Chondrosarcoma
Chondrosarcoma (CS) is the second most common primary bone malignancy. It represents a collection of heterogeneous bone cancers characterized by hyaline cartilaginous neoplastic tissue. Genetically, mutant isocitratedehydrogenase (IDH) 1 and 2 genes have been identified in more than half of central and periosteal CS cases [130, 131]. Further studies found that the overall survival of CS patients with IDH1/IDH2 mutations was significantly lower than that of patients without IDH1/IDH2 mutations [132]. Gutteridge et al. identified mutant IDH in the plasma of chondrosarcoma patients [133]. The authors demonstrated that cell-free IDH1/IDH2 could be detected in high-grade chondrosarcoma and dedifferentiated chondrosarcoma patients during the preoperative period; moreover, a high level of mutant IDH correlated with poor prognosis [133]. Mutant IDH1 was detected in plasma prior to traditional radiological imaging and clinical changes in patients whose tumors relapsed or metastasized during follow-up periods [133].These results indicate that mutated IDH in circulation may be a potential non-invasive biomarker to monitor chondrosarcoma patients in the clinical setting. Accumulating evidence has demonstrated that the COL2A1 [134] and exostosin glycosyltransferase (EXT1 or EXT2) genes are also frequently altered in CS [134, 135]. Additionally, other sporadic mutations such as mutations in TP53, pRB, AKT1, and MDM2 have also been identified in CS [134, 136, 137]. However, the above altered genes have not been applied in liquid biopsy.
miRNAs are also involved in normal chondrogenesis [138], and some miRNAs play an oncogenic role in CS, such as miR-181a, which is overexpressed in high-grade CS and promotes the formation of vascular endothelial growth factor [139]. In contrast, miR-100 is a tumor suppressor that is downregulated in CS [140]. Other epigenetic alterations in CS including DNA methylation [138], such as hypermethylation of the promoter region of the gene encoding the transcription factor of RUNX3 and prompted proliferation and inhibited apoptosis in CS tumor cells [141]. However, epigenetics alterations in CS have not been used in liquid biopsy, and further research is needed.
Although chromosomal translocation is sporadic in CS, some fusion genes have been detected in CS, such as EWSR1-NR4A3, RBP56-NR4A3, and TCF12-NR4A3, which were detected in extraskeletal myxoid chondrosarcoma [138, 142-144]. Additionally, the HEY1-NCOA2 orIRF2BP2-CDX1 fusion genes resulting from t(1;5)(q42;q32) also exists in mesenchymal chondrosarcoma [144, 145]. To date, these fusion genes have not been applied in liquid biopsy.
Liposarcoma
Liposarcoma ( LPS) is the most common soft tissue sarcoma (STS) in adults and account for approximately 25% of all STSs [146]. LPS represents a heterogeneous group of adipocytic malignancies that are classified into four key subtypes: well-differentiated liposarcoma, dedifferentiated liposarcoma, myxoid liposarcoma, and pleomorphic liposarcoma. Subtypes can be distinguished by the presence of the FUS-CHOP fusion oncoprotein (myxoid LPS) and MDM2 overexpression (well-differentiated liposarcoma and dedifferentiated liposarcoma) [146]; therefore, the above mutations have the potential to be promising biomarkers in LPS. Recently, Braiget al. found that the levels of the breakpoint t(12:16) and TERT C228T mutation in plasma in myxoid liposarcomas were associated with tumor burden and tumor dynamics [60]. Although mutant TP53 is not a common somatic event [146], it has also been identified in plasma from dedifferentiated LPS patients during HDM2 inhibitor therapy [147]. HDM2 interacts with wild-type TP53 and subsequently inhibitsTP53 [148]. The circulating mutant TP53 level was increased in patients treated with the HDM2 inhibitor compared with those treated with other therapies, and the levels correlated with tumor size [147]. These results suggest that the circulating TP53 mutation burden can be used to evaluate the tumor response to targeted therapy.
In addition, the level of miR-3613-3p was significantly upregulated in whole-blood samples from dedifferentiated liposarcoma patients, and it may potentially serve as an independent diagnostic biomarker to distinguish dedifferentiated liposarcoma patients from healthy individuals and lipoma patients [149].
Synovial sarcoma
Synovial sarcoma(SS) is an aggressive soft tissue malignancy which characterized by the formation of the SYT-SSX fusion gene. Several researchers have attempted to validate the potential use of the circulating SYT-SSX fusion gene as a non-invasive biomarker in SS patients [65, 150-152]. In 2001, Hashimoto et al. detected the SYT-SSX fusion gene in peripheral blood by nested PCR in an SS patient [150]. Although only a single patient was analysed, these results suggest that the circulating SYT-SSX fusion gene may be a promising non-invasive biomarker for monitoring SS patients. Similar findings were shown in a gastric SS patient [151]. Based on these findings, subsequent studies investigated the potential of the circulating SYT-SSX fusion gene in liquid biopsy of SS patients. However, existing evidence revealed that the presence of the circulating SYT-SSX fusion gene was an infrequent event in SS patients and that the circulating SYT-SSX fusion gene was not an ideal marker to monitor tumor dynamics [65, 152, 153]. Therefore, the SYT-SSX fusion gene may not be a reliable circulating biomarker and further research is needed in SS patients. Other than translocation, SSs are mutationally quiet [154].
Recent studies have reported promising findings related to circulating miRNAs in SS. Uotani et al. evaluated miR-92b-3p in the serum of SS patients with a miRNA microarray assay and found that it was correlated with tumor burden and tumor dynamics [155]. Fricke et al. identified a panel of upregulated miRNAs, including miR-99a-5p, miR-146b-5p, miR-148b-3p, miR-195-5p, miR-223-3p, miR-500b-3p and miR-505-3p, in peripheral whole-blood samples using a gene chip miRNA array coupled with qRT-PCR [56]. The authors found that these miRNAs could be used as diagnostic biomarkers for distinguishing patients with SS from patients with other sarcoma subtypes and healthy controls (Table 4), and these miRNAs could also be used to monitor local recurrence and distant metastasis. This study not only distinguished a panel of miRNAs as biomarkers in liquid biopsy of SS patients but also provided a practical protocol to screen miRNAs in body fluid.
