Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(1):132-141. doi:10.7150/jca.35426 This issue Cite
Research Paper
Aberrant CDK9 expression within chordoma tissues and the therapeutic potential of a selective CDK9 inhibitor LDC000067
1. Precision Medicine Center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China.
2. Sarcoma Biology Laboratory, Department of Orthopaedic Surgery, David Geffen School of Medicine at University of Los Angeles, Los Angeles, CA 90095, USA.
Received 2019-4-2; Accepted 2019-9-16; Published 2020-1-1
Abstract
Objectives: Chordomas are slow-growing malignancies that commonly affect vital neurological structures. These neoplasms are highly resistant to current chemotherapeutic regimens and often recur after surgical intervention. Therefore, there is an urgent need to identify molecular targets and more robust drugs to improve chordoma patient outcomes. It is well accepted that cyclin-dependent protein kinase 9 (CDK9) has tumorigenic roles in various cancers; however, the expression and significance of CDK9 in chordoma remains unknown.
Methods: Expression of CDK9 in chordoma cell lines and tumor tissues was examined by Western blot and immunohistochemistry (IHC). The correlation between CDK9 expression in patient tissues and clinical prognosis was analyzed. The functional roles of CDK9 in chordoma were investigated after the addition of small interfering RNA (siRNA) and CDK9 inhibitor (LDC000067). Cell growth and proliferation were assessed with MTT and clonogenic assays. The effect of CDK9 inhibition on chordoma cells was further evaluated with a three-dimensional (3D) cell culture model which mimics the in vivo environment.
Results: CDK9 was expressed in both chordoma cell lines and chordoma tissues. High- expression of CDK9 correlated with recurrence and poor outcomes for chordoma patients. CDK9 silencing with siRNA decreased growth and proliferation of chordoma cells and lowered levels of Mcl-1 and RNA polymerase II (RNAP II) phosphorylation. Pharmacological inhibition of CDK9 with the small molecular inhibitor LDC000067 reduced cell growth, supported apoptosis, suppressed cell colony formation in a clonogenic assay, and decreased spheroid growth in 3D culture.
Conclusion: We demonstrate that CDK9 expression in chordoma correlates with patient outcome, and, when inhibited, chordoma cell growth and proliferation significantly decreases. Taken together, these results support CDK9 as an emerging potential target in chordoma therapy.
Keywords: CDK9, chordoma tissues, LDC000067
1. Introduction
Chordomas are primary bone malignancies that arise along the axial skeleton and account for approximately 20% of primary spinal tumors and 3% of all bone tumors [1]. Although chordomas are slow growing, they are aggressive and prone to metastasize. Their growth and pervasive nature can make complete surgical resection impractical, as they often compress critical neurological structures such as the brain, spinal cord, and nerves [2]. Despite these surgical obstacles, resection remains the mainstay treatment as chordomas are highly chemoresistant. Currently, with thorough pre- and post-operative radiation therapy, the three and five year survival rates remain poor with high rates of relapse and metastasis [3]. These limitations in current management highlight the urgent need to identify new molecular targets and chemotherapeutic strategies for chordoma.
Cyclin-dependent kinases (CDKs) are a family of serine/threonine protein kinases that regulate cell-cycle progression and DNA transcription [4]. Previous studies have reported aberrant CDK expression and activation in various cancers [5-8]. This has prompted a growing interest in novel CDK inhibitor drugs. One such drug, palbociclib (PD0332991), a CDK4/6 inhibitor, was approved by the Food and Drug Administration (FDA) for the treatment of breast cancer [9]. With this clinical success, there have been an increasing number of clinical trials analyzing CDK inhibitors in various cancers [10-12].
Recently, CDK9 has seen more recognition for its role in DNA transcription initiation. Mechanistically, this occurs through a phosphorylation-mediated activation of the largest subunit of RNA polymerase II (RNAP II). This CDK9 mediated tumorigenic step has seen increasing attention within the literature and has been implicated in pancreatic cancer, hypopharyngeal carcinoma, leukemia, breast cancer, and hepatocellular carcinoma [13-17]. However, the function and therapeutic potential of CDK9 in chordoma have not been elucidated. In this study, we show CDK9 is related to poor outcomes and promotes chordoma progression and inhibits apoptosis by regulating RNAP II and apoptosis-related proteins. These findings substantiate CDK9 as a potential novel target in chordoma treatment.
2. Materials and methods
2.1. Reagents
Human non-specific siRNA (5′-CUCUUAUCUACAUAAGGAU-3′) and CDK9 targeted siRNA (5'-GCUGCUAAUGUGCUUAUCA-3) were purchased from Sigma-Aldrich (St. Louis, MO). Lipofectamine® RNAiMAX was purchased from Thermo Fisher Scientific (Waltham, MA). The highly selective inhibitor of CDK9 (LDC000067) was purchased from Selleck Chemicals (Houston, TX). Monoclonal rabbit anti-CDK9, anti-p-RNAP II Ser-2 (13499S), anti-Survivin (2808S), anti-Bax (5023S), anti-Mcl-1(94296) and monoclonal mouse anti-α-tubulin (3874) antibodies were purchased from Cell Signaling Technology (Danvers, MA). The anti-RNAP II antibodies were purchased from Abcam (Cambridge, MA).
