Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(7):1869-1882. doi:10.7150/jca.39163 This issue Cite
Research Paper
Novel candidate biomarkers of origin recognition complex 1, 5 and 6 for survival surveillance in patients with hepatocellular carcinoma
1. Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Province, China.
2. Department of Medical Cosmetology, The Second Affiliated Hospital of Guangxi Medical University, Nanning 530000, Guangxi Province, China.
3. Department of Hepatobiliary Surgery, The Third Affiliated Hospital of Guangxi Medical University, Nanning 530031, Guangxi Province, China.
4. Department of Health Management and Division of Physical Examination, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Zhuang Autonomous Region, People's Republic of China.
5. Department of Hematology, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Province, China.
Received 2019-8-9; Accepted 2019-12-20; Published 2020-1-20
Abstract
Background: Hepatocellular carcinoma (HCC) has high morbidity and mortality and lacks effective biomarkers for early diagnosis and survival surveillance. Origin recognition complex (ORC), consisting of ORC1-6 isoforms, was examined to assess the potential significance of ORC isoforms for HCC prognosis.
Methods: Oncomine and Gene Expression Profiling Interactive Analysis (GEPIA) databases were used to examine differential isoform expression, stage-specific expression, calculate Pearson correlations and perform survival analysis. A human protein atlas database was utilized to evaluate the protein expression of ORCs in liver tissue. The cBioPortal database was used to assess isoform mutations and the survival significance of ORCs in HCC. Cytoscape software was employed to construct gene ontologies, metabolic pathways and gene-gene interaction networks.
Results: Differential expression analysis indicated that ORC1 and ORC3-6 were highly expressed in tumor tissues in the Oncomine and GEPIA databases, while ORC2 was not. All the ORCs were showed positive and statistically significant correlations with each other (all P<0.001). ORC1-2 and ORC4-6 expressions were associated with disease stages I-IV (all P<0.05), but ORC3 was not. Survival analysis found that ORC1 and ORC4-6 expressions were associated with overall survival (OS), and ORC1-3 and ORC5-6 expression were associated with recurrence-free survival (RFS; all P<0.05). In addition, low expression of these ORC genes consistently indicated better prognosis compared with high expression. Protein expression analysis revealed that ORC1 and ORC3-6 were expressed in normal liver tissues, whereas ORC2 was not. Enrichment analysis indicated that ORCs were associated with DNA metabolic process, sequence-specific DNA binding and were involved in DNA replication, cell cycle, E2F-enabled inhibition of pre-replication complex formation and G1/S transition.
Conclusions: Differentially expressed ORC1, 5 and 6 are candidate biomarkers for survival prediction and recurrence surveillance in HCC.
Keywords: origin recognition complex, ORC1, ORC5, ORC6, biomarker, hepatocellular carcinoma.
Introduction
Liver cancer is one of the most common cancers worldwide. Hepatocellular carcinoma (HCC) is the most frequent type, comprising roughly 70% to 85% of all primary liver cancers [1]. HCC ranks as the sixth most common malignancy in the world and accounts for approximately 7% of all malignancies [2]. HCC is diagnosed in more than 740 000 new cases each year and is the third leading cause of malignancy-related deaths, with increasing incidence each year [3]. Many risk factors have been identified for HCC, including chronic hepatitis B or C virus infection, type 2 diabetes mellitus, alcohol ingestion and metabolic syndrome, among others [4]. Although these factors are well known, the prevention and surveillance of HCC is still burdensome and the prognosis remains unsatisfactory. Even with early diagnosis of HCC, it is difficult to treat advanced stage HCC due to its extra- and intrahepatic characteristics, including metastatic potential [5] and resistance to chemotherapeutic approaches such as hepatic artery infusion chemoembolization [6], transcatheter arterial chemoembolization [7] and the drug sorafenib [8]. To improve the rate of early diagnosis of HCC and monitor recurrence after hepatectomy, many efforts have been made to develop new biomarkers for early diagnosis and recurrence prediction, including osteopontin, midkine, golgi protein-73 and α-fetoprotein-L3 [9], but almost none of which have been widely accepted for clinical application.
Origin recognition complex (ORC), encoded by the latheo gene, is involved in the initiation of DNA replication and is composed of six isoforms, ORC1-6 [10, 11]. The ORC1 gene, also called HsOrc1 in humans, is weakly expressed in quiescent cells, but can be upregulated by cell growth signals [11]. The transcription of ORC1 is dependent on the E2F transcription factor [11]. In contrast, ORC2 expression is regulated differently from ORC1 [11]. Cohen et al. examined the localization of ORC1 in human leukemia cells and found that it was localized to the pericentriolar region during anaphase [12]. Phosphorylation of ORC2 and HBO1 induced by polo-like kinase 1 leads to gemcitabine resistance in pancreatic cancer [13]. Phosphorylation of ORC2 by polo-like kinase 1 promotes DNA replication under stress conditions [14]. Knockdown of ORC3 increased the level of mRNP-bound Nxf1, while knockdown of ORC5 decreased the association between Nxf1 and mRNP [15]. ORC6 is crucial for the formation of pre-replicative complex interactions with ORC chaperone proteins that promote chromatin binding of ORC [16].
ORC and the mini-chromosome maintenance (mcm) complex are well known to be involved in DNA biosynthesis in the cell cycle pathway. Our previous results indicated that the MCM complex is not only associated with the overall survival (OS) of HCC, but can also serve as both a diagnostic and prognostic biomarker for HCC [17]. Therefore, we conducted the present study to explore potential significance of ORC in HCC.
Material and methods
Oncomine database analysis
The Oncomine database (https://www.oncomine.org/resource/main.html) was first used to profile ORC1-6 isoform expression in multiple cancer types, including liver cancer, bladder cancer, breast cancer, cervical cancer, colorectal cancer, gastric cancer and esophageal cancer. The Oncomine database was then used to compare ORC1-6 isoform expression in HCC and normal liver tissues in several datasets, including the Chen liver [18], Roessler liver and Roessler liver 2 [19], Mas liver [20], Wurmbach liver [21] and Guichard liver [22] datasets. Criteria for the inclusion and exclusion: all the enrolled objects were surgical resection and pathological diagnosed of HCC.
Gene Expression Profiling Interactive Analysis (GEPIA) database analysis
The GEPIA2 database (http://gepia2.cancer-pku.cn/#index) was used to explore transcripts per million and further determine ORC1-6 isoform expression in HCC and normal liver tissues [23]. Survival analysis, including OS and disease-free survival (DFS), was employed to examine the prognostic significance of ORC1-6 isoforms in HCC at median cutoff. In addition, Pearson correlation analysis identified correlations between each two ORC isoforms in tumor tissues, and analysis by stage determined the associations between ORC isoform expression and disease stage.