Expression level of miRNAs in active synovial sarcoma, synovial sarcoma with complete remission, healthy controls and others sarcoma subtypes
| miRNA associated with active SS (upregulated) | SS with CR | Healthy control | Active LMS | Active MPNST | Active ES | Active LPS |
|---|---|---|---|---|---|---|
| hsa-miR-99a-5p | √ | √ | √ | √ | ||
| hsa-miR-146b-5p | √ | √ | √ | √ | √ | √ |
| hsa-miR-148b-3p | √ | √ | √ | √ | √ | √ |
| hsa-miR-195-5p | √ | √ | √ | |||
| hsa-miR-223-3p | √ | √ | √ | √ | √ | √ |
| hsa-miR-500b-3p | √ | √ | √ | √ | √ | √ |
| hsa-miR-505-3p | √ | √ | √ | √ | √ | √ |
Abbreviation: CR: complete remission; LMS: leiomyosarcoma; MPNST: malignant peripheral nerve sheath tumor;ES: Ewing sarcoma; LPS: liposarcoma
Rhabdomyosarcoma
Rhabdomyosarcoma (RMS) is a small round cell malignant tumor that is frequently found in children. RMSs are mainly categorized into alveolar rhabdomyosarcoma (ARMS), embryonal rhabdomyosarcoma (ERMS) and pleomorphic rhabdomyosarcoma (PRMS) [156, 157]. Up to 90% of ARMSs present with t(1;13)(p36;q15) or t(2;13)(q35;q14), which results in the fusion of the PAX3 gene with FOXO1 or the PAX7 gene with FOXO1, respectively [156-158]. The specific known translocations in ARMS cases provide potential markers to monitor tumor dynamics. To detect sarcoma-specific translocations in cfDNA, translocation-specific sarcoma sequencing assays have been developed, and the PAX3/FOXO1 translocation was detected in all ARMS patients in research by Klega et al. [118]. Previous studies have demonstrated that patients with ARMS with PAX3/FOXO1 have worse outcomes than those without the translocation [157]. Subsequently, the PAX3-FOXO1 fusion was also identified in a plasma sample from a patient with ARMS and proved to be a reliable biomarker to detect early tumor progression and recurrence [159, 160].
Although the t(1;13)(p36;q15) and t(2;13) (q35;q14) translocations are rare in ERMS and PRMS, PAX3, PAX7 and FOXO1 are frequently overexpressed [161]. It is valuable to further explore the upregulated PAX and FOXO1 in liquid biopsy samples.
In addition to fusion genes and overexpressed PAX3, PAX7 and FOXO1, serum miRNAs (miR-1, miR-133a, miR-133b and miR-206) have been identified in RMS cases [162]. Additionally, miR-206 can be used as a diagnostic biomarker in distinguishing RMS from non-RMS tumors with a sensitivity of 1.0 and a specificity of 0.913 [162].
Leiomyosarcoma
Leiomyosarcoma (LMS) is a highly aggressive malignant neoplasm derived from smooth muscle tissue that makes up approximately 10% of STSs [163]. LMS commonly affects the uterus or retroperitoneum but also occurs throughout the body [164]. LMS presents with a highly complex karyotype with no specific mutations [52, 165]. Therefore, liquid biopsy in LMS is rare, and a recent study by Hemming et al. found that a higher level of ctDNA, including a panel of genes with copy number variations, detected by ultra-low-passage whole-genome sequencing was correlated with tumor size and tumor progression in LMS [166]. In addition to primary sarcoma, liquid biopsy has also been used in some metastatic sarcomas. Nicholas C. Eastley et al. identified 5 of 11 metastatic STS patients as having cancer-related mutations including mutations in TP53, PIK3CA and HRAS in plasma samples [50]. In addition, they found that these mutations correlated with tumor burden [50]. The 5 patients encompassed two patients with LMS and one patient each with undifferentiated pleomorphic sarcoma, soft tissue chondrosarcoma, and epithelioid angiosarcoma [50].
Increasing evidence indicates that the miRNA expression profile can be used to distinguish many tumor types, including sarcomas [54, 167]. miR-1 and miR-113a/b which are regulators of myogenesis, were significantly overexpressed in LMS [58, 168]. A study found that miR-21 was differentially expressed between LMS and leiomyoma [169]. Additionally, Guled et al. indicated that a collection of serum miRNAs, including miR-199b-5p, miR-320a, miR-199a-3p, miR-126, and miR-22, were differentially expressed between LMS and undifferentiated pleomorphic sarcoma [170]. These results indicated that these blood-borne miRNAs could be used in the differential diagnosis of different sarcoma subtypes. Additionally, further studies need to be performed to explore more miRNAs as diagnostic markers in sarcoma.
Gastrointestinal Stromal Tumor
Gastrointestinal stromal tumor (GIST) is the most common gastrointestinal sarcoma with an incidence of 15-20 cases per million per year [54, 171]. KIT or platelet-derived growth factor receptor alpha (PDGFRA) gain-of-function mutations [54, 171] are present in approximately 85-90% of GIST cases [172, 173]. Specific mutations in GIST have the potential to be markers in liquid biopsy. The application of liquid biopsy in GIST was first reported at the 2013 ASCO Annual Meeting [174]. In this study, Reichardt et al found 84% overall concordance between plasma and tumor tissue samples in the detection of primary KIT exon 9 and 11 mutations [174]. In the same year, Maier et al evaluated 291 plasma samples from 38 GIST patients using 25 different allele-specific ligation PCR assays covering KIT and PDGFRA alterations [175]. The authors found that ratio of mutant ctDNA to wild-type ctDNA for KIT and PDGFRA were positively correlated with disease status [175]. For instance, the ctDNA ratio was significantly higher in samples from patients with active disease than in samples from patients incomplete remission without residual disease. Taken together, the above studies indicated that mutant KIT or PDGFRA can be detected in the plasma of GIST patients and that the concentration of ctDNA correlates with tumor burden and therapy response. These findings suggest that ctDNA (KIT or PDGFRA) has the potential to be a reliable non-invasive biomarker for monitoring GIST. Subsequently, Bauer and collaborators reported promising results on liquid biopsy of GIST at the 2015 ASCO Annual Meeting [176]. They collected 30 plasma and 15 matched tumor samples from 22 metastatic GIST patients and identified 87 nonsynonymous Kit alterations in plasma samples by a custom-designed targeted sequencing panel for the Illumina MiSeq platform. Primary mutations and resistance mutations were found in 41% and 86% of GIST patients, respectively [176]. Nearly at the same time, Kang et al analysed mutations in KIT, PDGFRA and BRAF in GIST patient plasma samples via NGS [177]. They found that additional mutations in plasma emerged in GIST patients who were resistant to tyrosine kinase inhibitor (TKI) treatment [177]. These results indicate that blood-derived ctDNA can be used as a non-invasive marker for the prediction of treatment response in GIST patients. In the wake of the above research, Wanda and collaborators isolated plasma ctDNA before and after imatinib treatment from 4 imatinib-resistant GIST patients [178]. They found that a primary mutation in c-KIT exon11 mutated into secondary KIT exon 13 and 18 mutations [178]. The results indicated that the detection of secondary c-KIT mutations in ctDNA could be used to predict anticancer effects in GIST. More recently, studies exploring the role of primary mutations in GISTs have been initiated. Kang and coworkers analysed paired plasma-tissue samples in primary GIST patients and revealed 72% concordance between them [179]. In a subsequent study, Boonstra et al proved that ddPCR is an effective method to detect KIT exon 11 mutations in GISTs in both tumor tissue and ctDNA with a 100% specificity and a 77% sensitivity [180]. Furthermore, they also demonstrated that the level of ctDNA negatively correlated with treatment response; for instance, a decrease in KIT exon ctDNA corresponded to lesion remission or stable disease on radiological examination [180]. In summary, specific mutations, including mutations in KIT or PDGFRA, in GIST have been proven to be promising non-invasive biomarkers in liquid biopsy. To date, there are no reports on circulating non-coding RNAs in GIST, leaving many questions unanswered on their potential role in evaluating tumor dynamics and TKI therapy response [171].