2.2. Chordoma tissue microarray (TMA)
A retrospective study with 55 tissue samples was conducted for the TMA immunohistochemical (IHC) stain as reported previously [18]. These tissues were hematoxylin and eosin (H&E) stained to enable better evaluation and diagnosis of the chordoma tissue samples. All tissues were used in accordance with the policies of the institutional review board (IRB) of the hospital and common rules of the U.S. Department of Health and Human Services. Written informed consent was obtained from the patients for all specimens and clinical information used in this study. Histopathological and clinical information including patient age, gender, metastasis, recurrence, tumor location(s), months of follow-up, expression of brachyury, and disease status were collected (Table 1).
2.3. Immunohistochemistry (IHC)
The multi-tissue block was pre-heated for two hours at 60˚C. The sections were then deparaffinized in a series of xylene solutions, then rehydrated in graded ethanol and distilled water. Next, the slides underwent antigen retrieval with Target Retrieval Solution (Dako, North America, Inc., Carpinteria, CA) before being incubated in 3% hydrogen peroxide for 10 minutes to quench endogenous peroxidase activity. They were then blocked with 5% goat serum and incubated with human CDK9 primary antibody overnight in a 4°C humidified chamber (1:50 dilution, in 1% bovine serum albumin BSA. Cell Signaling Technology). After the slides were rinsed with Tris-buffered saline with Tween® 20 (TBST) three times, the residual array bound antibodies were detected with SignalStain® Boost Detection Reagent (Cell Signaling Technology) and SignalStain® DAB (Cell Signaling Technology). The DAB reaction was terminated upon optimal color visualization. Following the DAB reaction, the slides were counterstained with hematoxylin and mounted with VectaMount AQ (Vector Laboratories, CA) for long-term preservation.
The stained slides underwent microscopic examination (Nikon Instruments Inc., NY) with the percentage of cells with positive nuclear staining calculated by two independent investigators blinded to the clinical data and the other viewer's score. The nuclear staining of CDK9 was graded into the following six groups: 0, <15% of cells stained positive; 1+, 15-30% stained positive; 2+, 31-50% stained positive; 3+, 51-70% stained positive; 4+, 71-85% stained positive; 5+, >85% stained positive.
2.4. Human chordoma cell lines and cell culture
The human chordoma cell line UCH1 and UCH2 were established and kindly provided by Dr. Silke Bruderlein (University Hospitals of Ulm, Ulm, Germany) [19]. The cell line CH22 was established in our laboratory as previously reported [20]. CH19, another new chordoma cell line, was established in our laboratory as well. The UCH2, CH22, and CH19 cells were maintained in Dulbecco's Modified Eagle Medium (DMEM) medium (GIBCO, Grand Island, NY) and the UCH1 cells were maintained in RPMI1640 medium (GIBCO). These media were supplemented with 10% fetal bovine serum (Sigma-Aldrich, MO) and 1% penicillin/streptomycin (Life Technologies, CA). In their respective media, the cells were incubated at 37°C with 5% CO2.
2.5. Immunofluorescence (IF) assay
The expression of CDK9 in chordoma cells was visualized via immunofluorescence assay as described previously [21]. The UCH2 and CH22 cells were first seeded into 24-well plates before being incubated in 4% paraformaldehyde, fixed with ice-cold methanol, and blocked with 1% bovine serum albumin. Immunostaining was performed with CDK9 (1:200 dilution) and β-actin (1:500 dilution, Sigma-Aldrich, St. Louis, MO). After being washed three times with PBS, the cells were incubated with Alexa Fluor 594 (red) goat anti-mouse antibody and Alexa Fluor 488 (green) conjugated goat anti-rabbit antibody (Invitrogen) for one hour. Images were obtained with a Nikon Eclipse Ti-U fluorescence microscope equipped with a SPOT RT™ digital camera. Green color highlights CDK9 protein, and red color highlights the cytoplasm.
2.6. Western blotting analyses
Cell pellets and human chordoma tissues were lysed in protein lysis buffer. The protein concentrations were calculated with Protein Assay Reagents (Bio-Rad, CA) and a SpectraMax 340PC Microplate Reader from Molecular Devices (San Jose, CA). Equal amounts of protein were separated on 4-12% Bis-Tris gels (NuPAGE®, Thermo Fisher Scientific, CA) and transferred to nitrocellulose membranes, where they were incubated with specific primary antibodies at 4°C overnight (CDK9 1:1000 dilution; RNAP II at 1: 1000 dilution; p-RNAP II Ser-2 at 1: 1000 dilution; Mcl-1 at 1: 1000 dilution; Bax at 1: 1000 dilution; Survivin at 1: 1000 dilution α -Tubulin at 1: 1000 dilution). After being washed with TBST three separate times for five minutes, the membranes were further incubated with Goat anti-rabbit IRDye 800CW or Goat anti-mouse IRDye 680LT secondary antibody (LI-COR Biosciences, NE. Final images were obtained with Odyssey® CLx equipment (LI-COR Biosciences). The abundance of α-Tubulin was monitored to ensure equal loading.
2.7. MTT cell viability assay
Cell viability was determined by conventional 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay using a standard protocol as described previously. Treatment was performed with coincubation of UCH2 or CH22 cells with LDC000067 or siRNA for 120 hours. The color change was determined by a photometer set to a 490nm wavelength. The results were normalized and used to generate cell survival graphs with GraphPad Prism 7 software (GraphPad Software, CA).
2.8. Cell clonogenic assay
UCH2 and CH22 cell lines were seeded at 300 cells/well in 6-well plates with various concentrations of LDC000067 and 2mL medium. The medium was replaced every five days for fresh medium until there were visible colonies. These colonies were then Giemsa stained and manually counted. Pictures of the stained colonies were captured with a digital camera (Olympus, Tokyo, Japan).