Human protein atlas (HPA) and cBioPortal database analysis
The HPA database (http://www.proteinatlas.org/) was utilized to identify protein expression of ORC isoforms in liver tissue in a tissue-based map of the human genome [24]. Brown color indicated higher expression compared with nattier blue. The cellular location of the ORCs was identified using the HPA database with a subcellular map [25]. Meanwhile, the cBioPortal database (http://www.cbioportal.org/) was used to identify the types of ORC isoform mutations, including mutation, amplification, and deep deletion, and to determine prognosis significance of isoforms in the presence and absence of mutations [26, 27]. An interaction network between ORC isoforms and other genes was constructed using the cBioPortal database as well.
Gene ontology (GO), KEGG pathway enrichment analysis and interaction network construction
ORC1-6 isoforms underwent enrichment analysis of GO terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. GO terms, including biological processes (BPs), cellular components (CCs) and molecular functions (MFs), were enriched and visualized using the BinGO plugin in Cytoscape software [28, 29]. KEGG pathways were enriched and visualized using the ClueGO plugin in Cytoscape software [30]. Gene-gene interaction and protein-protein interaction networks were constructed using the geneMANIA plugin in Cytoscape software [31] and the STRING website (https://string-db.org/) [32].
Statistical analysis
Gene expression analysis by stage and differential analysis utilized one-way analysis of variance. Survival analysis was performed using the log-rank test and Cox test. Cox proportional hazard ratios and 95% confidence intervals were included in the survival plots. A p-value ≤ 0.05 was considered statistically significant.
Results
Gene expression analysis
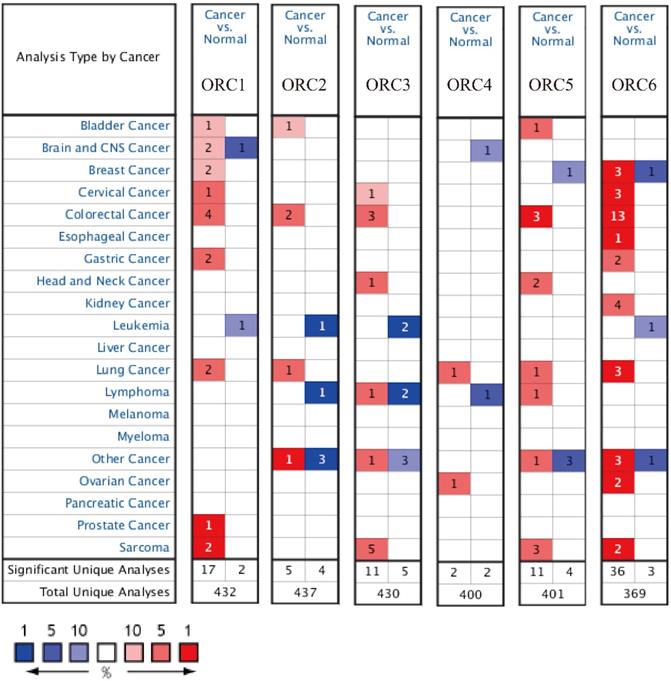
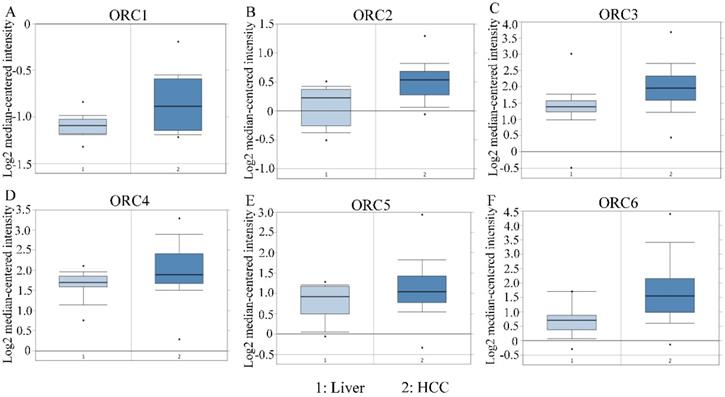
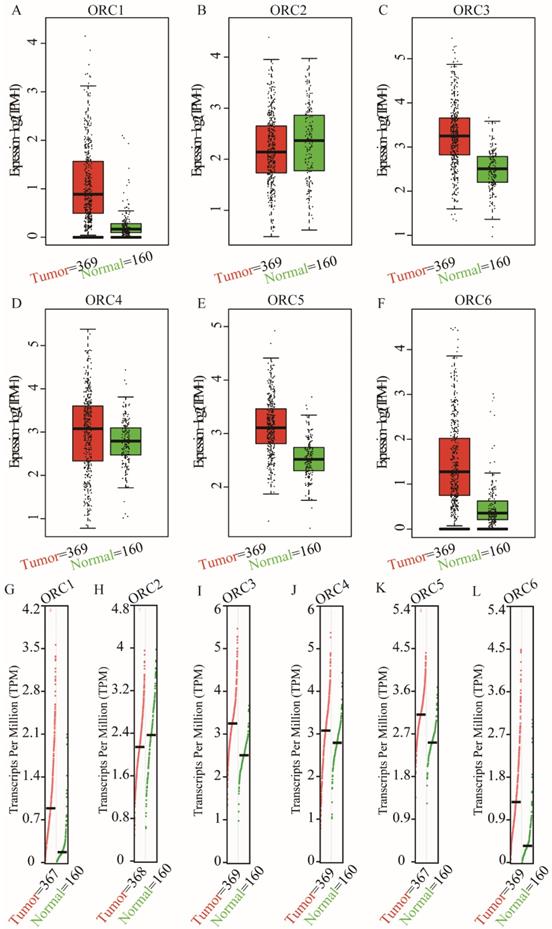
Studies about ORC isoforms in multiple cancers are shown in Figure 1. The significance of ORCs has been explored in many types of cancers, especially breast cancer, colorectal cancer, leukemia and lung cancer (Figure 1). The expression levels of ORC isoforms in HCC and normal liver tissues are shown in Figure 2. All isoforms were highly expressed in HCC tissues compared with normal liver tissues (Figure 2A-F). Expression level changes and the p-values of changes in ORCs were subsequently analyzed. Detailed changes between HCC and normal liver tissues in multiple datasets are presented in Table 1. Validation studies of the expression levels of ORCs in HCC were performed in the GEPIA database, which indicated that ORC1 and ORC3-6 presented high levels of expression in tumor tissues, while ORC2 showed the opposite results (Figure 3).