Other sarcoma subtypes
Although STSs are a heterogeneous group of malignant solid neoplasms that include approximately 50 subtypes, there are several subtypes of STSs that have been understudied in relation to liquid biopsy [167].
Fibrosarcoma comprises a rare subtype of STS derived from fibrous connective tissue [167, 181]. Fibrosarcoma with a complex karotype lacks of specific alterations such as recurrent point mutations and chromosomal translocation; therefore, there are very few reports on ctDNA in liquid biopsy for fibrosarcoma. Research on miRNAs in fibrosarcoma is limited. The miR-29 family activates MMP-2 to play tumor-suppressive roles in the HT-1080 human fibrosarcoma cell line [182]. Moreover, miRNA-520c and miRNA-373 have also been identified in the above cell line, and they activate the Ras/ Raf/ MEK/ Erk signalling pathway and NF-kB to promote the tumor cell migration and invasion [183]. The application of these miRNAs in liquid biopsy in fibrosarcoma requires further validation.
Angiosarcoma is a highly aggressive malignancy that originates from vascular or lymphatic tissues [184]. Due to the rarity of angiosarcoma, few comprehensive studies of genetic changes in angiosarcoma have been reported [184]. The most common aberrations in angiosarcoma are mutations in TP53 (29%-35%) and losses of CDKN2A(26%) [146, 185, 186]. Accordingly, Murali et al reported that more than 50% of angiosarcomas carry some genetic alterations affecting the MAPK signalling pathway, including mutations in HRAS, KRAS, NRAS, MAPK, BRAF and NF-1 or amplifications ofMAPK1/CRKL, CRAF or BRAF [186]. Moreover, mutations in FLT4, PTPRB, PLCG1, CIC and KDR were also detected in angiosarcoma [187]. These karyotype aberrations in angiosarcoma have the potential to be monitored in ctDNA in the future. Additionally, many miRNAs were identified in angiosarcoma, and miR-515-3p and miR-517c may be the most valuable diagnostic biomarkers. Sarver and collaborators identified that the above miRNAs were overexpressed by 12-fold in angiosarcoma relative to GIST, undifferentiated pleomorphic sarcoma and epithelioid sarcoma [188]. Furthermore, the miR-515-5p, miR-517a, miR-518b, miR-519a, miR-522 and miR-17-92 clusters are also upregulated in angiosarcoma [188, 189]. Further functional validation of these miRNAs in liquid biopsy will be required.
Malignant peripheral nerve sheath tumors (MPNSTs) constitute a rare subtype of STS and arise in large peripheral nerves [61]. Accumulating evidence has identified non-synonymous mutations of the polycomb regressive complex2 (PRC2) subunits SUZ112 and EED in approximately 80% of MPNSTs [190, 191]. Additionally, many miRNAs have been found to be overexpressed in MPNSTs, such as miR-210, miR-339-5p, miR-10b and the miR-199a/214 cluster [192-194]. Further, more extensive studies in liquid biopsy need to be performed.
Undifferentiated pleomorphic sarcoma (UPS) is an aggressive subtype of STS that frequently occurs in adults over the age of 40 [146, 181]. Due to its complex genetic features and difficult diagnosis, genetic alterations in UPS are also missing [146]. Recently, Demoret et al found that ctDNA comprehensive genomic profiling (CGP) of UPS was poorly concordant with solid tumor CGP [195]. The data shows that approximately 30% (2/6) of subjects had complete concordance and 50% (3/6) of subjects had complete or partial concordance [195]. Given the poor concordance between ctDNA CGP and tumor CGP in UPS, more extensive studies need to be performed. Guled et al investigated the differential expression level of microRNAs between LMS and UPS and found that miR-199-5p was overexpressed in UPS [170]. In addition, miR-138 was overexpressed in UPS and was negatively correlated with distant metastasis-free survival [196]. In summary, the application of ctNAs in liquid biopsy for sarcomas shows promise. However, further research in different subtypes of STSs will need to be performed in subsequent studies.
Conclusions and Perspectives
Although the application of liquid biopsy in bone and soft tissue sarcoma is still in its infancy, the field has shown important progress worthy of discussion, especially regarding ctNAs. Moreover, accumulating evidence has indicated that circulating tumor non-coding RNAs are also promising biomarkers. A panel of sarcoma-specific mutated genes can be detected in blood circulation and used as a non-invasive biomarker to monitor sarcoma patients. Furthermore, many types of non-coding RNAs, especially circulating miRNAs, have been indicated as specific biomarkers for diagnosing sarcoma, predicting prognosis and revealing chemotherapy resistance. Together these studies demonstrate the utility of circulating tumor nucleic acids in bone and soft tissue sarcomas as promising non-invasive biomarkers. Sarcomas with specific genetic mutations or translocations, such as GIST, CS, ES and RMS, have potential to be monitored with ctDNA in liquid biopsy. Conversely, some sarcomas harbour complex and irregular genomic changes, such as OS, LMS, LPS and UPS, and circulating non-coding RNA seems to be a promising surrogate in liquid biopsy in these diseases. Additionally, other epigenetic alterations, such as DNA methylation and histone modification, also need to be explored in liquid biopsy in the future. Thus, liquid biopsy opens up a new approach for effective and real-time tumor monitoring.
Despite these achievements, the application of ctNAs is still in the preclinical stage. Many studies include only few samples, and randomized clinical trials to support the preliminary results are lacking. Moreover, the sample and detection methods are inconsistent among different studies. Furthermore, the sensitivity and specificity of ctDNA as a biomarkers in liquid biopsy of peripheral blood are lower than expected. Further studies are required to explore the value of ctNAs in sarcoma patients.
Abbreviations
OS: osteosarcoma; ES: ewing sarcoma; CS: chondrosarcoma; LPS: liposarcoma; SS: synovial sarcoma; RMS: rhabdomyosarcoma; ARMS: alveolar rhabdomyosarcoma; ERMS: embryonal rhabdomyosarcoma; PRMS: pleomorphic rhabdomyosarcoma; LMS: leiomyosarcoma; GIST: gastrointestinal stromal tumor; MPNST: malignant peripheral nerve sheath tumor; UPS: undifferentiated pleomorphic sarcoma; STS: soft tissue sarcoma; ctNAs: circulating tumor nucleic acids; cfDNA: cell-free DNA; ctDNA: circulating tumor DNA; mtDNA: mitochondrial DNA; CTCs: circulating tumor cells; lncRNA: long non-coding RNA; PCR: polymerase chain reaction; dPCR: digital polymerase chain reaction; ddPCR: droplet digital polymerase chain reaction; RT-qPCR: quantitative reverse-transcription polymerase chain reaction; NGS: next generation sequencing; IDH: isocitrate dehydrogenase; EXT: exostosin glycosyltransferase; PDGFRA: plate-derived growth factor receptor alpha; TKIs: tyrosine kinase inhibitors; PRC2: polycomb regressive complex 2.
Acknowledgements
This work was supported by the Key Nature Science Foundation of Tianjin (grant nos. 18YFZCSY00550 to Jilong Yang).