2.9. 3D culture
CH22 and UCH2 cells were cultured in VitroGelTM 3D (The Well Bioscience Inc., NJ) in 24-well plates at a density of 2×104 cells/well, according to the manufacturer's protocol. Different cell culture media (with or without 5 µM of LDC000067) was added to cover the hydrogel. The plates were then placed in the incubator and had their cover media changed every 48 hours. Following a 21-day period, the spheroids were imaged on the Nikon Eclipse Ti-U fluorescence microscope equipped with a SPOT RT™ digital camera.
2.10. Statistical analysis
The data was analyzed using GraphPad PRISM 7 software. Statistical significance between groups was determined with a log-rank test for the Kaplan-Meier analysis, and a Wilcoxon signed-rank test for group comparisons. Errors were SD of averaged results and p values <0.05 were considered statistically significant between means.
3. Results
3.1. Correlations between CDK9 expression and clinical prognosis
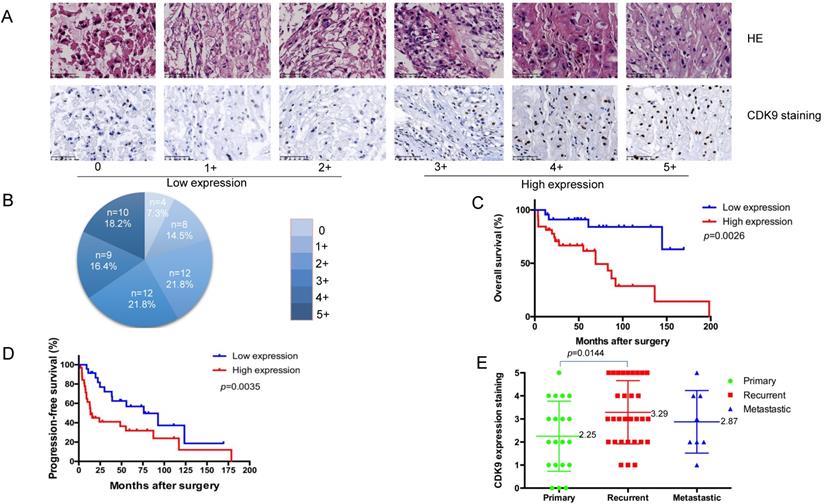
To evaluate the clinical relevance of CDK9 expression in chordoma, we analyzed its expression in a human chordoma TMA. The CDK9 protein was predominantly localized within the nucleus of chordoma cells (Figure 1A). The patient characteristics of our cohort are summarized in Table 1. We analyzed 55 total patients who had an average age of 57.6 years (median age 59). Among these tissue samples, 20 (36.4%) were localized, 32 (58.2%) tissue samples were locally recurred only, and 3(5.4%) were metastatic without local recurrence. Of note, there were 8 total metastatic tissue samples, 5 of which had local recurrence and metastasis. The recurrent tissue samples were obtained from the primary chordoma site. IHC was conducted to assess the expression profile of CDK9. The samples were divided into two subgroups as follows: a low-expression group including 24 total patients with scores of 0 (4 of 55, 7.3%), 1+ (8 of 55, 14.5%), and 2+(12 of 55, 21.8%), and a high-expression group including 31 total patients with scores of 3+ (12 of 55, 21.8%), 4+ (9 of 55, 16.4%), and 5+ (10 of 55, 18.2%) (Figure 1A, 1B and Table 1).
Kaplan-Meier survival analysis revealed patients in the CDK9 low-expression group to have significantly better outcomes than those in the CDK9 high-expression group (p= 0.0026). Specifically, patients with a low-expression of CDK9 had a significantly longer overall survival (OS) time (mean OS = 78.4 months) compared to those with high-expression (mean OS = 50.2 months) (Figure 1C). Furthermore, CDK9 expression was inversely correlated with progression-free survival (PFS) (Figure 1D). The patients with low-expression of CDK9 had a longer PFS (mean PFS= 57.5 months) compared to the high-expression group (mean PFS= 33.0 months), which was statistically significant (p= 0.0035). The average CDK9 expression for patients with primary versus locally recurrent chordomas was statistically significant as well, at 2.25 and 3.29, respectively (p= 0.0144). The mean expression of CDK9 for the metastatic disease group was 2.87. There was no significant difference between the groups in terms of age, sex, location, or margin of resection (Table 2). There was also no significant difference between the metastatic disease group and the primary tumor or locally recurrent groups (Figure 1E). Taken together, these data outline how elevated CDK9 expression is a potential prognostic marker and agent of chordoma progression.
