Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(1):251-259. doi:10.7150/jca.35382 This issue Cite
Research Paper
A 6-Membrane Protein Gene score for prognostic prediction of cytogenetically normal acute myeloid leukemia in multiple cohorts
1. Hubei Bioinformatics & Molecular Imaging Key Laboratory, Department of Bioinformatics and Systems Biology, Key Laboratory of Molecular Biophysics of the Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, 430074, China.
2. Institute of Hematology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, China.
* Equal contribution authors
Received 2019-4-1; Accepted 2019-9-27; Published 2020-1-1
Abstract
Background: Cytogenetically normal acute myeloid leukemia (CN-AML) is a large proportion of AMLs with diverse prognostic outcomes. Identifying membrane protein genes as prognostic factors to stratify CN-AML patients will be critical to improve their outcomes.
Purpose: This study aims to identify prognostic factors to stratify CN-AML patients to choose better treatments and improve their outcomes.
Methods: CN-AML data were from TCGA cohort (n = 79) and four GEO datasets. We identified independent prognostic genes by Cox regression and Kaplan-Meier methods, and constructed linear regression model using LASSO algorithm. The prediction error curve was calculated using R package “pec”.
Results: Based on independent prognostic membrane genes, we constructed a regression model for CN-AML prognosis prediction: score = (0.0492 * CD52) - (0.0018 * CD96) + (0.0131 * EMP1) + (0.2058 * TSPAN2) + (0.0234 * STAB1) - (0.3658 * MBTPS1), which was named as MPG6 (6-Membrane Protein Gene) score. Tested in multiple CN-AML datasets, consistent results showed that CN-AML patients with high MPG6 score had poor survival, higher WBC count and shorter EFS. Comparing with other reported scoring models, the benchmark result of MPG6 achieved better association with survival in multiple cohorts. Moreover, by combining with other clinical indicators in CN-AML, MPG6 could improve the performance of survival prediction and serve as a robust prognostic factor.
Conclusions: We identified the MPG6 score as a stable indicator with great potential for clinical application in risk stratification and outcome prediction in CN-AML.
Keywords: Cytogenetically normal acute myeloid leukemia, membrane protein genes, MPG6 (6-Membrane Protein Gene) score, risk stratification, outcome prediction.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by expansion of undifferentiated myeloid precursors, resulting in impaired hematopoiesis [1]. Currently, chromosomal aberrations are well established as the diagnostic and prognostic markers in AML [2]. Patients with abnormal cytogenetic chromosomes such as PML-RARA, RUNX1-RUNX1T1 or MYH11-CBFB fusions are associated with favorable prognosis, whereas patients with monosomy karyotype or complex alterations are associated with high prognosis risk [3]. However, nearly half of AML patients are cytogenetically normal (CN-AML) with an intermediate prognosis and markedly diverse outcomes [4]. Therefore, it is very important to identify prognostic factors to stratify CN-AML patients to choose better treatments and improve their outcomes.
Recently, DNA mutations and aberrant RNA expression profiles were proposed as prognostic indicators for the treatment outcome in CN-AML. Notably, recurrent lesions in NPM1, FLT3-ITD and CEBPA mutations were identified to improve risk stratification for CN-AML patients [5]. Aberrant expression level of single gene such as DNMT3B [6], BAALC [7], and ERG [8] has also been reported to be associated with CN-AML patient outcome. Meanwhile, prognostic indicators based on multiple genes were also integrated to define CN-AML subgroups. In 2011, an integrative prognostic risk score based on clinical and molecular markers for gene expression and mutation was proposed for outcome prediction of CN-AML patients [9]. Recently, high expression of stem cell-associated genes was validated with negative prognostic impact in primary CN-AML [10]. However, these prognostic signatures lack consistency in different CN-AML cohorts and are not easy to use because they refer to many genes and mutations. Thus, there was still no stable and easy-used prognostic gene signatures applied into risk classifications in CN-AML. Currently, large scale datasets in TCGA [11] and GEO [12], and their analysis tools such as GSCALite webserver [13] made the analysis feasible.
Membrane proteins are essential for many biological processes such as cell signaling, transporting and cell adhesion and approximately occupy 20-30% of genes in human genome [14, 15]. Moreover, membrane proteins represent 60% of the known drug targets for therapeutics [16]. Thus, they are ideal biomarkers as an independent predictor for CN-AML patients' prognosis and classification. In this study, we generated a 6-Membrane Protein Gene (MPG6) score that was highly correlated to survival outcomes. The MPG6 score was confirmed to be independent in five CN-AML datasets in overall survival (OS) models. More importantly, we showed that the MPG6 score could improve the predictive performance to predict patients' survival and function as a good prognostic factor.