Protein expression and cellular locations
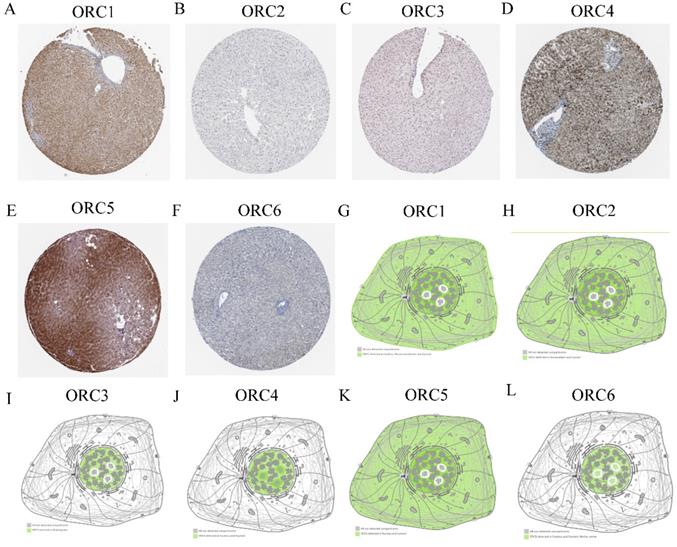
Protein expression and cellular locations of ORCs were analyzed in the HPA database. Protein expression analysis found that ORC1 and ORC3-5 showed moderately positive expression in normal liver tissues (Figure 4A, C-E); ORC6 showed weakly positive expression (Figure 4F), whereas ORC2 was not expressed in liver tissue (Figure 4B). ORC1 was detected in the nucleus, plasma membrane and cytosol (Figure 4G); ORC2 was detected in nucleoplasm and cytosol (Figure 4H); ORC3 was detected in nucleoplasm (Figure 4I); ORC4 was detected in the nucleus and nucleoli (Figure 4J); ORC5 was detected in nucleus and cytosol (Figure 4K); and ORC6 was detected in the nucleus and nucleoli fibrillar center (Figure 4L).
Analysis of ORC isoform expressions in HCC and NR.
| Gene | Reporter | Types | FC | P value | T test | Reference |
|---|---|---|---|---|---|---|
| ORC1 | IMAGE:194236 | HCC vs. NR | 1.477 | 1.34E-4 | 3.724 | Chen liver (Reference 55) |
| 205085 _at | HCC vs. NR | 1.156 | 0.001 | 3.287 | Roessler liver (Reference 56) | |
| 205085 _at | HCC vs. NR | 1.133 | 3.21E-9 | 5.933 | Roessler liver 2 (Reference 56) | |
| 205085 _at | HCC vs. NR | 1.052 | 0.021 | 2.109 | Mas liver (Reference 57) | |
| 205085 _at | HCC vs. NR | 1.271 | 0.007 | 2.613 | Wurmbach liver (Reference 58) | |
| ORC2 | 204853 _at | HCC vs. NR | 1.322 | 2.78E-5 | 4.620 | Mas liver (Reference 57) |
| ORC3 | IMAGE:260336 | HCC vs. NR | 1.789 | 6.71E-14 | 8.037 | Chen liver (Reference 55) |
| 210028_s_at | HCC vs. NR | 1.511 | 1.80E-31 | 12.701 | Roessler liver 2 (Reference 56) | |
| 210028_s_at | HCC vs. NR | 1.249 | 3.77E-4 | 3.609 | Mas liver (Reference 57) | |
| 210028_s_at | HCC vs. NR | 1.316 | 0.002 | 3.083 | Roessler liver (Reference 56) | |
| ORC4 | 203351_s_at | HCC vs. NR | 1.366 | 2.25E-19 | 9.367 | Roessler liver 2 (Reference 56) |
| 203351_s_at | HCC vs. NR | 1.287 | 0.011 | 2.417 | Roessler liver (Reference 56) | |
| 203352 _at | HCC vs. NR | 1.082 | 1.22E-5 | 4.268 | Roessler liver 2(Reference 56) | |
| ORC5 | 204957 _at | HCC vs. NR | 1.445 | 2.97E-22 | 10.185 | Roessler liver 2 (Reference 56) |
| 211212_at | HCC vs. NR | 1.217 | 2.02E-13 | 7.513 | Roessler liver 2 (Reference 56) | |
| 07-103594861 | HCC vs. NR | 1.053 | 1.71E-7 | 5.439 | Gurchard liver (Reference 59) | |
| 204957 _at | HCC vs. NR | 1.293 | 0.015 | 2.245 | Roessler liver (Reference 56) | |
| 211212_at | HCC vs. NR | 1.132 | 0.042 | 1.797 | Roessler liver (Reference 56) | |
| 204957 _at | HCC vs. NR | 1.275 | 0.009 | 2.575 | Wurmbach liver (Reference 58) | |
| ORC6 | 219105_x_at | HCC vs. NR | 1.690 | 4.12E-5 | 4.728 | Roessler liver (Reference 56) |
| 219105_x_at | HCC vs. NR | 2.002 | 1.26E-6 | 4.166 | Wurmbach liver (Reference 58) | |
| 219105_x_at | HCC vs. NR | 1.476 | 6.75E-23 | 10.623 | Roessler liver 2 (Reference 56) |
Note: HCC: hepatocellular carcinoma; NR: normal; FC: fold change; ORC1: origin recognition complex 1; ORC2: origin recognition complex 2; ORC3: origin recognition complex 3; ORC4: origin recognition complex 4; ORC5: origin recognition complex 5; ORC6: origin recognition complex 6.
Analysis of ORC1-6 isoforms in multiple cancers.
Expression of ORC1-6 isoforms in normal liver and HCC tissues. A-F: Expression of ORC1-6 isoforms in normal liver and HCC tissues.
Expression and transcripts of ORC1-6 isoforms in normal liver and tumor tissues. A-F: Expression of ORC1-6 isoforms in normal liver and tumor tissues; G-L: Transcripts of ORC1-6 isoforms in normal liver and tumor tissues.