3.2. Expression of CDK9 in chordoma cell lines and tissues
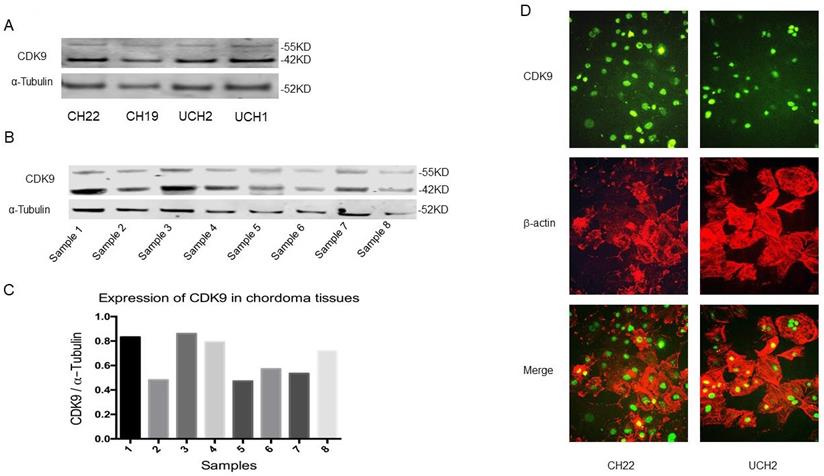
CDK9 is expressed in two isoforms; a lighter 42KD isoform, and a heavier 55KD isoform which is transcribed from an upstream transcriptional start site which extends the shared mRNA sequence. This larger 55KD protein has an additional 117 amino acids at the N-terminus [22]. Western blot analysis revealed both CDK9 isoforms are expressed in CH22, CH19, UCH2 and UCH1 cell lines (Figure 2A). To establish the existence of these two isoforms in primary cancer as well, we examined CDK9 expression in eight primary chordoma specimens. The tissues had variable levels of CDK9 expression (Figure 2B, 2C). Clinical information of the eight patients is shown in Supplementary Table S1.
Correlations between CDK9 expression and clinicopathological features
| Parameter | Number of cases (%) | CDK9 low expression (n, %) | CDK9 high expression (n, %) | |
|---|---|---|---|---|
| Total | 55 | 24(43.6%) | 31(56.4%) | |
| Age | Average | 57.6 | 61.22 | 55.00 |
| Median age | 59 | 62 | 52 | |
| Sex | Male | 41 | 18(43.9%) | 23(56.1%) |
| Female | 14 | 6(42.9%) | 8(57.1%) | |
| Location | Mobile Spine | 21 | 8(38.1%) | 13 (61.9%) |
| Sacral | 34 | 16(47.1%) | 18(52.9%) | |
| Stage | Primary | 20 | 11(55.0%) | 9 (45.0%) |
| Metastatic | 8 | 4(50.0%) | 4(50.0%) | |
| Recurrent | 32 | 11(34.4%) | 21(65.6%) | |
| Status | NED | 22 | 10(45.5%) | 12(54.5%) |
| AWD | 8 | 7(87.5%) | 1(12.5%) | |
| DOD | 23 | 5(21.7%) | 18(78.3%) | |
| DOO | 2 | 2(100.0%) | 0(0.00%) |
NED: No evidence of disease, AWD: Alive with disease, DOD: Dead of disease, DOO: Dead of other disease.
The clinical parameters of chordoma tissue microarray
| Variables | Total number of patient events | Non-survival n (%) | Survival (month) Mantel-Cox analysis |
|---|---|---|---|
| Age at diagnosis, year | 0.4296 | ||
| <50 | 19 | 8(42.1%) | |
| 50 | 36 | 14(38.9%) | |
| Sex | 0.5404 | ||
| Male | 41 | 16(39.0%) | |
| Female | 14 | 6(42.8%) | |
| Location | 0.7308 | ||
| Mobile Spine | 21 | 11(52.4%) | |
| Sarcral | 34 | 12(35.3%) | |
| Margin | 0.0101 | ||
| Intralesional | 23 | 15(65.2%) | |
| Marginal | 3 | 2(66.7%) | |
| Attempted wide with contanminated margin | 10 | 1(10.0%) | |
| Wide | 16 | 3(18.8%) | |
| N/A | 3 | 1(33.3%) | |
| Radiation | 0.4430 | ||
| No | 12 | 6(50.0%) | |
| Yes | 43 | 16(37.2%) | |
| Brachyury | 0.5367 | ||
| Low | 30 | 14(46.7%) | |
| High | 18 | 6(33.3%) | |
| Unknown | 7 | 2(28.6%) | |
| CDK9 expression | 0.0026* | ||
| Low | 24 | 4(16.7%) | |
| High | 31 | 18(58.1%) |
N/A: not applicable. *Statistically significant
Correlation between CDK9 expression and clinicopathological outcomes for chordoma patients. (A) Representative images of different immunohistochemical stain intensities of CDK9. On the basis of the percentage of cells with positive nuclear staining, CDK9 staining patterns were categorized into 6 groups: 0, no nuclear staining; 1+: <10% of positive cells; 2+, 10%-25% of positive cells; 3+, 26%-50% of positive cells; 4+, 51%-75% of positive cells; 5+, >75% of positive cells. Original magnification 200×. (B) Pie chart representing relative frequency of different CDK9 staining patterns in chordoma tissue microarrays. (C) Kaplan-Meier curves depicting overall survival rates in the two groups of chordoma patients by CDK9 staining pattern. Low expression (number=24) mean OS = 78.4 months, high expression (number=31) mean OS = 50.2 months, p=0.0026. (D) Kaplan-Meier curves depicting progression-free survival rates in the two groups of chordoma patients by CDK9 staining pattern. Low expression (number=24) mean PFS= 57.5 months, high expression (number=31) mean PFS= 33.0 months, p=0.0035 (E) Levels of CDK9 expression in chordoma patients with primary chordoma (number=20, mean score=2.25), patients who developed recurrence (number=32, mean score=3.29) or metastasis (number=8, mean score=2.87).
To further confirm the expression of CDK9 and its subcellular localization, we tested CDK9 expression with immunofluorescence assay in CH22 and UCH2 cell lines. The CDK9 protein was localized in the nucleus in both chordoma cell lines (Figure 2D), which is consistent with the chordoma tissue IHC results (Figure 1A).