Material and methods
Patient's clinical information
We downloaded the gene expression data and clinical information of 79 CN-AML from the TCGA LAML dataset (https://tcga-data.nci.nih.gov/tcga/). Four other CN-AML microarray datasets (GSE311602 (n = 79) [11], GSE71014 (n = 104) [17], GSE12417 (n = 163) [11] and GSE6891 (n = 187) [18] were downloaded from the GEO database and were normalized as described by Metzeler et al [11]. The GSE311602 was a test cohort consisted of 79 adult German patients who were diagnosed with CN-AML in 2004. The GSE71014 cohort consisted of 104 CN-AML patients from the National Taiwan University Hospital. The GSE12417 was a training cohort consisted of 163 adult German patients. The patients in GSE6891 were diagnosed as CN-AML younger than or equal to 60 years. We should note that the clinical information such as age, sex, white blood cell (WBC) count, mutation status and FAB classification in TCGA and GSE6891 datasets were more complete than other three cohorts, which only have the OS information in GEO portal.
Survival analysis and membrane protein gene identification
High and low gene expression was defined using the median expression level of all CN-AML samples as threshold in that cohort. OS was defined as the time from AML diagnosis until death from any cause or last clinical follow-up. Clinical variables such as age, sex, WBC count, mutation status and FAB classification were assessed in the univariate analysis using the Kaplan-Meier method with the R package “survival” and variables with p-value <0.1 were remained for further analysis. After univariate analysis, we combined those significant prognostic factors in a multivariate analysis. The log-rank test was used to assess statistical significance. For multivariate analysis, multivariate Cox regression model [19] was used to study the association between gene expression levels and OS in the presence of other known clinical covariates such as age, sex, WBC count, mutation status and FAB classification in R package. Hazard ratios (HR) with relative 95% confidence interval (CI) were shown in multivariate analysis. The list of membrane protein genes were from Membranome 2.0 webserver [20].
Prognosis signature training
The dataset TCGA CN-AML (n = 79) was used for prognosis signature training. The correlation analysis of gene expressions was using R package “psych” (p <0.01, r <0.5) and genes with expression value ≤5 (RPKM) were removed as low expression. Using all genes' expression as features, we performed the linear regression analysis for gene expression against survival based on the Least absolute shrinkage and selection operator (LASSO) algorithm. Next, Sequential Backward Selection (SBS) method was used to choose the optimal feature for the model. Finally, an optimized linear regression model that made up of six membrane protein genes (MPG6) was constructed, which was highly correlated to survival outcomes in the training cohort.
Prognosis signature testing
We tested the robustness and practice performance of the MPG6 score in other four CN-AML cohorts: GSE311602 (n = 79), GSE71014 (n = 104), GSE12417 (n = 163) and GSE6891 (n = 187). For each cohort, we performed survival analysis with the median threshold of the MPG6 score to separate samples into high and low groups. Specifically, we counted the number of samples whose OS >2 years and OS >3 years in the high and low score groups. Then, we investigated the association between MPG6 score and other clinical indicators such as age, sex, WBC count, FAB classification, the presence of FLT3-ITD mutation and NPM1 mutation in TCGA CN-AML (n = 79) and GSE6891 (n = 187) cohorts.
Predictive performance test and independent prognostic analysis
The performance of the MPG6 score to predict survival of CN-AML patients was conducted by logistic regression. First, we used the score as the single variant to predict survival. Then, we combined multiple variants including age, WBC count, FLT3-ITD mutation and NPM1 mutation to predict survival and compared the result with that only using the score. Next, we used the multivariate logistic regression models that combined score and the above clinical indicators to assess the predictive performance. Finally, to test whether the MPG6 score could function as an independent prognostic factor, a multivariate survival analysis combined the above factors with multivariate Cox regression model was performed. The multivariate Cox regression analysis was performed in TCGA (n = 79) and GSE6891 (n = 187) CN-AML cohorts that had more detailed clinical information. Furthermore, we calculated the prediction error curve of the multivariate Cox regression model with R package “pec” [21]. The prediction error curve was defined via Brier's score [22] and designed to estimate the performance of a risk prediction model.
Benchmark the performance of MPG6
To evaluate the performance of MPG6, we benchmarked it with other three AML scoring models including the LSC17 score [23], the 7-gene score [24] and the 6-gene score [25] on CN-AML cohorts mentioned above. Survival analysis was performed using median score of each model as the threshold. For each model, absent genes in datasets were discarded.
