3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(18):5281-5288. doi:10.7150/jca.46150 This issue Cite
Research Paper
Genetic Variants in DNA Mismatch Repair Pathway predict prognosis of Lung Cancer patients with receiving Platinum-Based Chemotherapy
1. Department of Orthopaedics, The First Affiliated Hospital of the University of South China, Hengyang 421001, China.
2. National Institution of Drug Clinical Trial, Xiangya Hospital, Central South University, Changsha, Hunan, P.R.China.
3. Departments of Clinical Pharmacology, Xinagya Hospital, Central South University, Changsha 410008, China.
4. Institute of Clinical Pharmacology and Hunan Key Laboratory of Pharmacogenetics, Central South University, Changsha 410078, China.
5. Department of Medical Oncology, Lung cancer and Gastrointestinal unit, Hunan Cancer Hospital, Affiliated Cancer Hospital of Xiangya School of Medicine, Changsha 410013, China.
6. Hunan clinical research center in gynecologic cancer, Hunan Cancer Hospital, Affiliated Cancer Hospital of Xiangya School of Medicine, Changsha 410013, China.
7. Department of Pharmacy, Xinagya Hospital, Central South University, Changsha 410008, China.
Received 2020-3-19; Accepted 2020-6-15; Published 2020-7-9
Abstract
Objective: To investigate the relationships between genetic variants in DNA mismatch repair pathway genes and the prognosis of platinum-based chemotherapy in lung cancer patients.
Methods: 346 lung cancer patients who received at least two cycles of platinum-based chemotherapy were recruited in this study. A total of 35 single nucleotide polymorphisms in 7 DNA mismatch repair genes were genotyped to investigate their associations with platinum-based chemotherapy prognosis.
Result: The results revealed that patients carried MSH2 rs4608577 TT genotype had a significantly shorter progression free survival than patients with GG or GT genotypes (Additive model: P=0.003, OR =0.94, 95% CI =0.33-1.57). Patients with SAPCD1 rs707937 TT genotype had a significantly longer overall survival than patients with GG or GT genotypes (Additive model: P=0.0003, OR=0.75, 95% CI =0.35-1.14). Eight SNPs and fourteen SNPs were related to progression free survival and overall survival in subgroup analyses, respectively.
Conclusion: Our findings suggest that the MSH2 rs4608577 and SAPCD1 rs707937 may be potential clinical biomarkers for predicting platinum-based chemotherapy prognosis in lung cancer patients.
Keywords: lung cancer, platinum-based chemotherapy prognosis, genetic polymorphisms, MSH2, SAPCD1, DNA mismatch repair
Introduction
Lung cancer is the main causes of cancer-related mortality in the worldwide [1]. Two major pathologic types of lung cancer are non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), and NSCLC counted for about 85% of all cases [2, 3]. Many new therapeutic agents such as gefitinib and erlotinib for target therapy were used for NSCLC patients, but it based on the genomic mutations of the patients. Thus, platinum-based chemotherapy regimens were still widely used for treatment of NSCLC in clinic [4-7]. However, the prognosis of the lung cancer patients is still dissatisfactory and obstacle the treatment [8, 9]. The tumor microenvironment state and clinical stage are the traditional prognostic factors [10-13], and many factors including genetic polymorphism, histology type, clinical stage, gender, age, and so on have been reported to be associated with prognosis of platinum-based chemotherapy [14-20]. It is important to discern some new biomarkers that can be used to predict the prognosis of platinum-based chemotherapy.
The DNA repair has four major repair pathways including base excision repair (BER), nucleotide excision repair (NER), double-strand break repair (DSBR) and DNA mismatch repair (MMR). DNA mismatch repair (MMR) is a very important repair system which actuates DNA damage response [21, 22]. It can correct errors during DNA replication. MMR proteins have been collected together to form protein complexes that recognize and digest mismatch pieces of DNA and eventually fill in the mismatches [23-27]. Informally, MutLα plays a key role in the DNA mismatch repair system, and the endonuclease activity is related to DNA mismatch repair [28]. The complexes of MutLα and the MUTSα complex can guide an act on repairing the single-base and small-loop mismatches, and the MutSβ complex also guides the repair of small-to large-loop mismatches [29]. Therefore, MutSα and MutLα are important proteins in the MMR system and are used to cooperate the mismatch detection and repair events. In previous study, we found that MutSα and MutLα genes were associated with the toxicity of platinum-based chemotherapeutic in lung cancer [30, 31].
Recent study showed that mutations in DNA repair genes are related to the prognosis of platinum-based chemotherapy in NSCLC patients [32, 33]. MMR plays a key role in removing cisplatin-DNA adducts that the formation of platinum/DNA adducts blocks replication and transcription of DNA, and the mutations of MMR genes can decrease the response to platinum-based therapies and influence the prognosis through the highly conserved biological pathway [34-37]. The genetic polymorphisms of DNA mismatch repair genes were useful clinical biomarkers to predict chemotherapy response and prognosis in lung cancer patients [38].
In the current study, we genotyped 35 single-nucleotide polymorphisms (SNPs) in 7 MMR genes (MLH1, MSH2, MSH3, MSH4, MSH5, SAPCD1 and MSH6) in lung cancer patients, and evaluate the relationships between these genetic polymorphisms and platinum-based chemotherapy prognosis.
Materials and Methods
Subjects and follow up
All patients were selected by the following inclusion criteria: (1) patients newly diagnosed with lung cancer by histological examination at the Affiliated Cancer Hospital or Xiangya Hospital of Central South University (Changsha, Hunan, China) between August 2009 and January 2013; (2) patients who received at least 2 cycles of platinum-based chemotherapy; (3) patients with no history of surgery before chemotherapy. All patients provided written informed consent before they participated in this study. The study performed in accordance with the principles of Declaration of Helsinki and was approved by the Ethics Committee of Xiangya School of Medicine, Central South University.