3.3. siRNA CDK9 inhibition in chordoma cells
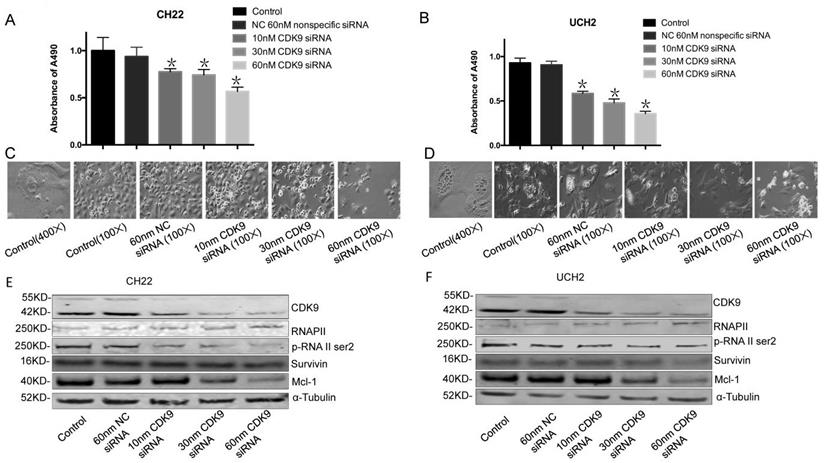
To evaluate the functional role of CDK9 in chordoma, we used synthetic RNA interference (RNAi) to disrupt CDK9 expression in chordoma cells lines. The MTT assay showed CDK9 specific siRNA significantly inhibits cell growth and proliferation while non-specific siRNA has no significant effect (Figure 3A, 3B).
To evaluate how CDK9 silencing affects CDK9 regulated proteins, we analyzed the expression of RNAP II within CDK9 siRNA transfected chordoma cells. Additionally, we tested the efficacy of CDK9-siRNA in CH22 and UCH2 cell lines by Western blot. In brief, cells were incubated with 10 nM, 30 nM, and 60 nM CDK9 siRNA for 48 hours. As expected, CDK9-siRNA silencing occurred in a dose-dependent manner, while the untreated and nonspecific siRNA treated samples showed no significant effect (Figure 3E, 3F). Furthermore, there was a strong decrease of p-RNAP II Ser-2 expression in a dose-dependent manner (Figure 3E, 3F). Downregulation of Mcl-1 was also observed in the CDK9 siRNA transfected chordoma cells (Figure 3E, 3F). We found inhibition of CDK9 in chordoma cell lines to decrease p-RNAP II Ser-2 and Mcl-1 while also suppressing proliferation and inducing apoptosis. Collectively, these results suggest that inhibition of CDK9 suppresses growth and proliferation of chordoma cells.
3.4. Pharmacological CDK9 inhibition in chordoma cells
To investigate the effects of CDK9 inhibition at the protein level, we treated cells with the CDK9 inhibitor drug LDC000067. MTT assay confirmed a significant decrease in cell growth after LDC000067 treatment (Figure 4A, 4B). The half-maximal inhibitory concentrations (IC50) values for UCH2 and CH22 were 2.211μM and 1.354 μM, respectively.
CDK9 expression in chordoma cell lines and tissues. (A) CDK9 expression in chordoma cell lines. (B and C) CDK9 expression in chordoma tissues. There are 2 isoforms of the CDK9 protein: the 42 KD CDK9 isoform and the 55 KD isoform. The smaller 42 KD isoform was the first identified isoform. Expression of CDK9 (55KD band) was normalized to α-Tubulin. (D) Cellular localization of CDK9 in UCH2 and CH22 cells was assessed by immunofluorescence with antibodies to CDK9 and actin.
Effect of CDK9 inhibition by siRNA on chordoma cell lines. (A and B) After transfected with increasing concentrations of CDK9 specific siRNA or nonspecific siRNA for 3 days, cell viability was determined by MTT assay after siRNA transfection in both UCH2 and CH22 cells. *p<0.05. (C and D) The morphology of UCH2 and CH22 cell lines changed after 3 days of siRNA transfection. α-Tubulin was used as a loading control. (E and F) The proteins of CDK9 and downstream proteins p-RNA II ser2 and Mcl-1 in cells were examined by Western blot after 3 days of siRNA transfection.
Effect of CDK9 inhibitor treatment on chordoma cell lines. (A and B) After exposure to increasing concentrations of CDK9 inhibitor for 120 h, cell viability was decreased in a dose-dependent manner in both UCH2 and CH22 cells, with the IC50 values for LDC000067 at 3.22 µM and 4.47 µM, respectively. (C) After incubation of UCH2 and CH22 cell lines with 1.25 µM, 2.5µM, 5.0µM, and 10.0 µM LDC000067 for 48 h, they showed a strong decrease of p-RNA II ser2 and Mcl-1 expression with increasing LDC000067 concentration. α-Tubulin was used as a loading control.