Author Contributions
Junqiang Wei drafted the work. Xinyue Liu, Ting Li, Peipei Xing, Chao Zhang substantively revised it. Jilong Yang designed this work.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ng VY, Scharschmidt TJ, Mayerson JL, Fisher JL. Incidence and survival in sarcoma in the United States: a focus on musculoskeletal lesions. Anticancer Res. 2013;33:2597-604
2. Hui JY. Epidemiology and Etiology of Sarcomas. Surg Clin North Am. 2016;96:901-14
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34
4. Weitz J, Antonescu CR, Brennan MF. Localized extremity soft tissue sarcoma: improved knowledge with unchanged survival over time. J Clin Oncol. 2003;21:2719-25
5. Stiller CA, Trama A, Serraino D, Rossi S, Navarro C, Chirlaque MD. et al. Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. European journal of cancer (Oxford, England: 1990). 2013;49:684-95
6. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
7. Yang Z, Zheng R, Zhang S, Zeng H, Li H, Chen W. Incidence, distribution of histological subtypes and primary sites of soft tissue sarcoma in China. Cancer Biol Med. 2019;16:565-74
8. Fukushima T, Ogura K, Akiyama T, Takeshita K, Kawai A. Descriptive epidemiology and outcomes of bone sarcomas in adolescent and young adult patients in Japan. BMC musculoskeletal disorders. 2018;19:297-305
9. Jacobs AJ, Lindholm EB, Levy CF, Fish JD, Glick RD. Racial and ethnic disparities in treatment and survival of pediatric sarcoma. J Surg Res. 2017;219:43-9
10. Alamanda VK, Song Y, Schwartz HS, Holt GE. Racial Disparities in Extremity Soft-Tissue Sarcoma Outcomes: A Nationwide Analysis. American journal of clinical oncology. 2015;38:595-9
11. Lazarides AL, Visgauss JD, Nussbaum DP, Green CL, Blazer DG 3rd, Brigman BE. et al. Race is an independent predictor of survival in patients with soft tissue sarcoma of the extremities. BMC cancer. 2018;18:488-94
12. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722-35
13. Raimondi L, De Luca A, Costa V, Amodio N, Carina V, Bellavia D. et al. Circulating biomarkers in osteosarcoma: new translational tools for diagnosis and treatment. Oncotarget. 2017;8:100831-51
14. Hegde V, Burke ZDC, Park HY, Zoller SD, Johansen D, Kelley BV. et al. Is Core Needle Biopsy Reliable in Differentiating Between Aggressive Benign and Malignant Radiolucent Bone Tumors? Clinical orthopaedics and related research. 2018;476:568-77
15. Filippiadis DK, Charalampopoulos G, Mazioti A, Keramida K, Kelekis A. Bone and Soft-Tissue Biopsies: What You Need to Know. Seminars in interventional radiology. 2018;35:215-20
16. Li X, Seebacher NA, Hornicek FJ, Xiao T, Duan Z. Application of liquid biopsy in bone and soft tissue sarcomas: Present and future. Cancer Lett. 2018;439:66-77
17. Namlos HM, Zaikova O, Bjerkehagen B, Vodak D, Hovig E, Myklebost O. et al. Use of liquid biopsies to monitor disease progression in a sarcoma patient: a case report. BMC cancer. 2017;17:29-35
18. Krumbholz M, Hellberg J, Steif B, Bauerle T, Gillmann C, Fritscher T. et al. Genomic EWSR1 Fusion Sequence as Highly Sensitive and Dynamic Plasma Tumor Marker in Ewing Sarcoma. Clin Cancer Res. 2016;22:4356-65
19. Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med. 2017;15:75-87
20. Bardelli A, Pantel K. Liquid Biopsies, What We Do Not Know (Yet). Cancer cell. 2017;31:172-9
21. Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med. 2018;379:1754-65
22. Husain H, Velculescu VE. Cancer DNA in the Circulation: The Liquid Biopsy. JAMA. 2017;318:1272-4
23. Masaoutis C, Korkolopoulou P, Theocharis S. Exosomes in sarcomas: Tiny messengers with broad implications in diagnosis, surveillance, prognosis and treatment. Cancer Lett. 2019;449:172-7
24. Tellez-Gabriel M, Brown HK, Young R, Heymann MF, Heymann D. The Challenges of Detecting Circulating Tumor Cells in Sarcoma. Frontiers in oncology. 2016;6:202-10
25. Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368-73
26. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature reviews Cancer. 2011;11:426-37
27. Alix-Panabières C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer discovery. 2016;6:479-91
28. Fidler IJ. Metastasis: quantitative analysis of distribution and fate of tumor emboli labeled with 125 I-5-iodo-2'-deoxyuridine. Journal of the National Cancer Institute. 1970;45:773-82
29. Nicolazzo C, Gradilone A. Significance of circulating tumor cells in soft tissue sarcoma. Analytical cellular pathology (Amsterdam). 2015;2015:697395-400
30. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clinical chemistry. 2015;61:112-23
31. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:1-25
32. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
33. Orozco AF, Lewis DE. Flow cytometric analysis of circulating microparticles in plasma. Cytometry Part A: the journal of the International Society for Analytical Cytology. 2010;77:502-14
34. Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends in cell biology. 2009;19:43-51
35. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654-9
36. Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F. et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Molecular cancer. 2015;14:155
37. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003-8
38. Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic acids research. 2011;39:7223-33
39. Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nature cell biology. 2011;13:423-33
40. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145-56
41. Luder Ripoli F, Mohr A, Conradine Hammer S, Willenbrock S, Hewicker-Trautwein M, Hennecke S. et al. A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay. Int J Mol Sci. 2016;17:724-35
42. Mittempergher L, de Ronde JJ, Nieuwland M, Kerkhoven RM, Simon I, Rutgers EJ. et al. Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PloS one. 2011;6:e17163-78
43. Narrandes S, Xu W. Gene Expression Detection Assay for Cancer Clinical Use. J Cancer. 2018;9:2249-65
44. Barata PC, Koshkin VS, Funchain P, Sohal D, Pritchard A, Klek S. et al. Next-generation sequencing (NGS) of cell-free circulating tumor DNA and tumor tissue in patients with advanced urothelial cancer: a pilot assessment of concordance. Ann Oncol. 2017;28:2458-63
45. Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548-54
46. Shukla NN, Patel JA, Magnan H, Zehir A, You D, Tang J. et al. Plasma DNA-based molecular diagnosis, prognostication, and monitoring of patients with EWSR1 fusion-positive sarcomas. JCO Precis Oncol. 2017;2017:1-17
47. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57-63
48. Li JR, Sun CH, Li W, Chao RF, Huang CC, Zhou XJ. et al. Cancer RNA-Seq Nexus: a database of phenotype-specific transcriptome profiling in cancer cells. Nucleic acids research. 2016;44:D944-51
49. Hudecova I. Digital PCR analysis of circulating nucleic acids. Clinical biochemistry. 2015;48:948-56
50. Eastley NC, Ottolini B, Neumann R, Luo JL, Hastings RK, Khan I. et al. Circulating tumour-derived DNA in metastatic soft tissue sarcoma. Oncotarget. 2018;9:10549-60
51. Weiser DA, West-Szymanski DC, Fraint E, Weiner S, Rivas MA, Zhao CWT. et al. Progress toward liquid biopsies in pediatric solid tumors. Cancer Metastasis Rev. 2019;38:553-71