The terminate date for patient follow-up was July 15, 2019. Survival data were collected by telephone follow-up or residence registration. Overall survival (OS) time was defined as the period between the date diagnosed of lung cancer and the date of last follow-up or death. Progression-free survival (PFS) was calculated from the date diagnosed of lung cancer until the date of the first local recurrence or metastases in the last follow-up. Patients without progression were censored at the date of the last contact.
SNP selection, DNA extraction and genotyping
All of the common genetic variants in MLH1, MSH2, MSH3, MSH4, MSH5, SAPCD1 and MSH6 were selected by Haploview (Broad Institute, Cambridge, MA, USA) using pair-wise tagging with default settings (pairwise r2 threshold = 0.8). SNPs with a minor allele frequency (MAF) ≥ 5% in the Han Chinese population were selected. Finally, Thirty-five SNPs were genotyped in the patients (Table 1). In our previous studies, the SNPs were investigated to be associated with platinum-based chemotherapy toxicity [30].
ALL blood samples were collected in the morning and stored in EDTA tube. Genomic DNA was isolated using a Genomic DNA Purification Kit (Promega, Madison, WI, USA) and stored at -20°C before use. Genotyping was analyzed using a Sequenom Mass ARRAY Genotyping Platform (Sequenom, San Diego, CA, USA) through polymerase chain reaction (PCR) system.
Statistical analysis
The covariates for logistic regression included sex, age, smoking status, histology and clinical stage. We applied three genetic models (dominant, additive, and recessive) to evaluate the associations between the single-nucleotide polymorphisms and prognosis of the patients. The cumulative PFS and OS were estimated by using the log-rank test or univariate COX regression. All analyses were performed using PLINK (version 1.9, http://pngu.mgh.harvard.edu/purcell/plink/) and SPSS 18.0 software (SPSS Inc, Chicago, IL, USA). Odds ratios (OR) and their 95% confidence intervals (95% CI) were used to assess the associations between gene polymorphisms and prognosis. P<0.05 was considered statistically significant.
The 35 single nucleotide polymorphisms examined in this study
| Genes | SNPs | Alleles | Call rate (%)# | MAF# |
|---|---|---|---|---|
| MLH1 | rs1800734 | G/A | 98.55 | 0.42 |
| rs1540354 | A/T | 97.98 | 0.32 | |
| MSH2 | rs2303428 | T/A, C, G | 98.84 | 0.36 |
| rs10191478 | G/T | 98.27 | 0.19 | |
| rs1981929 | G/A,C | 97.11 | 0.13 | |
| rs4608577 | T/G | 97.40 | 0.13 | |
| rs6544991 | A/C | 95.09 | 0.35 | |
| rs7602094 | T/C | 95.09 | 0.32 | |
| rs13019654 | G/A, T | 97.11 | 0.28 | |
| rs12999145 | A/G, T | 99.13 | 0.46 | |
| rs4952887 | C/T | 98.84 | 0.15 | |
| MSH3 | rs6151627 | A/G | 99.13 | 0.26 |
| rs394592 | G/A | 99.13 | 0.49 | |
| rs26784 | A/G | 98.27 | 0.37 | |
| rs7709909 | C/T | 99.13 | 0.29 | |
| rs1650665 | C/T | 99.71 | 0.04 | |
| rs245340 | G/A, T | 97.69 | 0.27 | |
| rs245346 | A/G | 98.27 | 0.42 | |
| rs26778 | A/T | 96.53 | 0.42 | |
| rs3816729 | A/G | 95.66 | 0.31 | |
| rs6151670 | C/G | 98.55 | 0.26 | |
| rs6151892 | T/A | 99.13 | 0.33 | |
| rs6151914 | C/T | 98.27 | 0.10 | |
| MSH4 | rs1146642 | A/C, G | 98.27 | 0.03 |
| rs3806162 | T/G | 99.13 | 0.23 | |
| rs5745532 | C/T | 98.84 | 0.25 | |
| MSH5 | rs707939 | C/A | 100 | 0.38 |
| rs1150793 | A/G | 99.42 | 0.04 | |
| rs3117572 | A/C, G | 99.13 | 0.28 | |
| rs707938 | A/G | 97.69 | 0.30 | |
| rs409558 | T/C | 100 | 0.13 | |
| MSH6 | rs2020910 | T/A | 98.55 | 0.18 |
| rs2348244 | T/C | 99.42 | 0.37 | |
| rs3136329 | C/T | 97.98 | 0.13 | |
| SAPCD1 | rs707937 | C/G | 96.53 | 0.43 |
MAF: Minor allele frequency;
#Data in this study.
Distribution of characteristics in lung cancer patients and prognosis analysis
| Variables | Patients (N%) | Death (N%) | MST-OS (year) | MST-PFS (year) |
|---|---|---|---|---|
| Age | ||||
| ≤55 | 154 (44.5) | 127 (45.5) | 4.52 | 3.09 |
| >55 | 192 (55.5) | 152 (54.5) | 4.35 | 3.21 |
| Gender | ||||
| Male | 286 (82.7) | 229 (82.1) | 4.42 | 2.93 |
| Female | 60 (17.3) | 50 (17.9) | 4.77 | 4.47 |
| Histology | ||||
| NSCLC | 234 (67.6) | 189 (67.7) | 4.58 | 3.25 |
| SCLC | 100 (28.9) | 80 (28.7) | 4.35 | 3.10 |
| Smoking status | ||||
| Non-smoker | 122 (35.3) | 97 (34.8) | 4.83 | 4.60 |
| Smoker | 224 (64.7) | 182 (65.2) | 4.13 | 2.61 |
| Stage | ||||
| I/II/LD | 47 (13.6) | 39 (14.0) | 3.80 | 3.19 |
| III/IV/ED | 294 (85.0) | 240 (86.0) | 4.53 | 3.16 |
MST, median survival time; LD, Limitation Disease; ED, Extensive Disease.