OS and DFS analysis
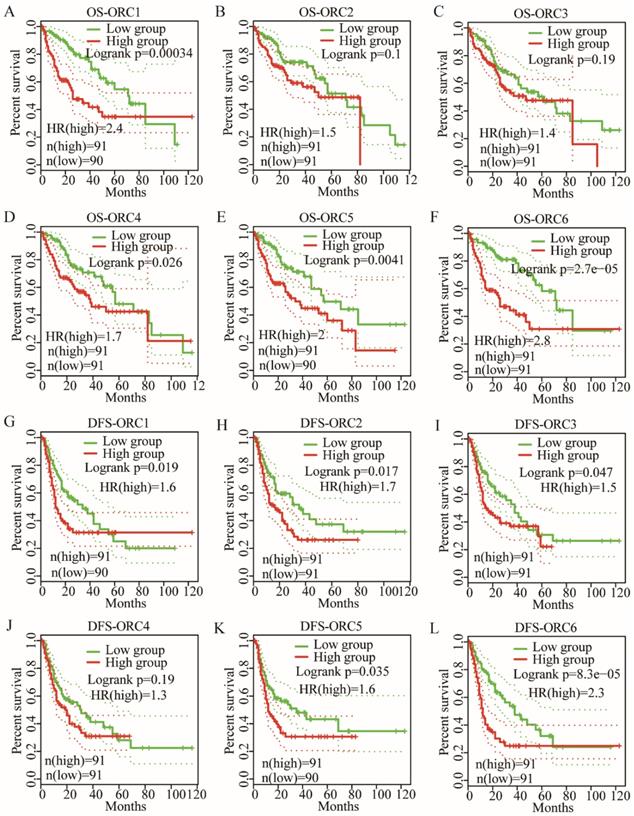
OS analysis indicated that ORC1 and ORC4-6 expression were associated with prognosis (Log-rank test results: P<0.001, P=0.026, P=0.004, and P<0.001 for Figure 5A, D-F, respectively), whereas ORC2 and ORC3 were not associated with prognosis (Log-rank test results: P=0.1 and P=0.19 for Figure 5B-C, respectively). Moreover, low expression of ORC1 and ORC4-6 were consistently associated with better prognosis compared with higher expression.
DFS analysis indicated that ORC1-3 and ORC5-6 expression were associated with prognosis (Log-rank test results: P=0.019, P=0.017, P=0.047, P=0.035 and P<0.001, respectively, for Figure 5G-I, K-L), but ORC4 expression was not associated with prognosis (Log-rank test, P=0.1; Figure 5J). Moreover, all five ORCs consistently revealed that low expression was associated with better prognosis compared with high expression. These results suggested that ORC isoforms may act as oncogenes in HCC.
Pearson correlation and stage analysis
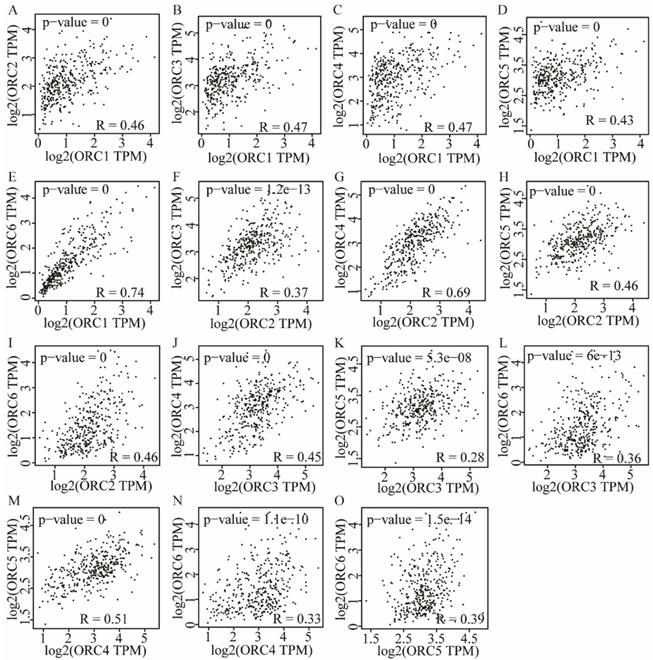
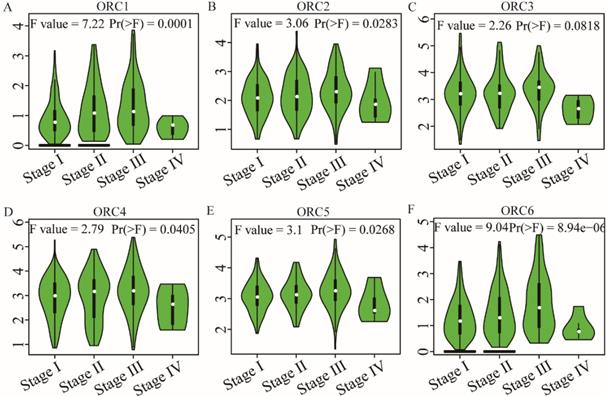
Pearson correlation analysis was performed for ORC isoforms in HCC samples. Analysis findings demonstrated that each two ORC isoforms were positively correlated (Figure 6). In addition, all correlations were statistically significant (P<0.05). Furthermore, analysis by disease stage found that all ORCs exhibited higher expression with progression from stage I to stage III (Figure 7). However, stage IV always induced the lowest expression compared with stages I, II and III for all the ORCs. To summarize, ORC3 was not significantly different among stages I-IV (P=0.0818, Figure 7C), but the other isoforms were significantly associated with stages I-IV (P=0.0001, P=0.00283, P=0.0405, P=0.0268 and P=8.94e-6, respectively; Figure 7A-B, D-F).
Mutation analysis and interaction network
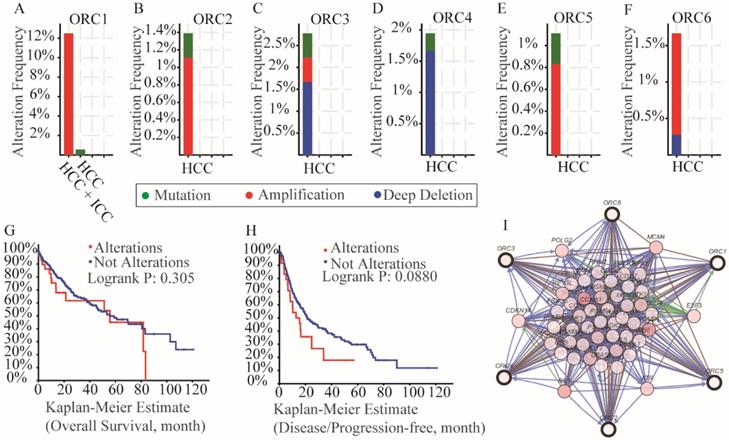
Mutation analysis in the cBioPortal database indicated that all of the ORCs had mutations (Figure 8A-F). Specifically, ORC1 had amplification more than 12% in HCC and intrahepatic cholangiocarcinoma (ICC) and mutation less than 2% HCCs (Figure 8A); ORC2 had amplification more than 1% and mutation less than 1% (Figure 8B); ORC3 had amplification, mutation and deep deletion more than 2.5% together (Figure 8C); ORC4 had mutation and deep deletion more than 1.5% together (Figure 8D); ORC5 had mutation and amplification more than 1% together (Figure 8E); ORC6 had amplification and deep deletion more than 1.5% together (Figure 8F). In addition, survival analysis including OS and DFS with and without mutations of ORC isoforms suggested that mutations did not associate with either OS or DFS (Figure 8G-H). We then constructed an interaction network using the ORC isoforms, which demonstrated that ORCs acted in concert with PSM family members such as PSME1, PSMB6 and PSMC4 to control expression of RRM2, POLE, POLE2, E2F3 and TK1, and showed controlling state changes of REV3L, CDC7, MCM7, ATR, PSMB3 and others (Figure 8I).