52. Soini Y. Epigenetic and genetic changes in soft tissue sarcomas: a review. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2016;124:925-34
53. Sadikovic B, Pare G. Genomics and epigenomics in pediatric oncology and clinical laboratory genetics. Clinical biochemistry. 2014;47:731-2
54. Kosela-Paterczyk H, Paziewska A, Kulecka M, Balabas A, Kluska A, Dabrowska M. et al. Signatures of circulating microRNA in four sarcoma subtypes. J Cancer. 2020;11:874-82
55. Asano N, Matsuzaki J, Ichikawa M, Kawauchi J, Takizawa S, Aoki Y. et al. A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat Commun. 2019;10:1299
56. Fricke A, Ullrich PV, Heinz J, Pfeifer D, Scholber J, Herget GW. et al. Identification of a blood-borne miRNA signature of synovial sarcoma. Molecular cancer. 2015;14:151-64
57. McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ. et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer. 2010;49:1062-9
58. Kohama I, Kosaka N, Chikuda H, Ochiya T. An Insight into the Roles of MicroRNAs and Exosomes in Sarcoma. Cancers. 2019;11:428-49
59. Shulman DS, Klega K, Imamovic-Tuco A, Clapp A, Nag A, Thorner AR. et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children's Oncology Group. British journal of cancer. 2018;119:615-21
60. Braig D, Becherer C, Bickert C, Braig M, Claus R, Eisenhardt AE. et al. Genotyping of circulating cell-free DNA enables noninvasive tumor detection in myxoid liposarcomas. International journal of cancer. 2019;145:1148-61
61. Network TCGAR. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell. 2017;171:950-65
62. Movva S, Wen W, Chen W, Millis SZ, Gatalica Z, Reddy S. et al. Multi-platform profiling of over 2000 sarcomas: identification of biomarkers and novel therapeutic targets. Oncotarget. 2015;6:12234-47
63. Hayashi M, Chu D, Meyer CF, Llosa NJ, McCarty G, Morris CD. et al. Highly personalized detection of minimal Ewing sarcoma disease burden from plasma tumor DNA. Cancer. 2016;122:3015-23
64. Barris DM, Weiner SB, Dubin RA, Fremed M, Zhang X, Piperdi S. et al. Detection of circulating tumor DNA in patients with osteosarcoma. Oncotarget. 2018;9:12695-704
65. Fricke A, Ullrich PV, Cimniak AF, Follo M, Nestel S, Heimrich B. et al. Synovial Sarcoma Microvesicles Harbor the SYT-SSX Fusion Gene Transcript: Comparison of Different Methods of Detection and Implications in Biomarker Research. Stem Cells Int. 2016;2016:6146047-58
66. Nugent M. MicroRNA function and dysregulation in bone tumors: the evidence to date. Cancer Manag Res. 2014;6:15-25
67. Demarez C, Gerard C, Cordi S, Poncy A, Achouri Y, Dauguet N. et al. MicroRNA-337-3p controls hepatobiliary gene expression and transcriptional dynamics during hepatic cell differentiation. Hepatology (Baltimore, Md). 2018;67:313-27
68. Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H. et al. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. Journal of hepatology. 2015;63:874-85
69. Eastlack SC, Dong S, Ivan C, Alahari SK. Suppression of PDHX by microRNA-27b deregulates cell metabolism and promotes growth in breast cancer. Molecular cancer. 2018;17:100-16
70. Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nature reviews Molecular cell biology. 2014;15:565-76
71. Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends in genetics: TIG. 2004;20:617-24
72. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews Molecular cell biology. 2005;6:376-85
73. Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Molecular cancer. 2017;16:15-32
74. Fujiwara T, Uotani K, Yoshida A, Morita T, Nezu Y, Kobayashi E. et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget. 2017;8:33375-92
75. Yu H, Guan Z, Cuk K, Zhang Y, Brenner H. Circulating MicroRNA Biomarkers for Lung Cancer Detection in East Asian Populations. Cancers. 2019;11:415-36
76. Di Cosimo S, Appierto V, Pizzamiglio S, Tiberio P, Iorio MV, Hilbers F. et al. Plasma microRNA levels for predicting therapeutic response to neoadjuvant treatment in HER2-positive breast cancer: Results from the NeoALTTO trial. Clin Cancer Res. 2019;25:2507-20
77. Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254-65
78. Li H, Liu H, Pei J, Wang H, Lv H. miR-542-3p overexpression is associated with enhanced osteosarcoma cell proliferation and migration ability by targeting Van Gogh-like 2. Molecular medicine reports. 2015;11:851-6
79. Li Q, Song S, Ni G, Li Y, Wang X. Serum miR-542-3p as a prognostic biomarker in osteosarcoma. Cancer biomarkers: section A of Disease markers. 2018;21:521-6
80. Yang Z, Zhang Y, Zhang X, Zhang M, Liu H, Zhang S. et al. Serum microRNA-221 functions as a potential diagnostic and prognostic marker for patients with osteosarcoma. Biomed Pharmacother. 2015;75:153-8
81. Wang T, Ji F, Dai Z, Xie Y, Yuan D. Increased expression of microRNA-191 as a potential serum biomarker for diagnosis and prognosis in human osteosarcoma. Cancer biomarkers: section A of Disease markers. 2015;15:543-50
82. Zhou S, Wang B, Hu J, Zhou Y, Jiang M, Wu M. et al. miR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumour biology. 2016;37:9001-7
83. Cong C, Wang W, Tian J, Gao T, Zheng W, Zhou C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer biomarkers: section A of Disease markers. 2018;21:449-54
84. Yoshida A, Fujiwara T, Uotani K, Morita T, Kiyono M, Yokoo S. et al. Clinical and Functional Significance of Intracellular and Extracellular microRNA-25-3p in Osteosarcoma. Acta medica Okayama. 2018;72:165-74
85. Yuan J, Chen L, Chen X, Sun W, Zhou X. Identification of serum microRNA-21 as a biomarker for chemosensitivity and prognosis in human osteosarcoma. The Journal of international medical research. 2012;40:2090-7
86. Allen-Rhoades W, Kurenbekova L, Satterfield L, Parikh N, Fuja D, Shuck RL. et al. Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med. 2015;4:977-88
87. Wang SN, Luo S, Liu C, Piao Z, Gou W, Wang Y. et al. miR-491 Inhibits Osteosarcoma Lung Metastasis and Chemoresistance by Targeting αB-crystallin. Molecular therapy. 2017;25:2140-9
88. Cai H, Zhao H, Tang J, Wu H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. The Journal of surgical research. 2015;194:505-10
89. Yao ZS, Li C, Liang D, Jiang XB, Tang JJ, Ye LQ. et al. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer biomarkers: section A of Disease markers. 2018;22:127-33
90. Pang PC, Shi XY, Huang WL, Sun K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. European review for medical and pharmacological sciences. 2016;20:3765-9
91. Liu W, Zhao X, Zhang YJ, Fang GW, Xue Y. MicroRNA-375 as a potential serum biomarker for the diagnosis, prognosis, and chemosensitivity prediction of osteosarcoma. J Int Med Res. 2018;46:975-83
92. Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2014;7:4194-203
93. Ouyang L, Liu P, Yang S, Ye S, Xu W, Liu X. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Med Oncol. 2013;30:340-7
94. Monterde-Cruz L, Ramirez-Salazar EG, Rico-Martinez G, Linares-Gonzalez LM, Guzman-Gonzalez R, Delgado-Cedillo E. et al. Circulating miR-215-5p and miR-642a-5p as potential biomarker for diagnosis of osteosarcoma in Mexican population. Hum Cell. 2018;31:292-9