Results
Demographic characteristics of patient characteristics
A total of 346 patients who received first-line platinum-based chemotherapy were collected for this study. As shown in Table 2, the median age was 52 years old (a range of 21 to 77 years old). Among them, 286 (82.7%) were males and 60 (17.3%) were females. There were 224 (64.7%) patients who had ever smoked, and the rest 122 (35.3%) patients never smoked. For histology, 234 (67.6%) patients were diagnosed with NSCLC, and 110 (28.9%) with NSCLC. 294 (85%) of the patients were in an advanced stage (stage III/IV/ED). The fundamental clinical information, outcomes of progression-free survival (PFS) and Overall survival (OS) of these lung cancer patients are summarized in Table 2.
Association of the MSH2 rs4608577 polymorphisms and PFS in lung cancer patients
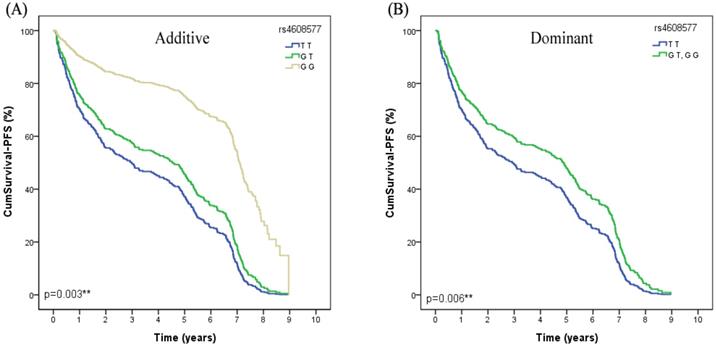
In the univariate Cox regression analysis, we found that MSH2 rs4608577 was significantly associated with PFS of lung cancer patients received platinum-based chemotherapy. After adjusted by age, smoking status, gender, histological type and stage, rs4608577 still significantly contributed to PFS of lung cancer patients (Additive model: P=0.003, OR =0.94, 95% CI =0.33-1.57; Dominant model: P=0.006, OR=0.94, 95%CI=0.27-1.61) (Table 3). Patients carrying TT genotype had a significantly shorter PFS than patients carrying GG or GT genotypes (Figure 1).
Association of the SAPCD1 rs707937 polymorphisms and OS in lung cancer patients
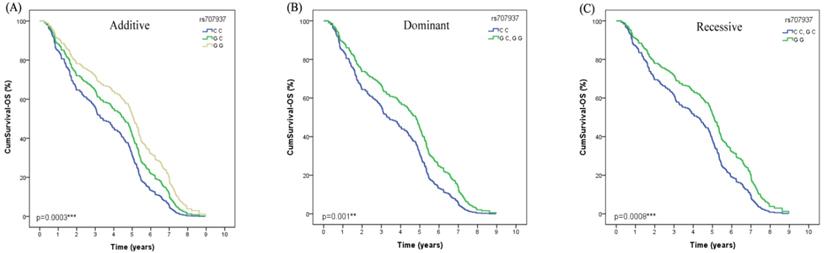
Both in the univariate Cox regression analysis and after adjusted by age, gender, smoking status, histological type and stage, we found that SAPCD1 rs707937 was significantly associated with OS of lung cancer patients received platinum-based chemotherapy (Additive model: P=0.0003, OR=0.75, 95% CI =0.35-1.14; Dominant model: P=0.001, OR=0.97, 95% CI =0.38-1.56; recessive model: P=0.008, OR=0.98, 95% CI =0.26-1.70) (Table 4). In additive analysis model, patients who carried the rs707937 GG genotype had a significantly longer OS than patients with CC genotype. In dominant analysis model, patients who carried rs707937 GG and GC genotypes had a significantly longer OS than patients with CC genotype. In recessive analysis model, patients who carried rs707937 GG genotype had a significantly longer OS than patients with CC and GC genotypes (Figure 2).
Association of the MSH2 rs4608577 polymorphisms and PFS in lung cancer patients
| Gene | Polymorphism | Genotype | MPFS (year) | Additive | Dominant | Recessive | |||
|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI) | p value | OR (95%CI) | p value | ||||
| MSH2 | rs4608577 | GG | 7.97 | 0.94 (0.33-1.57) | 0.003** | 0.94 (0.27-1.61) | 0.006** | 2.48 (-0.20-5.17) | 0.071 |
| GT | 4.94 | ||||||||
| TT | 2.67 | ||||||||
MPFS, median progression-free survival; OR, odds ratio; CI, confidence interval; Additive model: comparison between minor allele subjects and major allele subjects. Dominant model: comparison between minor allele carriers and major homozygous subjects. Recessive model: comparison between major allele carriers and minor homozygous subjects. ** P < 0.01.
The MSH2 rs4608577 is significantly associcated with PFS in lung cancer patients treated with platinum-based chemotherapy. (A) Patients carry the genotype of MSH2 rs4608577 GG have a longer surival time than GT than TT in Additive model (p=0.003**). (B) Patients carry the genotype of MSH2 rs4608577 GG or GT have a longer surival time than TT in Dominant model (p=0.006**).