Protein expression and localization of ORC1-6 isoforms in liver tissue and cell lines. A-F: Protein expression of ORC1-6 isoforms in liver tissues; G-L: Localization of ORC1-6 isoforms in cell lines.
OS and RFS plots of patients expressing ORC1-6 isoforms in HCC. A-F: OS plots of patients expressing each of the ORC1-6 isoforms in HCC; G-L: RFS plots of patients expressing each of the ORC1-6 isoforms in HCC.
Enrichment analysis and interaction network of gene-gene and protein-protein interactions
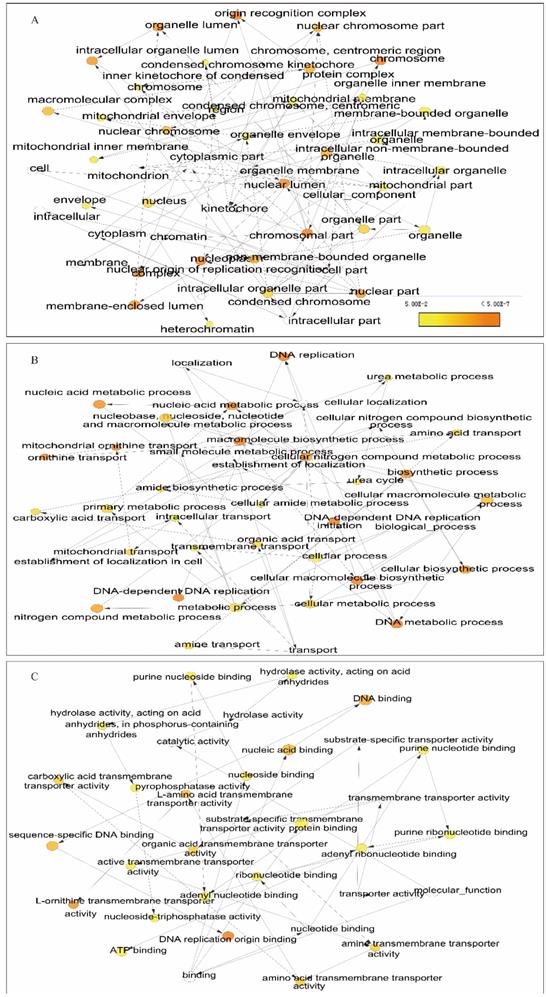
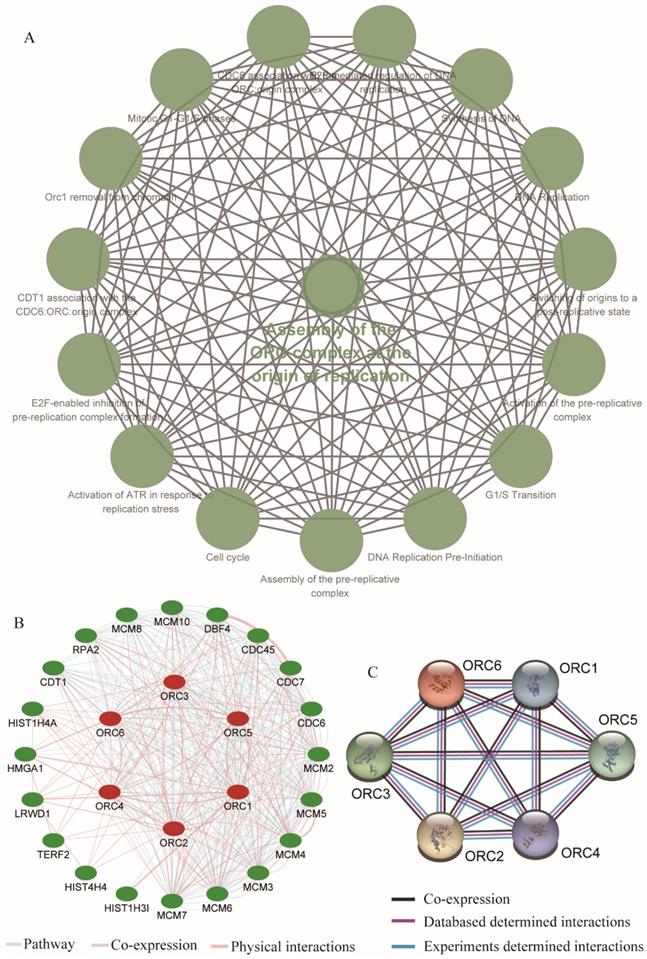
We further explored metabolic pathways and GO terms of ORCs. Enrichment of GO terms indicated that ORCs were involved in chromosomal part, the nuclear origin of ORC, the nuclear lumen and macromolecular complexes, etc. in CCs (Figure 9A); DNA-dependent DNA replication, DNA metabolic processes and cellular biosynthetic processes, etc. in BPs (Figure 9B); DNA binding, sequence-specific DNA binding and L-ornithine transmembrane transporter activity, etc. in MFs (Figure 9C). Pathway results indicated that ORCs were involved in DNA replication, cell cycle, E2F-enabled inhibition of pre-replication complex formation and the G1/S transition, etc. (Figure 10A). The gene-gene interaction network suggested that ORCs were co-expressed interactively with MCM2-8, MCM-10, CDC6-7 and CDC45, etc. (Figure 10B). ORCs showed interactions in the pathway with MCM members and CDC members. The protein-protein interaction network suggested that all of the ORCs were co-expressed and have determined interactions in experiments and databases (Figure 10C).
Pearson correlation plots among ORC1-6 isoforms in HCC tissues. A-E: Pearson correlation plots between ORC1 and ORC2-6 isoforms; F-I: Pearson correlation plots between ORC2 and ORC3-6 isoforms; J-L: Pearson correlation plots between ORC3 and ORC4-6 isoforms; M-N: Pearson correlation plots between ORC4 and ORC5-6 isoforms; O: Pearson correlation plots between ORC5 and ORC6.
Analysis of the expression of ORC1-6 isoforms at different disease stages. A-F: Analysis of ORC1-6 isoform expression at each disease stage.