Results
Identification of the independent prognostic MPG6 score in CN-AML
To identify potential independent prognostic markers in CN-AML, we initiated our study on the TCGA CN-AML cohort (n = 79). By performing Kaplan Meier analysis, we identified eight prognostic clinical indicators (age, sex, WBC count, mutation status and FAB classification) (p <0.1) (Table S1) and 1301 genes whose expression were significantly associated to OS. Then, after univariate and multivariate Cox regression analysis, we identified 203 genes independent of the eight clinical indicators in the TCGA CN-AML cohort (p <0.05) (Table S2). Among them, 23 were membrane protein genes, which are AGPAT4, AMICA1, B4GALT7, BAIAP3, CCT6B, CD1C, CD1E, CD2, CD33, CD52, CD7, CD96, CRIM1, EMP1, GPR125, GPR153, HRH2, LTB, MBTPS1, SMAGP, STAB1, TREML2 and TSPAN2. After correlation analysis and linear regression analysis, we finally constructed the regression model: score = (0.0492 * CD52) - (0.0018 * CD96) + (0.0131 * EMP1) + (0.2058 * TSPAN2) + (0.0234 * STAB1) - (0.3658 * MBTPS1), which was named as MPG6 score (6-Membrane Protein Gene score). The multivariable analysis of six membrane protein genes and clinical variables in TCGA were list in Table 1. Meanwhile, those six membrane protein genes are well reported. Among them, CD52, a small glycoprotein that is linked by a glycosylphosphatidylinositol (GPI) anchor to the surface membrane and was reported as a prognostic marker in hematological malignancies [26]. CD96 is a membrane bound receptor of the immunoglobulin superfamily and belongs to a network of interactions that manipulates in a multifaceted fashion adhesion, activation, and inhibition of participating cells[27]. EMP1 is an integral transmembrane glycoprotein, which has been identified as a poor prognostic factor in human cancers such as pediatric acute lymphoblastic leukemia, gliomas, gastric cancer, etc.[28]. TSPAN2 is a cell surface membrane protein of the tetraspanin superfamily and is involved in tumor metastasis and invasiveness in human malignancy [29]. STAB1 encodes a multifunctional type I transmembrane protein, which was identified as a prognostic factor for CN-AML in our recent work [30]. MBTPS1 (also known as Golgi-resident site-1 protease, S1P) acts as the inactive type I membrane precursor protein and serve as a crucial component that catalyzing the initial, sterol-regulated cut in the luminal loops of sterol regulatory element (SRE)-binding proteins [31].
High MPG6 score was correlated to poor survival in multiple independent datasets
Survival analysis using the median of MPG6 score as threshold indicated that CN-AML patients with high MPG6 score generally had lower survival rate in the TCGA training cohort (Fig. 1a) as well as other four independent cohorts (Fig. 1b). Furthermore, the OS median value of high MPG6 score group was much lower than that of low score group in all five cohorts, as also did the numbers of patients with OS >2 years and OS >3 years (Table 2). Especially in the GSE71014 cohort (n = 104), patients in high MPG6 score group had significantly shorter OS than patients in low MPG6 score group (p = 0.0095) (Fig. 1b). In GSE6891 (n = 187) cohort, the median OS of low MPG6 score group was five times higher than that of high MPG6 score group (11.99 vs 65.25) (log rank test p <0.0001) (Table 2). The results indicated that the MPG6 score could serve as a prognostic factor in CN-AML.
Multivariable analysis of six membrane protein genes in TCGA cohort.
| HR (95% CI)/(p-value) | ||||||
|---|---|---|---|---|---|---|