The polymorphism of SAPCD1 rs707937 is greatly related to OS in lung cancer patients of platinum-based chemotherapy treatment. (A) Patients with GG genotype have a longer overall survival time than GC than CC in Additive model (p=0.0003***). (B) Patients with GG or GC genotype have a longer overall survival time than CC in Dominant model in lung cancer patients (p=0.001**). (C) Patients with GG genotype have a longer overall survival time than GC or CC in Recessive model in lung cancer patients (p=0.0008***).
Stratification analyses
Stratification analyses were used to investigate the associations of all 35 SNPs with PFS and OS in the subgroups stratified by age, smoking status, gender, histological type and stage. For PFS subgroup analyses, we found the SNPs were related to PFS of lung cancer patients received platinum-based chemotherapy as follows: MSH2 rs4608577 and FBXO11 rs2710163 in patients with age >55 years old; MSH3 rs6151914 in patients with age ≤55 years old; MSH2 rs4608577 and MSH6 rs3136329 in male patients; CLIC1 rs2293852 in patients with smoking; MSH2 rs4608577, MSH3 rs6151914, MSH4 rs3806162, CLIC1 rs2293852 and rs805304 in NSCLC patients; MSH3 rs1650665, rs26784 and rs6151670, and MLH1 rs1800734 in SCLC patients; MSH2 rs4608577, MSH6 rs3136329 and CLIC1 rs805304 in lung cancer patients with advanced or extensive stage (Table 5). For OS subgroup analyses, we found the SNPs were associated with OS of lung cancer patients received platinum-based chemotherapy in different subgroups as follows: MSH2 rs2302428, MSH3 rs6151914 and rs245346, MSH4 rs5745532 and SAPCD1 rs707937 in patients with ≤55 years old; MSH3 rs245346 in patients with >55 years old; MSH2 rs6544991, MSH3 rs245346 and rs6151892 and MSH6 rs2020910 in male patients; MSH3 rs6151892 and rs3816729, and MSH4 rs5745532 in patients with non-smoking; MSH3 rs26778, FBXO11 rs2710163 and SAPCD1 rs707937 in NSLCL patients; MSH2 rs2303428, MSH3 rs6151914, MSH5 rs707938 and rs31175772 in SCLC patients; MSH3 rs6151914, MSH4 rs3806162 and FBXO11 rs3732190 in patients with early or limited stage; MSH2 rs6544991 and rs13019654, and SAPCD1 rs707939 in patients with advanced or extensive stage (Table 6).
Discussion
In this study, we emphasize to investigate whether polymorphisms of 7 MMR genes (MLH1, MSH2, MSH3, MSH4, MSH5, MSH6 and SAPCD1) were related to the prognosis of platinum-based chemotherapy in 346 lung cancer patients. Our results showed that MSH2 rs4608577 and SAPCD1 rs707937 were significantly related to the prognosis of lung cancer patients received platinum-based chemotherapy. Patients with GG genotype of MSH2 rs4608577 had better PFS, and patients with GG genotype of SAPCD1 rs707937 would have better OS.
In the subgroup analyses for PFS, MSH2 rs4608577, MSH3 rs1650665, rs26784, rs6151670, rs6151914; MSH4 rs3806162, MSH6 rs3136329, MLH1 rs1800734 were significantly associated with PFS in different subgroups. In the patients with older age (>55), we founded that the GG and GT genotype carriers in rs4608577 had significantly longer PFS when compared with TT genotype. For male patients, the GG and GT genotype also had significantly longer PFS. In adenocarcinoma subgroup, the hazard ratio for death was 1.29. Those patients with advanced stage carrying GG or GT genotype had longer PFS. For MSH3 rs1650665, patients with squamous cell carcinoma carrying GA or GG had longer PFS. For MSH3 rs26784, patients with squamous cell carcinoma carrying CC had longer PFS when compared with CT and TT. For MSH3 rs6151670, patients with squamous cell carcinoma carrying GG or GC had shorter PFS when compared with CC. For MSH3 rs6151914, patients with age ≤55 years old carrying CC or CT had longer PFS. Patients with adenocarcinoma subgroup carrying CC or CT had longer PFS. For MSH4 rs3806162, Patients with adenocarcinoma subgroup carrying GG or GT had shorter PFS. For MSH6 rs3136329, in male patients carrying CC or CT had longer PFS when compared with TT genotype. Patients with advanced or extensive stage carrying TT had longer PFS. For MLH1 rs1800734, patients with squamous cell carcinoma carrying GG or GA had longer PFS.