95. Nakka M, Allen-Rhoades W, Li Y, Kelly AJ, Shen J, Taylor AM. et al. Biomarker significance of plasma and tumor miR-21, miR-221, and miR-106a in osteosarcoma. Oncotarget. 2017;8:96738-52
96. Li H, Zhang K, Liu LH, Ouyang Y, Guo HB, Zhang H. et al. MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncology letters. 2015;10:1662-8
97. Russo F, Fiscon G, Conte F, Rizzo M, Paci P, Pellegrini M. Interplay Between Long Noncoding RNAs and MicroRNAs in Cancer. Methods in molecular biology. 2018;1819:75-92
98. Zare K, Shademan M, Ghahramani Seno MM, Dehghani H. CRISPR/Cas9 Knockout Strategies to Ablate CCAT1 lncRNA Gene in Cancer Cells. Biol Proced Online. 2018;20:21
99. Shi X, Wang X, Hua Y. LncRNA GACAT1 Promotes Gastric Cancer Cell Growth, Invasion And Migration By Regulating MiR-149-mediated Of ZBTB2 And SP1. J Cancer. 2018;9:3715-22
100. Zhang J, Hu SL, Qiao CH, Ye JF, Li M, Ma HM. et al. LncRNA-NEF inhibits proliferation, migration and invasion of esophageal squamous-cell carcinoma cells by inactivating wnt/beta-catenin pathway. Eur Rev Med Pharmacol Sci. 2018;22:6824-31
101. Huo X, Han S, Wu G, Latchoumanin O, Zhou G, Hebbard L. et al. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Molecular cancer. 2017;16:165-75
102. Yu HM, Wang C, Yuan Z, Chen GL, Ye T, Yang BW. LncRNA NEAT1 promotes the tumorigenesis of colorectal cancer by sponging miR-193a-3p. Cell Prolif. 2019;52:e12526
103. Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H. et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer research. 2015;35:1385-8
104. Zhang M, Li Y, Wang H, Yu W, Lin S, Guo J. LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer biology & therapy. 2019;20:524-36
105. Jiang Y, Lin L, Zhong S, Cai Y, Zhang F, Wang X. et al. Overexpression of novel lncRNA NLIPMT inhibits metastasis by reducing phosphorylated glycogen synthase kinase 3β in breast cancer. Journal of cellular physiology. 2019;234:10698-708
106. Raeisi F, Abolfathi M, Ahmadi-Naji R, Iranparast S, Noshadi E, Akbari A. et al. ARA lncRNA, is upregulated in liver and breast tumor tissues. Molecular biology reports. 2018;46:77-82
107. Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J. et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46-55
108. Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016;7:e2389
109. Cai L, Lv J, Zhang Y, Li J, Wang Y, Yang H. The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J Cell Mol Med. 2017;21:2654-62
110. Li T, Xiao Y, Huang T. HIF-1α-induced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncology reports. 2018;39:1072-80
111. Ma B, Li M, Zhang L, Huang M, Lei JB, Fu GH. et al. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016;37:4445-55
112. Wen JJ, Ma YD, Yang GS, Wang GM. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2017;21:498-503
113. Grunewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H. et al. Ewing sarcoma. Nature reviews Disease primers. 2018;4:5-27
114. Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M. et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162-5
115. Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML. et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326-41
116. Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A. et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342-53
117. Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S. et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS genetics. 2014;10:e1004475
118. Klega K, Imamovic-Tuco A, Ha G, Clapp AN, Meyer S, Ward A. et al. Detection of Somatic Structural Variants Enables Quantification and Characterization of Circulating Tumor DNA in Children With Solid Tumors. JCO Precis Oncol. 2018. 2018
119. West DC, Grier HE, Swallow MM, Demetri GD, Granowetter L, Sklar J. Detection of circulating tumor cells in patients with Ewing's sarcoma and peripheral primitive neuroectodermal tumor. J Clin Oncol. 1997;15:583-8
120. de Alava E, Lozano MD, Patino A, Sierrasesumaga L, Pardo-Mindan FJ. Ewing family tumors: potential prognostic value of reverse-transcriptase polymerase chain reaction detection of minimal residual disease in peripheral blood samples. Diagn Mol Pathol. 1998;7:152-7
121. Avigad S, Cohen IJ, Zilberstein J, Liberzon E, Goshen Y, Ash S. et al. The predictive potential of molecular detection in the nonmetastatic Ewing family of tumors. Cancer. 2004;100:1053-8
122. Allegretti M, Casini B, Mandoj C, Benini S, Alberti L, Novello M. et al. Precision diagnostics of Ewing's sarcoma by liquid biopsy: circulating EWS-FLI1 fusion transcripts. Ther Adv Med Oncol. 2018;10:1-9
123. Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in Liquid Biopsy - Current Status and Where We Need to Progress. Computational and structural biotechnology journal. 2018;16:190-5
124. Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM. et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629-38
125. Okochi O, Hibi K, Uemura T, Inoue S, Takeda S, Kaneko T. et al. Detection of mitochondrial DNA alterations in the serum of hepatocellular carcinoma patients. Clin Cancer Res. 2002;8:2875-8
126. Yu M, Wan YF, Zou QH. Cell-free circulating mitochondrial DNA in the serum: a potential non-invasive biomarker for Ewing's sarcoma. Arch Med Res. 2012;43:389-94
127. Nie CL, Ren WH, Ma Y, Xi JS, Han B. Circulating miR-125b as a biomarker of Ewing's sarcoma in Chinese children. Genetics and molecular research: GMR. 2015;14:19049-56
128. Li J, You T, Jing J. MiR-125b inhibits cell biological progression of Ewing's sarcoma by suppressing the PI3K/Akt signalling pathway. Cell Prolif. 2014;47:152-60
129. Iida K, Fukushi J, Matsumoto Y, Oda Y, Takahashi Y, Fujiwara T. et al. miR-125b develops chemoresistance in Ewing sarcoma/primitive neuroectodermal tumor. Cancer Cell Int. 2013;13:21-35
130. Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F. et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. The Journal of pathology. 2011;224:334-43
131. Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F. et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nature genetics. 2011;43:1262-5
132. Lugowska I, Teterycz P, Mikula M, Kulecka M, Kluska A, Balabas A. et al. IDH1/2 Mutations Predict Shorter Survival in Chondrosarcoma. J Cancer. 2018;9:998-1005
133. Gutteridge A, Rathbone VM, Gibbons R, Bi M, Archard N, Davies KEJ. et al. Digital PCR analysis of circulating tumor DNA: a biomarker for chondrosarcoma diagnosis, prognostication, and residual disease detection. Cancer Med. 2017;6:2194-202
134. Tarpey PS, Behjati S, Cooke SL, Van Loo P, Wedge DC, Pillay N. et al. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nature genetics. 2013;45:923-6
135. Totoki Y, Yoshida A, Hosoda F, Nakamura H, Hama N, Ogura K. et al. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome research. 2014;24:1411-20
136. Samuel AM, Costa J, Lindskog DM. Genetic alterations in chondrosarcomas - keys to targeted therapies? Cellular oncology (Dordrecht). 2014;37:95-105