Analysis of mutations, survival plots and interaction networks of ORC1-6 isoforms in HCC and ICC. A-F: Analysis of mutations of ORC1-6 isoforms in HCC and ICC; G-H: OS and RFS in patients expressing ORC1-6 isoform mutations; I: Interaction network among ORC1-6 isoforms.
Discussion
Our present study explored the potential prognostic significance of ORC1-6 isoforms in HCC. We discovered that ORC1-6 were highly expressed in HCC tissues compared to normal liver tissues. In addition, we found that ORC1 and ORC4-6 expression were associated with OS and that ORC1-3 and ORC5-6 expressions were associated with DFS. Low expression of these genes consistently indicated better prognosis compared with higher expression. These results suggest that ORC isoforms may act as oncogenes in HCC. Protein expression analysis revealed that ORC1 and ORC3-6 were also expressed in normal liver tissues. Combining ORC expression and survival analyses, we conclude that ORC1, 5 and 6 are candidate biomarkers for survival prediction and recurrence surveillance. Enrichment analysis indicated that ORCs were associated with DNA replication, chromosomal part, DNA metabolic process, DNA binding and sequence-specific DNA binding, and were also involved in DNA replication, cell cycle, E2F-enabled inhibition of pre-replication complex formation and the G1/S transition. Further investigation to validate the above findings is warranted.
ORC, consisting of six subunits, functions as the initiator in recognizing replication start sites and interacts with subsequent replication factors [33]. It was first discovered in Saccharomyces cerevisiae as a multi-protein complex that was connected to the autonomously replicating sequence [34]. The ORC plays a pivotal role in the initiation of DNA replication in all eukaryotic systems [16]. Molecular interactions within the eukaryotic replication origin occur within the overall structure [12]. In addition, melting of the DNA double helix facilitates a conformational change in the ORC-related post-replicative state which can prevent the re-initiation of replication at the specific binding site [35]. In eukaryotes, ORC is associated with chromatin at multiple sites [36-38] and these sites may be candidate replication origins, which initiate the assembly of the pre-replication complex at the G1 phase of the cell cycle [39, 40].
In addition to its main role in the formation of the pre-replication complex on chromosomes prior to DNA replication, subunits of ORC have been reported to be involved in several chromosome-associated processes [33, 41]. Hemerly et al. found that ORC1 controls centriole and centrosome re-duplication, as well as the initiation of DNA replication in human U2OS cells [42]. ORC2 and other ORC isoforms were first reported as essential for DNA replication initiation by connecting to replication origin sites to form the pre-replication complex at late G1 and early S phase in mammalian cells [41]. ORC2 was found localized to the centrosome and centromere, which was essential for proper chromatin segregation at the G2/M phase [43]. Wang et al. found that ORC2 was modified by the small ubiquitin-like modifier (SUMO) at the G2/M phase of the cell cycle and that SUMOylation of ORC2 was crucial for the smooth transition into mitosis [44]. ORC2 also localized to the telomeric region and plays a pivotal role in telomere homeostasis [45, 46]. Depletion of ORC2 leads to mitotic arrest because of defects in chromosome condensation [43].
Enriched biological processes, cellular components and molecular functions networks of ORC1-6 isoforms. A-C: Networks of ORC1-6 isoforms showing enriched cellular components (A), biological processes (B) and molecular functions (C).
Enriched KEGG pathways, gene-gene and protein-protein interaction networks of ORC1-6 isoforms. A: Enriched KEGG pathways of ORC1-6 isoforms; B: Gene-gene interaction network of ORC1-6 isoforms; C: Protein-protein interaction network of ORC1-6 isoforms.
ORC3 interacts with HP1 at the heterochromatin foci to promote the organization of higher chromatin structure [47]. ORC4 and ORC6 can directly interact with ENY2, which is bound to type C2H2 zinc fingers of insulator protein Su (Hw) and CTCF in Drosophila [48-50]. Moreover, protein Su (Hw) opens chromatin regions and accelerates the recruiting of the ORC to chromatin [51]. ORC5 interacts with the histone acetyltransferase GCN5/KAT2A, making origins of replication more accessible for activation [47]. Multi-mono-ubiquitylation of ORC5 facilitates this kind of interaction, enabling opening of the local origin chromatin environment and stimulating origin activation [52].
ORC6 is the most divergent and evolutionarily least conserved subunit among all the ORC proteins [16]. ORC6 interacts with other chaperone proteins, such as high mobility group protein A1a, which may be helpful for targeting ORCs to specific chromatin regions in addition to functioning in the assembly of the pre-replicative complex [53]. ORC6 binds to the outer kinetochore during mitosis and localizes to the midplane of cell division in anaphase where it is required for cytokines through a connection with septin protein [16]. Thomae et al. found that replication competence was significantly associated with the potential of mutant forms to cooperate with human ORCs, suggesting an active role of ORC6 in origin activation [16]. They also observed that ORC6 was abundantly expressed in correlation with ORC2 expression and contributes to the replication of the initiation process independent from ORC1-5 [16].
Diffley et al. reported that the main function of ORCs was to initiate replication origins in the G1 phase of the cell cycle [39, 40]. However, DePamphilis demonstrated that ORCs were related to chromatin at other stages of the cell cycle or in G0 cells [54]. These findings suggest that the functions of ORCs are not restricted to DNA replication. Sasaki et al. demonstrated that ORC also initiated non-replication functions [55, 56] such as transcription silencing [57], chromatid cohesion [58], neuron development [59] and cytokinesis [60]. Notably, previous studies have reported the association of ORC with human diseases, such as Meier-Gorlin syndrome, Epstein-Barr virus-infected diseases, American trypanosomiasis and African trypanosomiasis [55].
More specifically, De Munnik et al. observed mutations of ORC1, 4 and 6 in a study involving 35 individuals with Meier-Gorlin syndrome [61]. Young indicated that because ORC1 was significantly upregulated after irradiation for 6 and 24 hours in PC-3 cells, ORC1 and other DNA repair candidates may be potential targets for radiation sensitization and serve as predictive biomarkers for prostate cancer [62]. Chen et al. found that ORC1 participated in the XIST/miR-140-5p/ORC1 axis of progression in cervical cancer, which will shed new light on epigenetic diagnostics and therapeutics in cervical cancer [63]. Our present study found that ORC1 was involved in DNA binding and biosynthesis in the cell cycle, which is consistent with previous findings of the main functions of ORCs [39, 40, 54]. Moreover, our study indicated that ORC1 was not only associated with both OS and DFS, but was also a potential biomarker for HCC survival prediction and recurrence surveillance. This potential role of ORC1 in tumors is in accordance with previous reports of cervical cancer and prostate cancer [62, 63].