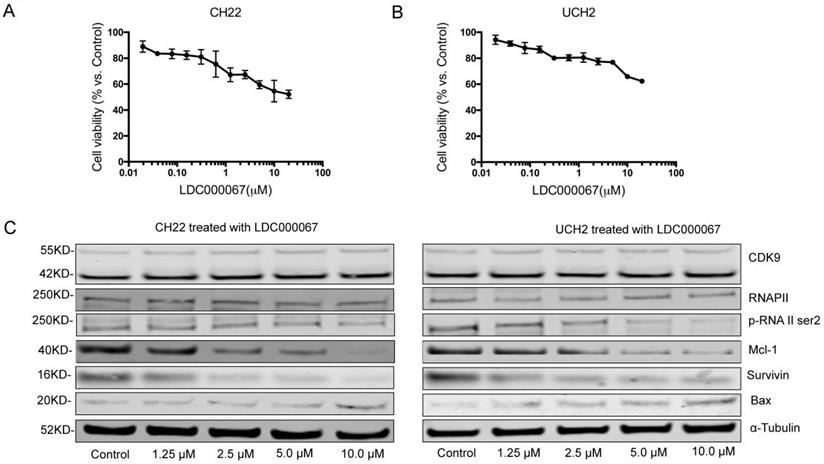
To evaluate how CDK9 inhibition alters downstream pathways, we observed several CDK9 related downstream markers after LDC000067 treatment. Here we reveal that increasing concentrations of LDC000067 over a 48 hour period reduce p-RNAP II Ser-2 in a dose-dependent manner in both CH22 and UCH2 cells, without significantly affecting the CDK9 or RNAP II proteins. This finding is consistent with previous work which reports LDC000067 inhibits CDK9 activity without affecting its overall expression [23].
We also examined apoptosis-associated proteins in LDC000067 treated cells. As shown in Figure 4C, following LDC000067 treatment, expression of the pro-apoptotic protein Bax increased while the anti-apoptotic proteins Survivin and Mcl-1 decreased (Figure 4C). Overall, these results support targeting CDK9 as a potential strategy in chordoma therapy.
3.5. Effect of CDK9 inhibition on clonogenic assay and 3D culture
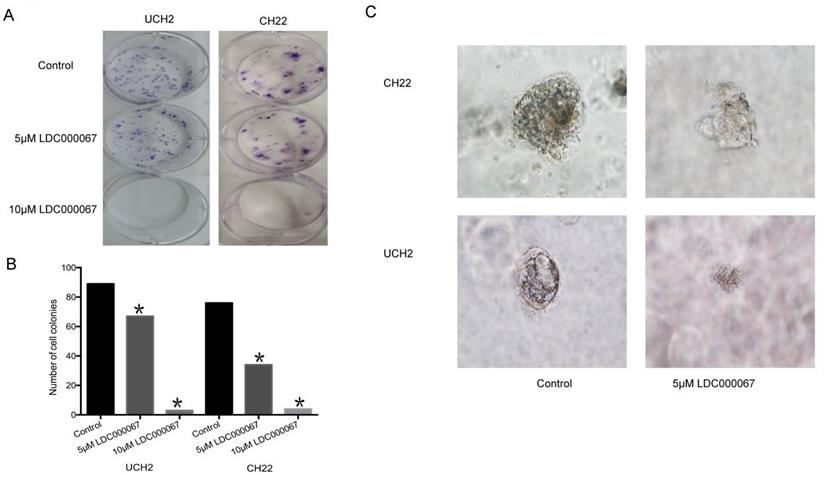
In order to assess the effect of CDK9 inhibition on colony formation, we implemented a clonogenic assay. The number and size of colonies in LDC000067 treated cells were significantly reduced, according to dose, compared to untreated cells (Figure 5A and 5B).
3D cell culture mimics the in vivo environment by permitting cancer cells to grow in all directions, similar to how they would in a living tissue. Given the advantage of this artificial environment, we investigated the effect of CDK9 inhibition on chordoma cell proliferation within the 3D culture. Over time, we observed the diameter of formed cancer spheroids from LDC000067 treated cells to be significantly smaller than the untreated cells (Figure 5C).
4. Discussion
Chordomas are resistant to radiotherapy and presently utilized drug regimens [24]. Therefore, revealing which chemotherapeutics effectively target chordoma is crucial for advancing past the barriers of current chordoma therapies. In the present study, we show CDK9 is aberrantly expressed in chordoma cell lines and tumor tissues. In addition, the expression of CDK9 in patients with recurrent disease was significantly higher compared to those with primary disease. It seems likely, therefore, that CDK9 expression may feature more prominently in recurrent chordoma tumorigenesis. Further work is required to establish the exact mechanism of this relationship. This finding is especially important clinically, as recurrence remains a major obstacle in management and complete resection is complicated by the close relationship of these tumors with nearby neurovascular structures.
Effects of CDK9 inhibition on clonogenic assay and 3D culture. (A and B) Representative results of colony formation of chordoma cell lines treated with CDK9 inhibitor. (C) Assessed spheroid formation of chordoma cell lines treated with CDK9 inhibitor in 3D cell culture.
We observed that patients with a high expression of CDK9 have significantly worse outcomes in terms of OS and PFS. This significant correlation between CDK9 expression and patient outcome is consistent with previous works on other cancers and adds to a growing body of evidence recognizing CDK9 as a potential prognostic marker [13, 25, 26]. For example, CDK9 can be highly expressed in pancreatic cancers, and when elevated, correlates to worse outcomes for these patients [13]. Similar results have been reported in breast and gastric cancer [27]. These findings, alongside our current work, provide increasing evidence that CDK9 may be an emerging biomarker with significant tumorigenic potential.