137. Schrage YM, Briaire-de Bruijn IH, de Miranda NF, van Oosterwijk J, Taminiau AH, van Wezel T. et al. Kinome profiling of chondrosarcoma reveals SRC-pathway activity and dasatinib as option for treatment. Cancer Res. 2009;69:6216-22
138. Chow WA. Chondrosarcoma: biology, genetics, and epigenetics. F1000Research. 2018;7:1826-35
139. Sun X, Charbonneau C, Wei L, Chen Q, Terek RM. miR-181a Targets RGS16 to Promote Chondrosarcoma Growth, Angiogenesis, and Metastasis. Molecular cancer research: MCR. 2015;13:1347-57
140. Zhu Z, Wang CP, Zhang YF, Nie L. MicroRNA-100 resensitizes resistant chondrosarcoma cells to cisplatin through direct targeting of mTOR. Asian Pacific journal of cancer prevention: APJCP. 2014;15:917-23
141. Jin Z, Han YX, Han XR. Loss of RUNX3 expression may contribute to poor prognosis in patients with chondrosarcoma. Journal of molecular histology. 2013;44:645-52
142. Nazeri E, Gouran Savadkoohi M, Majidzadeh AK, Esmaeili R. Chondrosarcoma: An overview of clinical behavior, molecular mechanisms mediated drug resistance and potential therapeutic targets. Critical reviews in oncology/hematology. 2018;131:102-9
143. Panagopoulos I, Mertens F, Isaksson M, Domanski HA, Brosjo O, Heim S. et al. Molecular genetic characterization of the EWS/CHN and RBP56/CHN fusion genes in extraskeletal myxoid chondrosarcoma. Genes Chromosomes Cancer. 2002;35:340-52
144. Nyquist KB, Panagopoulos I, Thorsen J, Haugom L, Gorunova L, Bjerkehagen B. et al. Whole-transcriptome sequencing identifies novel IRF2BP2-CDX1 fusion gene brought about by translocation t(1;5)(q42;q32) in mesenchymal chondrosarcoma. PloS one. 2012;7:e49705-12
145. Wang L, Motoi T, Khanin R, Olshen A, Mertens F, Bridge J. et al. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer. 2012;51:127-39
146. Thoenen E, Curl A, Iwakuma T. TP53 in bone and soft tissue sarcomas. Pharmacol Ther. 2019;202:149-64
147. Jung J, Lee JS, Dickson MA, Schwartz GK, Le Cesne A, Varga A. et al. TP53 mutations emerge with HDM2 inhibitor SAR405838 treatment in de-differentiated liposarcoma. Nat Commun. 2016;7:12609-16
148. Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes & development. 1993;7:1126-32
149. Fricke A, Cimniak AFV, Ullrich PV, Becherer C, Bickert C, Pfeifer D. et al. Whole blood miRNA expression analysis reveals miR-3613-3p as a potential biomarker for dedifferentiated liposarcoma. Cancer biomarkers: section A of Disease markers. 2018;22:199-207
150. Hashimoto N, Myoui A, Araki N, Asai T, Sonobe H, Hirota S. et al. Detection of SYT-SSX fusion gene in peripheral blood from a patient with synovial sarcoma. Am J Surg Pathol. 2001;25:406-10
151. Ogino S, Konishi H, Ichikawa D, Hamada J, Shoda K, Arita T. et al. Detection of fusion gene in cell-free DNA of a gastric synovial sarcoma. World journal of gastroenterology. 2018;24:949-56
152. Mihály D, Nagy N, Papp G, Pápai Z, Sápi Z. Release of circulating tumor cells and cell-free nucleic acids is an infrequent event in synovial sarcoma: liquid biopsy analysis of 15 patients diagnosed with synovial sarcoma. Diagnostic pathology. 2018;13:81-9
153. Przybyl J, van de Rijn M, Rutkowski P. Detection of SS18-SSX1/2 fusion transcripts in circulating tumor cells of patients with synovial sarcoma. Diagn Pathol. 2019;14:24
154. Hale R, Sandakly S, Shipley J, Walters Z. Epigenetic Targets in Synovial Sarcoma: A Mini-Review. Front Oncol. 2019;9:1078
155. Uotani K, Fujiwara T, Yoshida A, Iwata S, Morita T, Kiyono M. et al. Circulating MicroRNA-92b-3p as a Novel Biomarker for Monitoring of Synovial Sarcoma. Scientific reports. 2017;7:14634-45
156. Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nature genetics. 1993;3:113-7
157. Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM. et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol. 2002;20:2672-9
158. Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869-72
159. Eguchi-Ishimae M, Tezuka M, Kokeguchi T, Nagai K, Moritani K, Yonezawa S. et al. Early detection of the PAX3-FOXO1 fusion gene in circulating tumor-derived DNA in a case of alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2019;58:521-9
160. Tombolan L, Zin A, Bisogno G. Cell-Free DNA in Pediatric Rhabdomyosarcoma: Potential and Challenges. Methods Mol Biol. 2019;1909:165-75
161. Goldstein M, Meller I, Issakov J, Orr-Urtreger A. Novel genes implicated in embryonal, alveolar, and pleomorphic rhabdomyosarcoma: a cytogenetic and molecular analysis of primary tumors. Neoplasia (New York, NY). 2006;8:332-43
162. Miyachi M, Tsuchiya K, Yoshida H, Yagyu S, Kikuchi K, Misawa A. et al. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochemical and biophysical research communications. 2010;400:89-93
163. Danielson LS, Menendez S, Attolini CS, Guijarro MV, Bisogna M, Wei J. et al. A differentiation-based microRNA signature identifies leiomyosarcoma as a mesenchymal stem cell-related malignancy. The American journal of pathology. 2010;177:908-17
164. Beck AH, Lee CH, Witten DM, Gleason BC, Edris B, Espinosa I. et al. Discovery of molecular subtypes in leiomyosarcoma through integrative molecular profiling. Oncogene. 2010;29:845-54
165. Guo X, Jo VY, Mills AM, Zhu SX, Lee CH, Espinosa I. et al. Clinically Relevant Molecular Subtypes in Leiomyosarcoma. Clin Cancer Res. 2015;21:3501-11
166. Hemming ML, Klega KS, Rhoades J, Ha G, Acker KE, Andersen JL. et al. Detection of Circulating Tumor DNA in Patients With Leiomyosarcoma With Progressive Disease. JCO Precis Oncol. 2019;2019:1-11
167. Varshney J, Subramanian S. MicroRNAs as potential target in human bone and soft tissue sarcoma therapeutics. Frontiers in molecular biosciences. 2015;2:31-42
168. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature genetics. 2006;38:228-33
169. Nuovo GJ, Schmittgen TD. Benign metastasizing leiomyoma of the lung: clinicopathologic, immunohistochemical, and micro-RNA analyses. Diagn Mol Pathol. 2008;17:145-50
170. Guled M, Pazzaglia L, Borze I, Mosakhani N, Novello C, Benassi MS. et al. Differentiating soft tissue leiomyosarcoma and undifferentiated pleomorphic sarcoma: A miRNA analysis. Genes Chromosomes Cancer. 2014;53:693-702
171. Ravegnini G, Sammarini G, Serrano C, Nannini M, Pantaleo MA, Hrelia P. et al. Clinical relevance of circulating molecules in cancer: focus on gastrointestinal stromal tumors. Ther Adv Med Oncol. 2019;11:1-19
172. Joensuu H, Wardelmann E, Sihto H, Eriksson M, Sundby Hall K, Reichardt A. et al. Effect of KIT and PDGFRA Mutations on Survival in Patients With Gastrointestinal Stromal Tumors Treated With Adjuvant Imatinib: An Exploratory Analysis of a Randomized Clinical Trial. JAMA oncology. 2017;3:602-9
173. Subramanian S, West RB, Corless CL, Ou W, Rubin BP, Chu KM. et al. Gastrointestinal stromal tumors (GISTs) with KIT and PDGFRA mutations have distinct gene expression profiles. Oncogene. 2004;23:7780-90
174. Reichardt P, Demetri GD, Jeffers M, Kang YK, Blay JY, Rutkowski P. et al. Mutational analysis of plasma DNA from patients (pts) in the phase III GRID study of regorafenib (REG) versus placebo (PL) in tyrosine kinase inhibitor (TKI)-refractory GIST: Correlating genotype with clinical outcomes. 2013; 37: 58.