A study by Radojkovic et al. found a novel mutation A286V within ORC4 and reported that this mutation might be a favorable prognostic biomarker for B-cell lymphoproliferative disorders [64]. Our study revealed that ORC4 expression, highly expressed in tumor tissue, was associated with OS and might be a candidate biomarker for HCC. ORC2 and ORC5 were reported to be upregulated in microarray data originating from the cisplatin-sensitive T24 cell line and cisplatin-resistant T24R2 cell line [65]. However, this study did not report any prognostic value for ORC2 and ORC5. Our study indicated that ORC2 expression was correlated with DSF in HCC; furthermore, ORC5 expression was associated with both OS and DFS in HCC, suggesting it is a novel candidate biomarker for HCC survival. The role of ORC3 has not yet been documented in tumors. However, a single nucleotide polymorphism of ORC6 was reported as a breast cancer-related candidate gene [66]. Xi et al. found that ORC6 was highly expressed in tumors compared to paired normal tissues in colorectal cancer [67]. Thus, our results agree with Xi et al.'s finding of high expression of ORC6 in tumor tissues. These authors further demonstrated that ORC6 expression was associated with 5-fluorouracil-related resistance in human colorectal cell lines [68]. Moreover, they found that decreased ORC6 expression sensitized human colorectal cell lines to 5-fluorouracil and cisplatin, and thus, ORC6 may be a novel therapeutic target in colon cancer [69]. Our present study demonstrated that ORC6 was correlated with both OS and DFS of HCC, indicating that it may serve as a prognostic biomarker for HCC survival.
There are several limitations of our study, which should be recognized. First, our findings will require further large population and cohort validations, including clinical parameters and our medical center cohort validation. Second, our study results need to be further validated in Asian population cohorts, including our medical center cohort. Then, additional experiments are needed to explore the specific mechanisms underlying ORC involvement in HCC. Finally, the potential clinical application of ORCs should be explored in multi-race studies in multiple countries.
Acknowledgements
Funding
This work was supported in part by the National Natural Science Foundation of China (No.: 81560535, 81802874, 81072321, 30760243, 30460143 and 30560133), Natural Science Foundation of Guangxi Province of China (Grant No.2017JJB140189y, 2018GXNSFAA050119), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Natural Sciences Foundation (No.: GuiKeGong 1104003A-7), and Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018). The present study is also partly supported by Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (Z2016318), the Guangxi Key R & D Program (GKEAB18221019), Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (GKE2018-01, GKE2019-11), The Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (2018KY0110), Guangxi Key Laboratory for the Prevention and Control of Viral Hepatitis (No.GXCDCKL201902), 2018 Innovation Project of Guangxi Graduate Education (JGY2018037). As well as, the present study is also partly supported by Guangxi Key Laboratory for the Prevention and Control of Viral Hepatitis (No. GXCDCKL201902) and Research Institute of Innovative Think-tank in Guangxi Medical University (The gene-environment interaction in hepatocarcinogenesis in Guangxi HCCs and its translational applications in the HCC prevention). We would also acknowledge the supported by the Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education.
Authors' Contributions
Xiang-Kun Wang and Tao Peng designed this manuscript; Qiao-Qi Wang, Jian-Lu Huang, Lin-Bo Zhang, Xin Zhou, Jun-Qi Liu, Zi-Jun Chen, Xi-Wen Liao, Rui Huang, Cheng-Kun Yang, Guang-Zhi Zhu, Chuang-Ye Han and Xin-Ping Ye conducted the study and analyzed the data. Xiang-Kun Wang wrote the manuscript, and Tao Peng guided the writing.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Delgado TC, Barbier-Torres L, Zubiete-Franco I, Lopitz-Otsoa F, Varela-Rey M, Fernandez-Ramos D. et al. Neddylation, a novel paradigm in liver cancer. Transl Gastroenterol Hepatol. 2018;3:37
2. Noor EASM. Management of Hepatocellular Carcinoma: Bangladesh Perspective. Euroasian J Hepatogastroenterol. 2018;8:52-3
3. White DL, Thrift AP, Kanwal F, Davila J, El-Serag HB. Incidence of hepatocellular carcinoma in all 50 United States, from 2000 through 2012. Gastroenterology. 2017;152(e5):812-20
4. Gerbes A, Zoulim F, Tilg H, Dufour JF, Bruix J, Paradis V. et al. Gut roundtable meeting paper: selected recent advances in hepatocellular carcinoma. Gut. 2018;67:380-8
5. Wada F, Koga H, Akiba J, Niizeki T, Iwamoto H, Ikezono Y. et al. High expression of CD44v9 and xCT in chemoresistant hepatocellular carcinoma: Potential targets by sulfasalazine. Cancer Sci. 2018;109:2801-10
6. Nagamatsu H, Sumie S, Niizeki T, Tajiri N, Iwamoto H, Aino H. et al. Hepatic arterial infusion chemoembolization therapy for advanced hepatocellular carcinoma: multicenter phase II study. Cancer Chemother Pharmacol. 2016;77:243-50
7. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-55
8. Nakano M, Tanaka M, Kuromatsu R, Nagamatsu H, Tajiri N, Satani M. et al. Sorafenib for the treatment of advanced hepatocellular carcinoma with extrahepatic metastasis: a prospective multicenter cohort study. Cancer Med. 2015;4:1836-43
9. Sengupta S, Parikh ND. Biomarker development for hepatocellular carcinoma early detection: current and future perspectives. Hepat Oncol. 2017;4:111-22
10. Pinto S, Quintana DG, Smith P, Mihalek RM, Hou ZH, Boynton S. et al. latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron. 1999;23:45-54
11. Ohtani K, DeGregori J, Leone G, Herendeen DR, Kelly TJ, Nevins JR. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Molecular and cellular biology. 1996;16:6977-84
12. Coffman FD, Reyes M-L, Brown M, Lambert WC, Cohen S. Localization of ORC1 during the cell cycle in human leukemia cells. Analytical cellular pathology. 2011;34:355-61
13. Song B, Liu XS, Rice SJ, Kuang S, Elzey BD, Konieczny SF. et al. Plk1 phosphorylation of orc2 and hbo1 contributes to gemcitabine resistance in pancreatic cancer. Mol Cancer Ther. 2013;12:58-68
14. Song B, Liu XS, Davis K, Liu X. Plk1 phosphorylation of Orc2 promotes DNA replication under conditions of stress. Mol Cell Biol. 2011;31:4844-56
15. Kopytova D, Popova V, Kurshakova M, Shidlovskii Y, Nabirochkina E, Brechalov A. et al. ORC interacts with THSC/TREX-2 and its subunits promote Nxf1 association with mRNP and mRNA export in Drosophila. Nucleic acids research. 2016;44:4920-33
16. Thomae AW, Baltin J, Pich D, Deutsch MJ, Ravasz M, Zeller K. et al. Different roles of the human Orc6 protein in the replication initiation process. Cellular and Molecular Life Sciences. 2011;68:3741-56
17. Liao X, Liu X, Yang C, Wang X, Yu T, Han C. et al. Distinct Diagnostic and Prognostic Values of Minichromosome Maintenance Gene Expression in Patients with Hepatocellular Carcinoma. J Cancer. 2018;9:2357-73
18. Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J. et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929-39
19. Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202-12
20. Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L. et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85-94
21. Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M. et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938-47
22. Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB. et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694-8
23. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98-w102
24. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419
25. Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Blal HA. et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321
26. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401-4
27. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1
28. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448-9
29. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498
30. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A. et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091-3
31. Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL. et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927
32. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M. et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. 2017; 45: D362-d8.