Recently, several studies have contended CDK9 targeting to be a promising chemotherapeutic strategy [13, 16, 17]. Their promising results are likely a function of reducing the activity of CDK9 induced RNAP II transcription of various potentially tumorigenic genes. To verify the roles of CDK9 in chordoma cell growth and proliferation, we conducted a knockdown analysis using CDK9 specific siRNA. After this treatment, MTT revealed CDK9 knockdown to inhibit chordoma cell growth and proliferation. Furthermore, our Western blot data showed inhibition of CDK9 suppresses anti-apoptotic protein expression. Specifically, expression of Mcl-1 and Survivin decreased after genetic and chemical inhibition of CDK9.
CDK9 is known to transcriptionally upregulate the expression of Mcl-1 [28, 29]. In addition, previous works have shown inhibition of Mcl-1 to promote apoptosis and reduce tumor proliferation in Mcl-1 overexpressing osteosarcoma and melanoma cells [30, 31], prompting the preclinical development and clinical trials of Mcl-1 inhibitors [29, 32]. Consistent with our work, we observed CDK9 inhibition to induce a concomitant decrease of Mcl-1 and associated antiapoptotic factors, with an overall suppression of chordoma cell growth. Survivin is a member of the inhibitor of apoptosis protein family (IAP) and functionally suppresses the terminal effector enzymes caspase-3 and caspase-7 [33, 34]. Experimental findings have revealed inhibition of Survivin to prevent chordoma cell growth, and as expected, to induce apoptosis [35]. Collectively, the decrease in Mcl-1 and Survivin reasonably explains how inhibition of CDK9 may induce apoptosis in chordoma cell lines.
To expand the clinical application of our work and further characterize the role of CDK9 in chordoma cell growth and proliferation, we conducted an inhibition analysis using the specific CDK9 inhibitor LDC000067. This novel drug specifically binds to the ATP binding pockets of kinases. Importantly, the selectivity of LDC000067 for CDK9 is better than other CDK9 inhibitors such as flavopiridol and DRB, making it ideal for our investigation [23]. In our study, the addition of LDC000067 in chordoma cell lines showed encouraging results, as it inhibited tumor cell growth and proliferation. In accordance with our work, previous studies have demonstrated that pharmacological inhibition of CDK9 induces apoptosis in several human cancers [13, 23, 36]. For example, at concentrations of LDC000067 greater than 2 μM, breast cancer cell lines increase their percentage of apoptotic cells [23]. Our study shows the expression of Survivin to decrease and the expression of the pro-apoptotic protein Bax to increase in LDC000067 treated chordoma cells, suggesting this drug can disrupt aberrant CDK9 apoptotic signaling cascades. As an additional mechanism, phosphorylation of the RNAP II CTD residues serine 2 (p-RNA II ser2) was inhibited in a dose-dependent manner after treatment with CDK9 inhibitor, which is consistent with previous reports on colon carcinoma and adult T-cell leukemia/lymphoma [15, 37]. Overall, our results support LDC000067 mediated inhibition of CDK9 as a potential strategy in chordoma therapy.
Finally, we utilized a 3D cell culture platform to better connect our in vitro work with in vivo application. Permitting cancer to form spheroids allows for more accurate assessment of chemotherapeutic agents, as the cancer model better mimics live tissue drug diffusion, retention, and therapeutic outcome [38]. We successfully established 3D spheroids of chordoma cells and found that CDK9 inhibition significantly diminishes chordoma spheroid growth.
In summary, our study demonstrates that CDK9 is highly expressed in chordoma, and when elevated, correlates with recurrence and worse prognosis. We also show knockdown and inhibition of CDK9 decreases chordoma cell proliferation and growth. Although there is abundant room for further research and validation, these findings suggest CDK9 is a potential therapeutic target for chordoma treatment.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported, in part, by a Joint Research Fund (228282) devoted to clinical pharmacy and precision medicine (Z.D, Z. Y). Support has also been provided by the Gattegno and Wechsler funds. Dr. Duan is supported, in part, a Grant from National Cancer Institute (NCI)/National Institutes of Health (NIH), UO1, CA151452-01. Dr. Shen is supported by scholarship from the Henan Health Scholarship Council.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gröschel S. et al. Defective homologous recombination DNA repair as therapeutic target in advanced chordoma. Nat Commun. 2019;10(1):1635
2. Denaro L, Berton A, Ciuffreda M, Loppini M, Candela V, Brandi ML, Longo UG. Surgical management of chordoma: A systematic review. J Spinal Cord Med. 2018:1-16
3. Al-Mefty O. Chordoma. Acta Neurochir (Wien). 2017;159(10):1869-1871
4. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17(2):93-115
5. Liu W, Rodgers GP. Olfactomedin 4 expression and functions in innate immunity, inflammation, and cancer. Cancer Metastasis Rev. 2016;35(2):201-212
6. Li KK, Ng IO, Fan ST, Albrecht JH, Yamashita K, Poon RY. Activation of cyclin-dependent kinases CDC2 and CDK2 in hepatocellular carcinoma. Liver. 2002;22(3):259-268