175. Maier J, Lange T, Kerle I, Specht K, Bruegel M, Wickenhauser C. et al. Detection of mutant free circulating tumor DNA in the plasma of patients with gastrointestinal stromal tumor harboring activating mutations of CKIT or PDGFRA. Clin Cancer Res. 2013;19:4854-67
176. Bauer S, Herold T, Muehlenberg T, Reis AC, Grunewald S. Plasma sequencing to detect a multitude of secondary KIT resistance mutations in metastatic gastrointestinal stromal tumors (GIST). Annual Meeting of the American-Society-of-Clinical-Oncology (ASCO). 2015
177. Kang G, Bae BN, Sohn BS, Pyo JS, Kang GH, Kim KM. Detection of KIT and PDGFRA mutations in the plasma of patients with gastrointestinal stromal tumor. Targeted oncology. 2015;10:597-601
178. Wada N, Kurokawa Y, Takahashi T, Hamakawa T, Hirota S, Naka T. et al. Detecting Secondary C-KIT Mutations in the Peripheral Blood of Patients with Imatinib-Resistant Gastrointestinal Stromal Tumor. Oncology. 2016;90:112-7
179. Kang G, Sohn BS, Pyo JS, Kim JY, Lee B, Kim KM. Detecting Primary KIT Mutations in Presurgical Plasma of Patients with Gastrointestinal Stromal Tumor. Molecular diagnosis & therapy. 2016;20:347-51
180. Boonstra PA, Ter Elst A, Tibbesma M, Bosman LJ, Mathijssen R, Atrafi F. et al. A single digital droplet PCR assay to detect multiple KIT exon 11 mutations in tumor and plasma from patients with gastrointestinal stromal tumors. Oncotarget. 2018;9:13870-83
181. Smolle MA, Leithner A, Posch F, Szkandera J, Liegl-Atzwanger B, Pichler M. MicroRNAs in Different Histologies of Soft Tissue Sarcoma: A Comprehensive Review. Int J Mol Sci. 2017 18
182. Kim JH, Jeon S, Shin BA. MicroRNA-29 Family Suppresses the Invasion of HT1080 Human Fibrosarcoma Cells by Regulating Matrix Metalloproteinase 2 Expression. Chonnam medical journal. 2017;53:161-7
183. Liu P, Wilson MJ. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-kappaB factor in human fibrosarcoma cells. J Cell Physiol. 2012;227:867-76
184. Young RJ, Brown NJ, Reed MW, Hughes D, Woll PJ. Angiosarcoma. The Lancet Oncology. 2010;11:983-91
185. Megquier K, Turner-Maier J, Swofford R, Kim JH, Sarver AL, Wang C. et al. Comparative Genomics Reveals Shared Mutational Landscape in Canine Hemangiosarcoma and Human Angiosarcoma. Molecular cancer research: MCR. 2019;17:2410-21
186. Murali R, Chandramohan R, Moller I, Scholz SL, Berger M, Huberman K. et al. Targeted massively parallel sequencing of angiosarcomas reveals frequent activation of the mitogen activated protein kinase pathway. Oncotarget. 2015;6:36041-52
187. Huang SC, Zhang L, Sung YS, Chen CL, Kao YC, Agaram NP. et al. Recurrent CIC Gene Abnormalities in Angiosarcomas: A Molecular Study of 120 Cases With Concurrent Investigation of PLCG1, KDR, MYC, and FLT4 Gene Alterations. Am J Surg Pathol. 2016;40:645-55
188. Sarver AL, Phalak R, Thayanithy V, Subramanian S. S-MED: sarcoma microRNA expression database. Laboratory investigation; a journal of technical methods and pathology. 2010;90:753-61
189. Italiano A, Thomas R, Breen M, Zhang L, Crago AM, Singer S. et al. The miR-17-92 cluster and its target THBS1 are differentially expressed in angiosarcomas dependent on MYC amplification. Genes Chromosomes Cancer. 2012;51:569-78
190. De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N. et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247-51
191. Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M. et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nature genetics. 2014;46:1227-32
192. Presneau N, Eskandarpour M, Shemais T, Henderson S, Halai D, Tirabosco R. et al. MicroRNA profiling of peripheral nerve sheath tumours identifies miR-29c as a tumour suppressor gene involved in tumour progression. Br J Cancer. 2013;108:964-72
193. Chai G, Liu N, Ma J, Li H, Oblinger JL, Prahalad AK. et al. MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer science. 2010;101:1997-2004
194. Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic acids research. 2009;37:123-8
195. Demoret B, Gregg J, Liebner DA, Tinoco G, Lenobel S, Chen JL. Prospective Evaluation of the Concordance of Commercial Circulating Tumor DNA Alterations with Tumor-Based Sequencing across Multiple Soft Tissue Sarcoma Subtypes. Cancers. 2019;11:1829-38
196. Wong P, Hui A, Su J, Yue S, Haibe-Kains B, Gokgoz N. et al. Prognostic microRNAs modulate the RHO adhesion pathway: A potential therapeutic target in undifferentiated pleomorphic sarcomas. Oncotarget. 2015;6:39127-39
197. Chang L, Asatrian G, Dry SM, James AW. Circulating tumor cells in sarcomas: a brief review. Medical oncology (Northwood, London, England). 2015;32:430-7
Author contact
![]() Corresponding author: Prof. Jilong Yang, PhD, MD, Phone: +86-18622221626; Fax: +86-22-23340123-1081; E-mail: yangjilongcom
Corresponding author: Prof. Jilong Yang, PhD, MD, Phone: +86-18622221626; Fax: +86-22-23340123-1081; E-mail: yangjilongcom

 Global reach, higher impact
Global reach, higher impact