| OS Covariate | CD52 | CD96 | EMP1 | TSPAN2 | STAB1 | MBTPS1 |
| Age | 2.42 | 2.59 | 2.36 | 2.25 | 2.44 | 1.59 |
| (1.13-3.67) /(p=0.013) | (1.27-3.99)/(p=0.0011) | (1.07-3.51)/(p=0.0104) | (1.03-3.32)/(p=0.0172) | (1.15-3.62)/(p=0.0147) | (1.04-3.18)/(p=0.0077) | |
| DNMT3A | 1.45 | 2.63 | 2.21 | 2.53 | 2.20 | 2.07 |
| (0.69-2.45)/(p=0.1053) | (1.27-3.96)/(p=0.1008) | (1.07-3.43)/(p=0.018) | (1.08-4.16)/(p=0.0069) | (1.07-3.50)/(p=0.028) | (1.02-3.27)/(p=0.019) | |
| RUNX1 | 1.91 | 2.14 | 1.97 | 1.82 | 2.04 | 1.91 |
| (0.92-3.06)/(p=0.0329) | (1.09-5.82)/(p=0.0488) | (0.78-4.39)/(p=0.1763) | (0.79-4.83)/(p=0.0574) | (1.03-5.37)/(p=0.0417) | (0.88-4.57)/(p=0.0407) | |
| FLT3-ITD | -1.47 | 0.64 | 1.74 | 1.53 | 1.59 | 1.57 |
| (0.12-1.23)/(p=0.1377) | (0.67-2.48)/(p=0.5326) | (0.86-3.13)/(p=0.0517) | (0.79-3.01)/(p=0.1103) | (0.89-3.07)/(p=0.1124) | (0.77-2.97)/(p=0.1074) | |
| MT-CYB | 1.76 | 1.71 | 1.25 | 1.04 | 1.33 | 1.32 |
| (0.93-4.57)/(p=0.0729) | (0.82-10.94)/(p=0.1338) | (0.63-8.51)/(p=0.3494) | (0.53-7.42)/(p=0.2805) | (0.66-8.75)/(p=0.185) | (0.58-7.75)/(p=0.175) | |
| WT1 | 1.54 | 0.77 | 0.72 | 0.57 | 0.16 | 0.17 |
| (0.67-10.11)/(p=0.1203) | (0.58-3.92)/(p=0.4993) | (0.53-3.95)/(p=0.4718) | (0.48-3.60)/(p=0.5357) | (0.44-5.02)/(p=0.2461) | (0.43-4.92)/(p=0.2031) | |
| IDH2 | 0.07 | 0.021 | 0.43 | 0.52 | -0.47 | -0.46 |
| (0.43-2.26)/(p=0.9125) | (0.51-2.48)/(p=0.098) | (0.53-2.65)/(p=0.4344) | (0.56-2.57)/(p=0.5327) | (0.56-1.85)/(p=0.638) | (0.55-1.75)/(p=0.535) | |
| NPM1 | 0.68 | 0.19 | 0.16 | 0.19 | -0.05 | -0.04 |
| (0.52-3.79)/(p=0.4357) | (0.59-2.01)/(p=0.8976) | (0.51-1.86)/(p=0.817) | (0.57-2.07)/(p=0.7654) | (0.53-1.85)/(p=0.963) | (0.51-1.82)/(p=0.768) | |
| IDH1 | -0.23 | -1.47 | -1.31 | -1.43 | -1.28 | -1.18 |
| (0.41-1.67)/(p=0.7176) | (0.13-1.23)/(p=0.1287) | (0.2-1.28)/(p=0.144) | (0.12-1.27)/(p=0.145) | (0.16-1.4)/(p=0.2016) | (0.14-1.2)/(p=0.1789) | |
| Total | 2.4 | 2.5 | 2.2 ( | 1.8 | 1.98 | 0.56 |
| (1.2-4.8)/(p=0.013) | (1.2-5.2)/(p=0.016) | 1.2-4.1)/(p=0.018) | (1-3.3)/(p=0.041) | (1.01-3.03)/(p=0.0473) | (0.32-0.98)/(p=0.041) | |
The model was generated from a Cox regression model that included Age, gene mutation of DNMT3A, and RUNX1, FLT3-ITD, MT-CYB, WT1, IDH2, NPM1, IDH1 and expression level of each membrane protein gene. HR: Hazard Ratio.
Overall survival (OS) in high score and low score groups of five datasets.
| Dataset | Race | OS | HS group | LS group |
|---|---|---|---|---|
| TCGA CN-AML (n=79) | White/Africa | OS median (months) | 10.45 | 24.8 |
| Number of OS>2 years | 11 | 21 | ||
| Number of OS>3 years | 7 | 15 | ||
| GSE311602 (n=79) | White | OS median | 12.17 | 17.97 |
| Number of OS>2 years | 13 | 18 | ||
| Number of OS>3 years | 11 | 13 | ||
| GSE71014 (n=104) | White | OS median | 9 | 21.9 |
| Number of OS>2 years | 12 | 25 | ||
| Number of OS>3 years | 8 | 17 | ||
| GSE12417 (n=163) | Asian | OS median | 8.18 | 14.03 |
| Number of OS>2 years | 15 | 31 | ||
| Number of OS>3 years | 9 | 22 | ||
| GSE6891 (n=187) | White | OS median | 11.99 | 65.25 |
| Number of OS>2 years | 29 | 60 | ||
| Number of OS>3 years | 26 | 54 |
HS: High Score; LS: Low Score.
The MPG6 score correlated to OS and EFS in five independent CN-AML cohort. (a) Kaplan-Meier estimate of OS using median threshold of score in the TCGA CN-AML training dataset. (b) Kaplan-Meier estimate of OS using median threshold of score in other four array datasets (GSE311602, GSE71014, GSE6891 datasets). (c) Kaplan Meier-estimates of EFS using median threshold of score in TCGA CN-AML and GSE6891 datasets.