In the subgroup analyses for OS, MSH2 rs2303428, rs6544991, rs13019654 and MSH3 rs6151914, rs245346, rs6151892, rs26778, rs3816729; MSH4 rs5745532, rs3806162 and MSH5 rs707938 rs3117572; MSH6 rs2020910 and SAPCD1 rs707937 were significantly associated with OS. We found the SNPs were related to OS of lung cancer patients received platinum-based chemotherapy as follows: MSH2 rs2303428, patients with younger age (≤55) carrying TT or TC had longer OS when compared with CC. Patients with squamous cell carcinoma carrying TT or TC had longer OS. For rs6544991, in male patients carrying CC or CA had longer OS. Patients with advanced or extensive stage carrying CC or CA had shorter OS when compared with AA genotype. For rs13019654, patients with advanced or extensive stage carrying GG or GT had longer OS. For MSH3 rs6151914, patients with younger age (≤55) carrying CC or CT had longer OS. Patients with squamous cell carcinoma carrying CC or CT had longer OS when compared with TT genotype. Patients with early or limited stage carrying CC had longer OS. For MSH3 rs245346, patients with younger age (≤55) carrying TT or TC had shorter OS(recessive model OR=2.58, 95% CI=1.01,6.60, P=0.049) and older age (>55) carrying TT or TC had longer OS. In male patients carrying TT had longer OS. For rs6151892, in male patients carrying AA had longer OS when compared with AT or TT. Patients with non-smoking carrying AA or AT had longer OS. For rs26778, patients with adenocarcinoma subgroup carrying TT or TA had shorter OS when compared with AA genotype. For rs3816729, patients with non-smoking carrying CC or CT had longer OS. For MSH4 rs5745532, patients with younger age (≤55) carrying TT or TC had longer OS and patients with non-smoking carrying TT or TC had longer OS. For rs3806162, patients with early or limited stage carrying GG or GT had longer OS. For MSH5 rs707938, patients with squamous cell carcinoma carrying TT or TC had longer OS. For rs3117572, patients with squamous cell carcinoma carrying AA had longer OS when compared with AT or TT genotype. For MSH6 rs2020910, in male patients carrying AA had longer OS when compared with AT or TT genotype. For SPACD1 rs707937, patients with younger age (≤55) carrying GG or GC had longer OS. Patients with advanced or extensive stage carrying GG or GC had longer OS and also patients with adenocarcinoma subgroup carrying GG or GC had longer OS when compared with CC genotype.
Association of the SAPCD1 rs707937 polymorphisms and OS in lung cancer patients
| Gene | Polymorphism | Genotype | MST (year) | Additive | Dominant | Recessive | |||
|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI) | p value | OR (95%CI) | p value | ||||
| SAPCD1 | rs707937 | GG | 4.72 | 0.75 (0.35-1.14) | 0.0003*** | 0.97 (0.38-1.56) | 0.001** | 0.98 (0.26-1.70) | 0.008** |
| GC | 4.80 | ||||||||
| CC | 4.05 | ||||||||
MST, median survival time; OR, odds ratio; CI, confidence interval; Additive model: comparison between minor allele subjects and major allele subjects. Dominant model: comparison between minor allele carriers and major homozygous subjects. Recessive model: comparison between major allele carriers and minor homozygous subjects. ** P < 0.01; *** P < 0.001.
Stratification analyses of Association between the eight polymorphisms and PFS in lung cancer patients
| Genes | SNPs | Subgroups | Additive | Dominant | Recessive | |||
|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI) | p value | OR (95%CI) | p value | |||
| MSH2 | rs4608577 | Age (>55) | 1.04 (0.22-1.86) | 0.014* | 1.10 (0.20-2.00) | 0.017* | 1.80 (-1.38-4.98) | 0.268 |
| Male | 0.86 (0.19-1.54) | 0.013* | 0.83 (0.09-1.53) | 0.028* | 2.61 (-0.08-5.31) | 0.059 | ||
| NSCLC | 1.29 (0.57-2.02) | 0.001* | 1.21 (0.42-2.00) | 0.003* | 4.45 (1.40-7.49) | 0.005* | ||
| Stage (III/IV/ED) | 0.84 (0.18-1.51) | 0.014* | 0.81 (0.09-1.54) | 0.029* | 2.51 (-0.18-5.20) | 0.068 | ||
| MSH3 | rs1650665 | SCLC | 1.82 (0.10-3.54) | 0.041* | 1.82 (0.10-3.54) | 0.041* | Ref | |
| rs26784 | SCLC | -0.95 (-1.82- -0.08) | 0.034* | -1.06 (-2.19-0.07) | 0.068 | -1.53 (-3.46-0.40) | 0.123 | |
| rs6151670 | SCLC | -0.24 (-1.18-0.70) | 0.619 | 0.17 (-0.94-1.29) | 0.761 | -2.65 (-5.14--0.16) | 0.040* | |
| rs6151914 | Age (≤55) | -1.20 (-2.19--0.21) | 0.019* | -1.32 (-2.50--0.14) | 0.030* | -2.48 (-5.55-0.59) | 0.116 | |
| NSCLC | -0.89 (-1.72--0.07) | 0.034* | -0.96 (-1.87--0.04) | 0.042* | -1.70 (-4.77-1.38) | 0.280 | ||
| MSH4 | rs3806162 | NSCLC | -0.05 (-0.61-0.51) | 0.866 | -0.32 (-0.97-0.33) | 0.336 | 1.66 (0.01-3.31) | 0.049* |
| MSH6 | rs3136329 | Male | 0.72 (0.07-1.36) | 0.030* | 0.68 (-0.06-1.43) | 0.074 | 2.10 (0.05-4.14) | 0.045* |
| Stage (II/IV/ED) | 0.32 (-0.32-0.97) | 0.326 | 0.16 (-0.59-0.91) | 0.675 | 2.08 (0.04-4.13) | 0.047* | ||
| MLH1 | rs1800734 | SCLC | 0.91 (0.10-1.72) | 0.029* | 1.33 (0.17-2.50) | 0.027* | 0.95 (-0.58-2.48) | 0.225 |
Additive model: comparison between minor allele subjects and major allele subjects. Dominant model: comparison between minor allele carriers and major homozygous subjects. Recessive model: comparison between major allele carriers and minor homozygous subjects. OR- odds ratio; CI- confidence interval; Ref.- reference. * P < 0.05.