33. Bell SP, Dutta A. DNA replication in eukaryotic cells. Annual review of biochemistry. 2002;71:333-74
34. Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128
35. Lei M, Tye BK. Initiating DNA synthesis: from recruiting to activating the MCM complex. Journal of cell science. 2001;114:1447-54
36. Kim JC, Nordman J, Xie F, Kashevsky H, Eng T, Li S. et al. Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes & development. 2011;25:1384
37. Macalpine DM, Rodríguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes & development. 2004;18:3094
38. Macalpine HK, Gordan RPowell SK, Hartemink AJ, Macalpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Research. 2010;20:201-11
39. Diffley JF. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes & development. 1996;10:2819-30
40. Machida YJ, Hamlin JL, Dutta A. Right Place,Right Time,and Only Once: Replication Initiation in Metazoans. Cell. 2005;123:13-24
41. Scholefield G, Veening J-W, Murray H. DnaA and ORC: more than DNA replication initiators. Trends in cell biology. 2011;21:188-94
42. Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789-93
43. Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. The EMBO journal. 2004;23:2651-63
44. Huang C, Cheng J, Bawa-Khalfe T, Yao X, Chin YE, Yeh EH. SUMOylated ORC2 Recruits a Histone Demethylase to Regulate Centromeric Histone Modification and Genomic Stability. Cell Reports. 2016;15:147-57
45. Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Current Biology. 2007;17:1989-95
46. Tatsumi Y, Ezura K, Yoshida K, Yugawa T, Narisawa-Saito M, Kiyono T. et al. Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes to Cells. 2008;13:1045-59
47. Giri S, Chakraborty A, Sathyan KM, Prasanth KV, Prasanth SG. Orc5 induces large-scale chromatin decondensation in a GCN5-dependent manner. J Cell Sci. 2016;129:417-29
48. Yesu J, Stefan B, Neerja K, Philipp K, Srinka G, David MA. et al. Temporal profile of replication of human chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6419-24
49. Lee DG, Bell SP. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Molecular & Cellular Biology. 1997;17:7159-68
50. Franziska B, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519:321
51. Kurshakova M, Maksimenko O, Golovnin A, Pulina M, Georgieva S, Georgiev P. et al. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Molecular cell. 2007;27:332-8
52. Coulombe P, Nassar J, Peiffer I, Stanojcic S, Sterkers Y, Delamarre A. et al. The ORC ubiquitin ligase OBI1 promotes DNA replication origin firing. Nature communications. 2019;10:2426
53. Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026-31
54. Depamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell cycle. 2005;4:70-9
55. Shen Z. The Origin Recognition Complex in Human Diseases. Bioscience Reports. 2013;33:475-83
56. Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Current Opinion in Cell Biology. 2007;19:337-43
57. Bell SP, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844-9
58. Kenji S, Gasser SM. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell. 2007;128:85-99
59. Zhen H, Keling Z, Reichardt LF. The origin recognition core complex regulates dendrite and spine development in postmitotic neurons. Journal of Cell Biology. 2005;170:527-35
60. Maxim B, Huijbregts RPH, Igor C. Functional analysis of an Orc6 mutant in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10672-7
61. de Munnik SA, Bicknell LS, Aftimos S, Al-Aama JY, van Bever Y, Bober MB. et al. Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet. 2012;20:598-606
62. Young A, Berry R, Holloway AF, Blackburn NB, Dickinson JL, Skala M. et al. RNA-seq profiling of a radiation resistant and radiation sensitive prostate cancer cell line highlights opposing regulation of DNA repair and targets for radiosensitization. BMC cancer. 2014;14:808
63. Chen X, Xiong D, Ye L, Wang K, Huang L, Mei S. et al. Up-regulated lncRNA XIST contributes to progression of cervical cancer via regulating miR-140-5p and ORC1. Cancer Cell Int. 2019;19:45
64. Radojkovic M, Ristic S, Divac A, Tomic B, Nestorovic A, Radojkovic D. Novel ORC4L gene mutation in B-cell lymphoproliferative disorders. Am J Med Sci. 2009;338:527-9
65. Kim SH, Ho JN, Jin H, Lee SC, Lee SE, Hong SK. et al. Upregulated expression of BCL2, MCM7, and CCNE1 indicate cisplatin-resistance in the set of two human bladder cancer cell lines: T24 cisplatin sensitive and T24R2 cisplatin resistant bladder cancer cell lines. Investig Clin Urol. 2016;57:63-72
66. Koleck TA, Bender CM, Clark BZ, Ryan CM, Ghotkar P, Brufsky A. et al. An exploratory study of host polymorphisms in genes that clinically characterize breast cancer tumors and pretreatment cognitive performance in breast cancer survivors. Breast Cancer: Targets and Therapy. 2017;9:95
67. Yaguang X, Andrea F, Go N, Marko K, Jingfang J. Validation of biomarkers associated with 5-fluorouracil and thymidylate synthase in colorectal cancer. Oncology Reports. 2008;19:257-62
68. Xi Y, Nakajima G, Schmitz JC, Chu E, Ju J. Multi-level gene expression profiles affected by thymidylate synthase and 5-fluorouracil in colon cancer. Bmc Genomics. 2006;7:68 -
69. Gavin EJ, Song B, Wang Y, Xi Y, Ju J. Reduction of Orc6 expression sensitizes human colon cancer cells to 5-fluorouracil and cisplatin. Plos One. 2008;3:e4054
Author contact
![]() Corresponding author: Tao Peng, M.D., Professor, Director, Department of Hepatobiliary Surgery, the First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Province, China. Tel: (+86)-771-5356528. Fax: (+86)-771-5350031. E-mail: pengtaogmucom.
Corresponding author: Tao Peng, M.D., Professor, Director, Department of Hepatobiliary Surgery, the First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Province, China. Tel: (+86)-771-5356528. Fax: (+86)-771-5350031. E-mail: pengtaogmucom.

 Global reach, higher impact
Global reach, higher impact