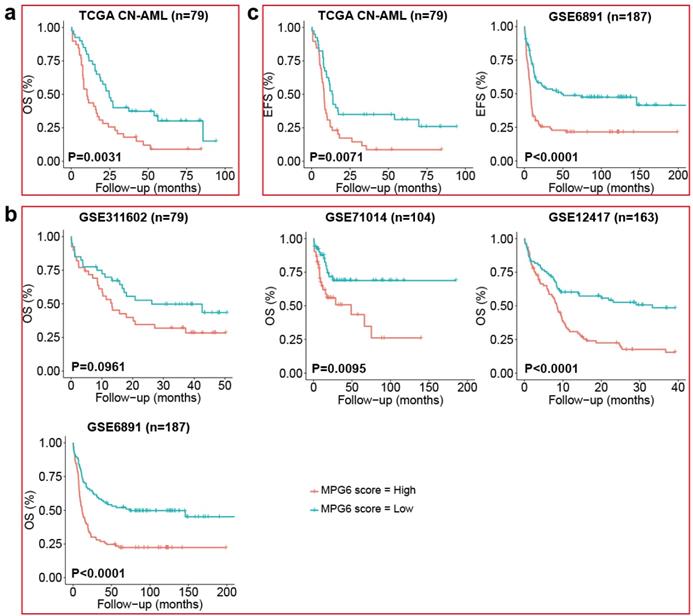
Furthermore, we benchmarked the performance of MPG6 score with other scoring models including LSC17 score [23], the 7-gene score [24] and the 6-gene score [25] as we mentioned in the method. The result indicated that other scoring models were correlated to survival in only one or two datasets (Fig. S1), while the MPG6 score achieved robust performance with a significant correlation of survival in four of five datasets (Fig. 1b).
The MPG6 score was significantly associated with patient clinical information
To further investigate the correlation of MPG6 score with other clinical indicators, we separated cohorts based on clinical information into high and low score groups, and the difference between two groups was measured by significance test for each clinical indicator. The result showed that except for OS, the MPG6 score was significantly associated with other clinical indicators such as WBC count (Wilcoxon rank-sum test p = 0.0053), EFS month (log rank test p = 0.0071 in TCGA CN-AML cohort and p <0.0001 in GSE6891 cohort) (Fig. 1c) and the presence of FLT3-ITD mutation (Fisher's exact test p = 0.0248) (Table 3). In addition, patients in high MPG6 score group was observed with higher WBC (median 50.64 vs 5) and shorter EFS than in low MPG6 score group (median 7.2 vs 13.4 in TCGA CN-AML cohort and 9 vs 14.39 in GSE6891 cohort) (log rank test p <0.0001) (Table 3). In GSE6891 cohort, the ratio of FLT3-ITD positive patients in high score group was 52.13% (49 out of 94) and 31.18% (29 out of 93) in low MPG6 score groups (Fisher's exact test p = 0.0248) (Table 3).
The MPG6 score can improve the predictive performance to predict survival of patients
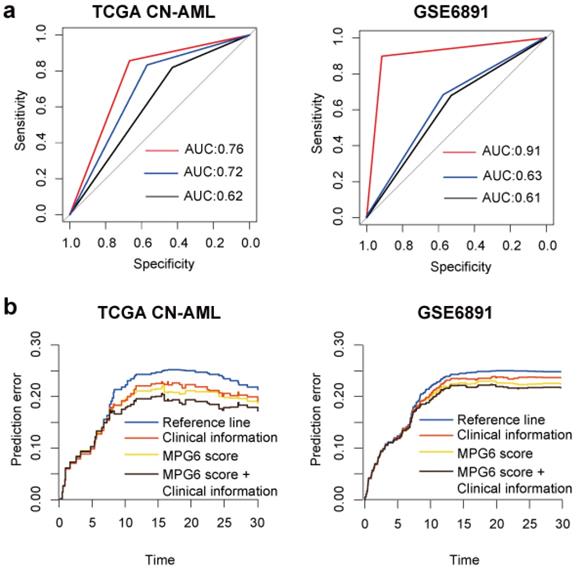
To investigate the performance of MPG6 score in predicting survival of patients, we performed logistic regression using MPG6 score as a single variant in the model and compared the result with models (detail in method). The result showed that in the training cohort, the predictive performance of MPG6 score as a single continuous variant outperformed the performance of combined variants including age, WBC count, FLT3-ITD and NPM1 mutation (AUC = 0.702 versus 0.624) (Fig. 2a). In multivariate logistic regression models that considered age, WBC count, FLT3-ITD mutation, NPM1 mutation and score as variants, we observed that the inclusion of MPG6 score greatly improved the predictive performance (AUC = 0.762 versus 0.624 in TCGA CN-AML cohort and AUC = 0.912 versus 0.614 in GSE6891 cohort) (Fig. 2a). The results demonstrated that the MPG6 score improved the performance to predict survival of CN-AML patients.