Stratification analyses of Association between the fourteen polymorphisms and OS in lung cancer patients
| Genes | SNPs | Subgroups | Additive | Dominant | Recessive | |||
|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p value | OR (95%CI) | p value | OR (95%CI) | p value | |||
| MSH2 | rs2303428 | Age (≤55) | 1.60 (0.95-2.70) | 0.079 | 2.19 (1.04-4.61) | 0.039* | 1.33 (0.48-3.72) | 0.583 |
| SCLC | 1.71 (0.86-3.42) | 0.127 | 3.33 (1.09-10.16) | 0.034* | 1.05 (0.31-3.56) | 0.938 | ||
| rs6544991 | Male | 1.28 (0.86-1.89) | 0.222 | 2.04 (1.16-3.59) | 0.013* | 0.62 (0.27-1.43) | 0.266 | |
| Stage (III/IV/ED) | 0.90 (0.60-1.33) | 0.586 | 1.24 (0.72-2.13) | 0.434 | 0.34 (0.13-0.89) | 0.027* | ||
| rs13019654 | Stage (III/IV/ED) | 0.64 (0.42-0.98) | 0.038* | 0.58 (0.34-0.98) | 0.041* | 0.57 (0.21-1.53) | 0.261 | |
| MSH3 | rs6151914 | Age (≤55) | 2.90 (1.10-7.67) | 0.032* | 2.90 (1.05-7.99) | 0.039* | Ref | |
| SCLC | 5.02 (1.58-15.93) | 0.006* | 5.21 (1.59-17.05) | 0.006* | Ref | |||
| Stage (I/II/LD) | 13.75 (1.39-136.4) | 0.025* | 13.75 (1.39-136.4) | 0.025* | Ref | |||
| rs245346 | Age (≤55) | 1.34 (0.80-2.27) | 0.269 | 1.00 (0.46-2.15) | 1.000 | 2.58 (1.01-6.60) | 0.049* | |
| Age (>55) | 1.38 (0.88-2.16) | 0.167 | 2.27 (1.10-4.67) | 0.026* | 0.94 (0.41.2.13) | 0.880 | ||
| Male | 1.50 (1.03-2.18) | 0.036* | 1.74 (0.97-3.14) | 0.064 | 1.70 (0.88-3.26) | 0.113 | ||
| rs6151892 | Male | 0.65 (0.43-0.99) | 0.043* | 0.61 (0.36-1.05) | 0.073 | 0.49 (0.18-1.29) | 0.148 | |
| Non-Smoker | 0.62 (0.33-1.16) | 0.135 | 0.41 (0.18-0.95) | 0.037* | 1.13 (0.30-4.17) | 0.860 | ||
| rs26778 | NSCLC | 1.21 (0.77-1.89) | 0.410 | 0.89 (0.48-1.64) | 0.706 | 2.66 (1.08-6.52) | 0.033* | |
| rs3816729 | Non-Smoker | 0.55 (0.29-1.08) | 0.082 | 0.39 (0.17-0.91) | 0.030* | 0.90 (0.23-3.59) | 0.881 | |
| MSH4 | rs5745532 | Age (≤55) | 2.16 (1.18-3.94) | 0.012* | 2.54 (1.22-5.27) | 0.012* | 2.61 (0.59-11.44) | 0.204 |
| Non-Smoker | 1.99 (1.05-3.77) | 0.035* | 2.86 (1.24-6.61) | 0.014* | 1.58 (0.40-6.29) | 0.517 | ||
| rs3806162 | Stage (I/II/LD) | 0.10 (0.01-0.84) | 0.034* | 0.08 (0.01-0.76) | 0.027* | Ref | ||
| MSH5 | rs707938 | SCLC | 0.41 (0.19-0.91) | 0.029* | 0.37 (0.14-0.94) | 0.036* | 0.25 (0.03-2.16) | 0.207 |
| rs3117572 | SCLC | 2.09 (1.02-4.26) | 0.043* | 2.35 (0.92-6.02) | 0.075 | 3.13 (0.69-14.20) | 0.139 | |
| MSH6 | rs2020910 | Male | 0.59 (0.35-0.98) | 0.044* | 0.61 (0.34-1.12) | 0.110 | 0.16 (0.02-1.25) | 0.080 |
| SAPCD1 | rs707937 | Age (≤55) | 1.90 (1.13-3.20) | 0.016* | 4.19 (1.67-10.55) | 0.002* | 1.38 (0.59-3.23) | 0.455 |
| Stage (III/IV/ED) | 1.54 (1.04-2.28) | 0.030* | 2.16 (1.18-3.96) | 0.013* | 1.36 (0.69-2.66) | 0.373 | ||
| NSCLC | 1.27 (0.83-1.95) | 0.278 | 2.01 (1.00-4.02) | 0.049* | 0.89 (0.43-1.84) | 0.748 | ||
Additive model: comparison between minor allele subjects and major allele subjects. Dominant model: comparison between minor allele carriers and major homozygous subjects. Recessive model: comparison between major allele carriers and minor homozygous subjects. OR- odds ratio; CI- confidence interval; Ref.- reference. * P < 0.05.
In MMR system, the loss of functional MMR proteins such as hMutSα (MSH2-MSH6) and hMutLα (MLH1-PMS2) generate the tolerance with DNA alkylating agents. Apoptosis ensues directed by phosphorylation of p53 also related to these MMR repair gene functional in many cases [39]. The functions of DNA mismatch repair act on DNA replication fidelity, mutation avoidance and genome stability, all of which contribute to the viability of organs and cells. The association of DNA mismatch repair gene polymorphisms and platinum-based chemotherapy toxicity in NSCLC patients have been reported [40, 41].
Gene variations in mutS homolog 2 (MSH2) have been founded to be related to tumor risk or survival in multiple kinds of tumors [42]. Polymorphism in ASCL1 target gene DDC is associated with clinical outcomes of small cell lung cancer patients [43, 44]. Nonetheless, there is no report about the association between OS or PFS and polymorphisms in the platinum resistance related MSH2 and SAPCD1 in Chinese patients. In our research, carrying GG or GT genotype in MSH2 rs4608577 had better progression-free survival in lung cancer patients. Therefore, molecular study was needed to elucidate the mechanism of rs4608577 in MSH2 that affects the MMR systems.
There are some limitations in this study. The samples were collected in the same region which also needed the multi-central study to overcome the analysis bias. Patients' sample size was not large enough to contribute to make results verification. The definite mechanism between MSH2 rs4608577, SAPCD1 rs707937 and the prognosis of platinum-based chemotherapy in lung cancer patients is still needed to be investigated. We hypothesized that rs4608577 and rs707937 were affect expression level of MSH2 and SAPCD1 respectively, and contributed to MMR activity.
In conclusion, our study showed that MSH2 rs4608577 was significantly related to progression-free survival and SAPCD1 rs707937 was significantly related to overall survival of platinum-based chemotherapy in lung cancer patients. MSH2 rs4608577 and SAPCD1 rs707937 perhaps could be biomarkers for predicting the prognosis in lung cancer patients with platinum-based chemotherapy.
Acknowledgements
This work was supported by the National Nature Science Foundation of China (81803640, 81603207), National Key Research and Development Program of China (2016YFC1306900), Hunan Provincial Natural Science Foundation of China (2019JJ50946), Technology Innovation Program of Hunan province (2018SK2077), and Youth Science Foundation of Xiangya Hospital (2017Q02, 2018Q014).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel R L, Miller K D, and Jemal A, Cancer statistics, 2019. CA Cancer J Clin, 2019; 69(1): 7-34
2. Chen W, Zheng R, Zeng H. et al. The updated incidences and mortalities of major cancers in China, 2011. Chin J Cancer. 2015;34(11):502-7
3. Shi Y, Sun Y, Medical management of lung cancer. Experience in China. Thorac Cancer. 2015;6(1):10-6
4. Qian C Y, Zheng Y, Wang Y. et al. Associations of genetic polymorphisms of the transporters organic cation transporter 2 (OCT2), multidrug and toxin extrusion 1 (MATE1), and ATP-binding cassette subfamily C member 2 (ABCC2) with platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Chin J Cancer. 2016;35(1):85
5. Herbst R S, Morgensztern D, and Boshoff C, The biology and management of non-small cell lung cancer. Nature, 2018; 553(7689): 446-454
6. Doles J, Oliver T G, Cameron E R. et al. Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A. 2010;107(48):20786-91
7. Moro-Sibilot D, Audigier-Valette C, Merle P. et al. Non-small cell lung cancer recurrence following surgery and perioperative chemotherapy: Comparison of two chemotherapy regimens (IFCT-0702: A randomized phase 3 final results study). Lung Cancer. 2015;89(2):139-45
8. Yin J Y, Li X, Zhou H H. et al. Pharmacogenomics of platinum-based chemotherapy sensitivity in NSCLC: toward precision medicine. Pharmacogenomics. 2016;17(12):1365-78
9. Yin J Y, Li X, Li X P. et al. Prediction models for platinum-based chemotherapy response and toxicity in advanced NSCLC patients. Cancer Lett. 2016;377(1):65-73
10. Xu X, Han L, Duan L. et al. Association between eIF3alpha polymorphism and severe toxicity caused by platinum-based chemotherapy in non-small cell lung cancer patients. Br J Clin Pharmacol. 2013;75(2):516-23
11. Lamba J K, Fridley B L, Ghosh T M. et al. Genetic variation in platinating agent and taxane pathway genes as predictors of outcome and toxicity in advanced non-small-cell lung cancer. Pharmacogenomics. 2014;15(12):1565-74
12. Altorki N K, Markowitz G J, Gao D. et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19(1):9-31
13. Yin J Y, Meng X G, Qian C Y. et al. Association of positively selected eIF3a polymorphisms with toxicity of platinum-based chemotherapy in NSCLC patients. Acta Pharmacol Sin. 2015;36(3):375-84
14. Lyu J, Hao X, Hui Z. et al. Prognosis of R1-resection at the bronchial stump in patients with non-small cell lung cancer. Chin Med J (Engl). 2014;127(16):2918-23
15. Fan H, Shao Z Y, Xiao Y Y. et al. Incidence and survival of non-small cell lung cancer in Shanghai: a population-based cohort study. BMJ Open. 2015;5(12):e009419
16. Xu Q, Lin D, Li X. et al. Association between single nucleotide polymorphisms of NOTCH signaling pathway-related genes and the prognosis of NSCLC. Cancer Manag Res. 2019;11:6895-6905
17. He C, Tu H, Sun L. et al. SNP interactions of Helicobacter pylori-related host genes PGC, PTPN11, IL1B, and TLR4 in susceptibility to gastric carcinogenesis. Oncotarget. 2015;6(22):19017-26
18. Wu Y L, Chien M H, Chou Y E. et al. Association of EGFR mutations and HMGB1 genetic polymorphisms in lung adenocarcinoma patients. J Cancer. 2019;10(13):2907-2914