Clinical characteristics of the TCGA CN-AML and GSE6891 cohorts.
| Clinical information | TCGA CN-AML cohort | GSE6891 cohort | ||||
|---|---|---|---|---|---|---|
| High score | Low score | p-value | High score | Low score | p-value | |
| OS median | 10.45 | 24.8 | 0.0031$ | 11.99 | 65.25 | 0.000015$ |
| Sex (number) | M: 18 | M: 21 | 0.8600+ | M: 15 | M: 20 | 0.0825+ |
| F: 22 | F:18 | F: 24 | F: 19 | |||
| Age (median) | 66 | 68 | 0.2675* | 48 | 45 | 0.7201* |
| BM blast (%) | 77.5 | 66 | 0.3798|| | - | - | - |
| WBC count | 50.54 | 5 | 0.0053|| | - | - | - |
| NPM1 mutation | Pos: 23 | Pos: 17 | 0.2635+ | Pos: 57 | Pos: 48 | 0.2399+ |
| Neg: 17 | Neg: 22 | Neg: 37 | Neg: 45 | |||
| FLT3 mutation | Pos: 17 | Pos: 12 | 0.3524+ | Pos: 49 | Pos: 29 | 0.0248+ |
| Neg: 23 | Neg: 27 | Neg: 45 | Neg: 64 | |||
| EFS month | 7.2 | 13.4 | 0.0071$ | 9.00 | 14.39 | <0.0001$ |
Pos: Positive; Neg: Negative; $: log rank test; *: Student's t-test; +: Fisher's exact test; ||: Wilcoxon rank-sum test; The “-” in table means clinical indicators were absent in GSE6891 cohort.
The MPG6 score can improve the performance to predict survival. (a) ROC curve of logistic regression model in TCGA CN-AML and GSE6891 cohorts. The red line represents the ROC curve of combining MPG6 score with clinical information including age, WBC count, FLT3-ITD mutation and NPM1 mutation as features; the blue line represents ROC curve of using MPG6 score as the only feature in logistic regression model, and the black line represents ROC curve using clinical information mentioned above as features. (b) Prediction error curve of multivariate Cox regression model with or without MPG6 score. Reference line indicates Kaplan-Meier estimation without additional variables. Lower curve (lower prediction error) indicates better predictive value.
Multivariate survival analysis of score and other known predictors of outcome in TCGA CN-AML and GSE6891 cohorts.
| Overall Survival Covariate | TCGA CN-AML cohort | GSE6891 cohort | ||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | p-value | Hazard Ratio (95% CI) | p-value | |
| Age (median) | 1.02 (1.00-1.04) | 0.0474 | 1.00 (0.99-1.02) | 0.6986 |
| WBC count | 1.00 (0.99-1.01) | 0.1361 | - | - |
| BM blast (%) | 0.99 (0.97-1.01) | 0.1707 | - | - |
| NPM1 mutation | 0.88 (0.46-1.66) | 0.6835 | 0.66 (0.45-0.97) | 0.0366 |
| FLT3 mutation | 2.03 (1.11-3.73) | 0.0216 | 1.55 (1.04-2.30) | 0.0296 |
| MPG6 score | 1.08 (1.03-1.12) | 0.0004 | 4.12 (2.04-8.30) | <0.0001 |
The “-” in table means clinical indicators were absent in GSE6891 cohort.
The MPG6 score can function as a good independent prognostic factor
To investigate whether the performance of MPG6 score affected by other known predictors of outcome, we performed multivariate survival analysis based on the multivariate Cox regression model. The result indicated that the score could serve as an independent prognostic factor in two tested cohorts with detailed clinical information available including TCGA and GSE6891 (Table 4). Besides, we observed that after the inclusion of score in the model, some known predictors of outcome turned to be not significant, such as the WBC count in TCGA CN-AML cohort. However, the presence of NPM1 mutation was not significant no matter with or without the score in the model (Table 4). The result demonstrated that MPG6 score can be an independent prognostic factor and outperformed other known predictors. In addition, the prediction error curve indicated that MPG6 score could improve the predictive performance of multivariate Cox regression model (Fig. 2b).
Discussion
Over the past decades, the high heterogeneity of CN-AML presents a considerable challenge in the risk stratification [32]. About 20-30% of genes encode membrane proteins, which have immense significance in pharmacological research. However, very few studies about their potential as prognostic indicators in leukemia were conducted. Therefore, our study aimed at searching membrane protein genes to stratify CN-AML patients and predict the outcome of CN-AML. In this study, we generated a 6-membrane protein gene (MPG6) score from the TCGA dataset and confirmed in four independent validation sets (Fig. 1). Among them, many were confirmed as the therapeutic targets in leukemia. CD52 has been developed as a drug target in chronic lymphocytic leukemia (CLL) [33] and predicted to be a prognostic marker in AML [34]. CD96 may serve as an LSC-specific therapeutic target [35]. EMP1 was identified as a potential drug target in acute lymphoblastic leukemia (ALL) [36]. Although the role of TSPAN2, STAB1 and MBTPS1were not confirmed in leukemia, they were reported to be involved in the progression of the tumor metastasis[37-39]. From other point of view, our strategy to identify MPG6 demonstrated a credible approach, which may also be applicable in identifying such gene signatures in other types of cancers.