19. Jiang M, Li X, Quan X. et al. Single Nucleotide Polymorphisms in HMGB1 Correlate with Lung Cancer Risk in the Northeast Chinese Han Population. Molecules. 2018 23(4)
20. Gong W J, Peng J B, Yin J Y. et al. Association between well-characterized lung cancer lncRNA polymorphisms and platinum-based chemotherapy toxicity in Chinese patients with lung cancer. Acta Pharmacol Sin. 2017;38(4):581-590
21. Huang M S, Liu J Y, Xia X B. et al. Hsa_circ_0001946 Inhibits Lung Cancer Progression and Mediates Cisplatin Sensitivity in Non-small Cell Lung Cancer via the Nucleotide Excision Repair Signaling Pathway. Front Oncol. 2019;9:508
22. Shen D, Bo W, Xu F. et al. Genetic diversity and population structure of the Tibetan poplar (Populus szechuanica var. tibetica) along an altitude gradient. BMC Genet. 2014;15(Suppl 1):S11
23. Li Z, Pearlman A H, and Hsieh P, DNA mismatch repair and the DNA damage response. DNA Repair (Amst), 2016; 38: 94-101
24. Roesner L M, Mielke C, Fahnrich S. et al. Stable expression of MutLgamma in human cells reveals no specific response to mismatched DNA, but distinct recruitment to damage sites. J Cell Biochem. 2013;114(10):2405-14
25. Zheng Y, Deng Z, Yin J. et al. The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy. Int J Cancer. 2017;141(11):2336-2347
26. Nogueira G A S, Costa E F D, Lopes-Aguiar L. et al. Polymorphisms in DNA mismatch repair pathway genes predict toxicity and response to cisplatin chemoradiation in head and neck squamous cell carcinoma patients. Oncotarget. 2018;9(51):29538-29547
27. Win A K, Hopper J L, Buchanan D D. et al. Are the common genetic variants associated with colorectal cancer risk for DNA mismatch repair gene mutation carriers? Eur J Cancer. 2013;49(7):1578-87
28. Kadyrova L Y and Kadyrov F A, Endonuclease activities of MutLalpha and its homologs in DNA mismatch repair. DNA Repair (Amst), 2016; 38: 42-49
29. Larrea A A, Lujan S A, Kunkel T A, SnapShot. DNA mismatch repair. Cell. 2010;141(4):730 e1
30. Liu J Y, Qian C Y, Gao Y F. et al. Association between DNA mismatch repair gene polymorphisms and platinum-based chemotherapy toxicity in non-small cell lung cancer patients. Chin J Cancer. 2017;36(1):12
31. Zhang L, Gao G, Li X. et al. Association between single nucleotide polymorphisms (SNPs) and toxicity of advanced non-small-cell lung cancer patients treated with chemotherapy. PLoS One. 2012;7(10):e48350
32. Chen J, Wu L, Wang Y. et al. Effect of transporter and DNA repair gene polymorphisms to lung cancer chemotherapy toxicity. Tumour Biol. 2016;37(2):2275-84
33. Lifson M A, Ozen M O, Inci F. et al. Advances in biosensing strategies for HIV-1 detection, diagnosis, and therapeutic monitoring. Adv Drug Deliv Rev. 2016;103:90-104
34. Iyama T and Wilson D M, 3rd, DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst), 2013; 12(8): 620-36
35. Xu X L, Yao Y L, Xu W Z. et al. Correlation of MSH3 polymorphisms with response and survival in advanced non-small cell lung cancer patients treated with first-line platinum-based chemotherapy. Genet Mol Res. 2015;14(2):3525-33
36. Ludovini V, Ricciuti B, Tofanetti F R. et al. KRAS mutation and DNA repair and synthesis genes in non-small-cell lung cancer. Mol Clin Oncol. 2018;9(6):689-696
37. Chen J, Wang Z, Zou T. et al. Pharmacogenomics of platinum-based chemotherapy response in NSCLC: a genotyping study and a pooled analysis. Oncotarget. 2016;7(34):55741-55756
38. Baretti M and Le D T, DNA mismatch repair in cancer. Pharmacol Ther, 2018; 189: 45-62
39. Edelbrock M A, Kaliyaperumal S, and Williams K J, Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat Res, 2013; 743-744: 53-66
40. Liu D, Keijzers G, and Rasmussen L J, DNA mismatch repair and its many roles in eukaryotic cells. Mutat Res, 2017; 773: 174-187
41. Cao S, Wang C, Ma H. et al. Genome-wide Association Study on Platinum-induced Hepatotoxicity in Non-Small Cell Lung Cancer Patients. Sci Rep. 2015;5:11556
42. Zhu X, Wang Z, Qiu X. et al. Rs2303428 of MSH2 Is Associated with Hepatocellular Carcinoma Prognosis in a Chinese Population. DNA Cell Biol. 2018;37(7):634-641
43. Kim J H, Lee S Y, Choi J E. et al. Polymorphism in ASCL1 target gene DDC is associated with clinical outcomes of small cell lung cancer patients. Thorac Cancer. 2020;11(1):19-28
44. Chakraborty U, Dinh T A, Alani E, Genomic Instability Promoted by Overexpression of Mismatch Repair Factors in Yeast. A Model for Understanding Cancer Progression. Genetics. 2018;209(2):439-456
Author contact
![]() Corresponding authors: Zhi-Wei Chen, Department of Orthopaedics, The First Affiliated Hospital of the University of South China, Hengyang 421001, Tel: 0086-734-8578571, E-mail: czw9915com; or Juan Chen, Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008, P. R. China. Tel: 0086-731-89753491, E-mail: cj1028edu.cn.
Corresponding authors: Zhi-Wei Chen, Department of Orthopaedics, The First Affiliated Hospital of the University of South China, Hengyang 421001, Tel: 0086-734-8578571, E-mail: czw9915com; or Juan Chen, Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008, P. R. China. Tel: 0086-731-89753491, E-mail: cj1028edu.cn.

 Global reach, higher impact
Global reach, higher impact