To our knowledge, this is the first report about predictive and/or prognostic biomarkers related to membrane protein gene in CN-AML. Although some prognostic biomarkers or factors of CN-AML patients were proven to be valuable, only a part of them was applied in clinical trial. Moreover, the prognostic values of many markers appear to be controversial because they were validated in limited samples. Compared with above issues, the MPG6 made up of only six genes and were easy testing using qPCR. Nevertheless, the limitation of MPG6 was that we detected mRNA expression here not protein. Therefore, a further detection of the membrane protein expression such as flow cytometry may be necessary.
It was reported that the outcome of AML was correlated to FLT3-ITD mutation and NPM1 mutation. The patients with FLT3-ITD mutation positive have a generally poor prognosis [40], on the contrary, patients with NPM1 mutation positive generally have a good outcome [41]. In this work, we expect that there are some correlations between gene mutations and MPG6. However, we only found that FLT3-ITD mutations were significantly associated with the high MPG6 score group in GSE6891 dataset (Table 3). Meanwhile, we observed that compared with OS, the correlation of MPG6 with other clinical indicators were not that strong, such as the difference of WBC count and FLT3-ITD mutation between high- and low-score groups were significant only in certain tested dataset. We consider that this may be caused by the following reason, the CN-AML was highly heterozygous, though, the model trained with genes' expression against survival performs good in predicting survival, when it came to the detail clinical indicators such as WBC count and FLT3-TID mutation, it worked not that well. In view of this problem, we suggest in the future work related to the prognostic model in CN-AML, clinical indicators related to the prognostic could be considered into the training model, which may lead to a more robust result.
Abbreviations
ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; CI: confidence interval; CLL: chronic lymphocytic leukemia; CN-AML: cytogenetically normal acute myeloid leukemia; CPH: Cox proportional hazards; OS: overall survival; MPG6: 6-Membrane Protein Gene; TCGA: The Cancer Genome Atlas; WBC: white blood cell; HR: Hazard ratios.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank Dr. Peter J.M. Valk at Erasmus University Medical Center for his kindly providing the clinical information of GSE6891 dataset and members working for the TCGA AML group. This work was supported by National Natural Science Foundation of China (NSFC Nos. 31822030, 31771458, 31801113).
Authors' contributions
Sheng-Yan Lin and Ya-Ru Miao analyzed the data and wrote the manuscript. Fei-Fei Hu, Hui Hu and Qiong Zhang participated in data analysis. Zhichao Chen and Qiubai Li provided specific knowledge on AML. An-Yuan Guo and Zhichao Chen designed the study and revised the manuscript. All authors read and approved the final manuscript and submission.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. Gan to Kagaku Ryoho Cancer & Chemotherapy. 2015;56:1136-1152
2. Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH. et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116:354-365
3. Islam M, Mohamed Z, Assenov Y. Differential Analysis of Genetic, Epigenetic, and Cytogenetic Abnormalities in AML. Int J Genomics. 2017;42:1495-1507
4. Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T. et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424-432
5. Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J. et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. N Engl J Med. 2012;366:1079-1086
6. Niederwieser C, Kohlschmidt J, Volinia S, Whitman SP, Metzeler KH, Eisfeld AK. et al. Prognostic and biologic significance of DNMT3B expression in older patients with cytogenetically normal primary acute myeloid leukemia. Leukemia. 2015;29:567-575
7. Langer C, Radmacher MD, Ruppert AS, Whitman SP, Paschka P, Mrózek K. et al. High BAALC expression associates with other molecular prognostic markers, poor outcome, and a distinct gene-expression signature in cytogenetically normal patients younger than 60 years with acute myeloid leukemia: a Cancer and Leukemia Group B (CALGB) stu. Blood. 2008;111:53-71
8. Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrózek K, Whitman SP. et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Onco. 2005;23:9234-9242
9. Damm F, Heuser M, Morgan M, Wagner K, Görlich K, Grosshennig A. et al. Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood. 2011;117:4561-4568
10. Metzeler KH, Maharry K, Kohlschmidt J, Volinia S, Mrózek K, Becker H. et al. A stem cell-like gene expression signature associates with inferior outcomes and a distinct microRNA expression profile in adults with primary cytogenetically normal acute myeloid leukemia. Leukemia. 2013;27:2023-2031
11. Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC. et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193-4201
12. Ron E, Michael D, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207-210
13. Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771-3772
14. Hayat M, Khan A. MemHyb: predicting membrane protein types by hybridizing SAAC and PSSM. J Theor Biol. 2012;292:93-102
15. Jones DT. Do transmembrane protein superfolds exist? Febs Letters. 1998;423:281-285
16. Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727-730
17. Chuang MK, Chiu YC, Chou WC, Hou HA, Tseng MH, Kuo YY. et al. An mRNA expression signature for prognostication inde novoacute myeloid leukemia patients with normal karyotype. Oncotarget. 2015;6:39098-39110
18. Roel GW, Bas JW, Claudia AJ, Erpelinck, Saman AH. Berna B. et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131-134
19. Li Z, Herold T, He C, Valk PJ, Chen P, Jurinovic V. et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol. 2013;31:1172-1181
20. Lomize AL, Hage JM, Pogozheva ID. Membranome 2.0: database for proteome-wide profiling of bitopic proteins and their dimers. Bioinformatics. 2017;34:1061-1062
21. Schumacher M, Binder H, Gerds T. Assessment of survival prediction models based on microarray data. Bioinformatics. 2007;23:1768-1774
22. Brier GW. Verification of forecasts expressed in terms of probability. Mon Weather Rev. 1950;78:1-3
23. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N. et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433-437
24. Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH. et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014;32:548-556
25. Zhao X, Li Y, Wu H. A novel scoring system for acute myeloid leukemia risk assessment based on the expression levels of six genes. Int J Mol Med. 2018;42:1495-1507
26. Blatt K, Herrmann H, Hoermann G, Willmann M, Cerny-Reiterer S, Sadovnik I. et al. Identification of campath-1 (CD52) as novel drug target in neoplastic stem cells in 5q-patients with MDS and AML. Clin Cancer Res. 2014;20:3589-3602
27. Georgiev H, Ravens I, Papadogianni G, Bernhardt G. Coming of Age: CD96 Emerges as Modulator of Immune Responses. Front Immuno. 2018;9:1072-1075
28. Wang YW, Cheng HL, Ding YR, Chou LH, Chow NH. EMP1, EMP 2, and EMP3 as novel therapeutic targets in human cancer. Rev Cancer. 2017;1868:199-211
29. Otsubo C, Otomo R, Miyazaki M, Matsushima-Hibiya Y, Kohno T, Iwakawa R. et al. TSPAN2 is involved in cell invasion and motility during lung cancer progression. Cell Rep. 2014;7:527-538
30. Lin SY, Hu FF, Miao YR, Hu H, Lei Q, Zhang Q. et al. Identification of STAB1 in multiple datasets as a prognostic factor for cytogenetically normal acute myeloid leukemia: mechanism and drug indications. Mol Ther-Nucl Acids. 2019 in press
31. Velho RV, De Pace R, Klunder S, Di Lorenzo G, Schweizer M, Braulke T. et al. Site-1 protease and lysosomal homeostasis. Mol Cell Res. 2017;1864:2162-2168
32. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND. et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209-2221
33. Jaglowski SM, Lapo A, Rosa L, Natarajan M, Byrd JC. The clinical application of monoclonal antibodies in chronic lymphocytic leukemia. Blood. 2010;116:3705-3714
34. Katharina B, Harald H, Gregor H, Michael W, Sabine CR, Irina S. et al. Identification of campath-1 (CD52) as novel drug target in neoplastic stem cells in 5q-patients with MDS and AML. Clin Cancer Res. 2014;20:3589-3602
35. Naoki H, Park CY, Naoya T, Yusuke O, Haruo S, Martin G. et al. CD96 is a leukemic stem cell-specific marker in human acute myeloid leukemia. PNAS. 2007;104:11008-11013
36. Aris IM, Jerchel IS, R E S R vdD, Berk LCJ, Van Den, Boer JM, Horstmann MA. et al. EMP1, a novel poor prognostic factor in pediatric leukemia regulates prednisolone resistance, cell proliferation, migration and adhesion. Leukemia. 2014;28:1828-1837
37. Yaseen IH, Monk PN, Partridge LJ. Tspan2: a tetraspanin protein involved in oligodendrogenesis and cancer metastasis. Biochem Soc T. 2017;45:465-475
38. Brekken RA, Puolakkainen P, Graves DC, Workman G, Lubkin SR, Sage EH. Enhanced growth of tumors in SPARC null mice is associated with changes in the ECM. J Clin Invest. 2003;111:487-495
39. Kennedy AJ, Mathews TP, Kharel Y, Field SD, Moyer ML, East JE. et al. Development of amidine-based sphingosine kinase 1 nanomolar inhibitors and reduction of sphingosine 1-phosphate in human leukemia cells. J Med Chem. 2011;54:3524-3548
40. Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD. et al. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res. 2001;61:7233-7239
41. Konstanze DH, Schlenk RF, Marianne H, Claudia S, Rücker FG, Andrea C. et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740-3746
Author contact
![]() Corresponding authors: Mailing address: College of Life Science and Technology, Huazhong University of Science and Technology, Luoyu Road 1037, Wuhan, China. Phone & Fax: 86-027-87793177; An-Yuan Guo, email: guoayedu.cn; Zhichao Chen, email: chenzhichaoedu.cn
Corresponding authors: Mailing address: College of Life Science and Technology, Huazhong University of Science and Technology, Luoyu Road 1037, Wuhan, China. Phone & Fax: 86-027-87793177; An-Yuan Guo, email: guoayedu.cn; Zhichao Chen, email: chenzhichaoedu.cn

 Global reach, higher impact
Global reach, higher impact