7. Zhou Y, Shen JK, Hornicek FJ, Kan Q, Duan Z. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer. Oncotarget. 2016;7(26):40846-40859
8. Whittaker SR, Mallinger A, Workman P, Clarke PA. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther. 2017;173:83-105
9. Walker AJ, Wedam S, Amiri-Kordestani L, Bloomquist E, Tang S, Sridhara R, Chen W, Palmby TR, Fourie Zirkelbach J, Fu W. et al. FDA Approval of Palbociclib in Combination with Fulvestrant for the Treatment of Hormone Receptor-Positive, HER2-Negative Metastatic Breast Cancer. Clin Cancer Res. 2016;22(20):4968-4972
10. Dickson MA, Tap WD, Keohan ML, D'Angelo SP, Gounder MM, Antonescu CR, Landa J, Qin LX, Rathbone DD, Condy MM. et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol. 2013;31(16):2024-2028
11. Dickson MA, Schwartz GK, Keohan ML, D'Angelo SP, Gounder MM, Chi P, Antonescu CR, Landa J, Qin LX, Crago AM. et al. Progression-Free Survival Among Patients With Well-Differentiated or Dedifferentiated Liposarcoma Treated With CDK4 Inhibitor Palbociclib: A Phase 2 Clinical Trial. JAMA Oncol. 2016;2(7):937-940
12. Lim JS, Turner NC, Yap TA. CDK4/6 Inhibitors: Promising Opportunities beyond Breast Cancer. Cancer Discov. 2016;6(7):697-699
13. Kretz AL, Schaum M, Richter J, Kitzig EF, Engler CC, Leithauser F, Henne-Bruns D, Knippschild U, Lemke J. CDK9 is a prognostic marker and therapeutic target in pancreatic cancer. Tumour Biol. 2017;39(2):1010428317694304
14. Cao S, Yu Y, Chen S, Lei D, Wang S, Pan X, Peng J. Inhibition of CDK9 induces apoptosis and potentiates the effect of cisplatin in hypopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2017;482(4):536-541
15. Narita T, Ishida T, Ito A, Masaki A, Kinoshita S, Suzuki S, Takino H, Yoshida T, Ri M, Kusumoto S. et al. Cyclin-dependent kinase 9 is a novel specific molecular target in adult T-cell leukemia/lymphoma. Blood. 2017;130(9):1114-1124
16. Mitra P, Yang RM, Sutton J, Ramsay RG, Gonda TJ. CDK9 inhibitors selectively target estrogen receptor-positive breast cancer cells through combined inhibition of MYB and MCL-1 expression. Oncotarget. 2016;7(8):9069-9083
17. Huang CH, Lujambio A, Zuber J, Tschaharganeh DF, Doran MG, Evans MJ, Kitzing T, Zhu N, de Stanchina E, Sawyers CL. et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28(16):1800-1814
18. Chen H, Garbutt CC, Spentzos D, Choy E, Hornicek FJ, Duan Z. Expression and Therapeutic Potential of SOX9 in Chordoma. Clin Cancer Res. 2017;23(17):5176-5186
19. Retraction note to. KDM3A confers metastasis and chemoresistance in epithelial ovarian cancer. J Mol Histol. 2015;46(6):511-518
20. Correction. Cdk5 Directly Targets Nuclear p21(CIP1) and Promotes Cancer Cell Growth. Cancer Res. 2017;77(6):1504
21. He Y, Chen X, Yu Y, Li J, Hu Q, Xue C, Chen J, Shen S, Luo Y, Ren F. et al. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Molecular carcinogenesis. 2018;57(6):772-783
22. Liu H, Herrmann CH. Differential localization and expression of the Cdk9 42k and 55k isoforms. J Cell Physiol. 2005;203(1):251-260
23. Albert TK, Rigault C, Eickhoff J, Baumgart K, Antrecht C, Klebl B, Mittler G, Meisterernst M. Characterization of molecular and cellular functions of the cyclin-dependent kinase CDK9 using a novel specific inhibitor. Br J Pharmacol. 2014;171(1):55-68
24. Stacchiotti S. et al. Building a global consensus approach to chordoma: a position paper from the medical and patient community. The Lancet Oncology. 2015;16(2):e71-83
25. Czudor Z, Balogh M, Banhegyi P, Boros S, Breza N, Dobos J, Fabian M, Horvath Z, Illyes E, Marko P. et al. Novel compounds with potent CDK9 inhibitory activity for the treatment of myeloma. Bioorg Med Chem Lett. 2018;28(4):769-773
26. Wang L, Gao W, Hu F, Xu Z, Wang F. MicroRNA-874 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting CDK9. FEBS Lett. 2014;588(24):4527-4535
27. Lu Y, Tang L, Zhang Q, Zhang Z, Wei W. MicroRNA-613 inhibits the progression of gastric cancer by targeting CDK9. Artif Cells Nanomed Biotechnol. 2018;46(5):980-984
28. Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19(12):1401-1415
29. Lyle L, Daver N. Current and emerging therapies for patients with acute myeloid leukemia: a focus on MCL-1 and the CDK9 pathway. Am J Manag Care. 2018;24(16 Suppl):S356-S365
30. Zhang Y, Shi C, Yin L, Zhou W, Wang H, Seng J, Li W. Inhibition of Mcl-1 enhances Pevonedistat-triggered apoptosis in osteosarcoma cells. Exp Cell Res. 2017;358(2):234-241
31. Lucas KM, Mohana-Kumaran N, Lau D, Zhang XD, Hersey P, Huang DC, Weninger W, Haass NK, Allen JD. Modulation of NOXA and MCL-1 as a Strategy for Sensitizing Melanoma Cells to the BH3-Mimetic ABT-737. Clinical Cancer Research. 2012;18(3):783-795
32. Eyvazi S, Hejazi MS, Kahroba H, Abasi M, Zamiri RE, Tarhriz V. Cdk9 as an Appealing Target for Therapeutic Interventions. Curr Drug Targets. 2018
33. Li DY, Hu CH, Li HB. Survivin as a novel target protein for reducing the proliferation of cancer cells (Review). Biomedical Reports. 2018;8(5):399-406
34. Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. IAP-family protein Survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Research. 1998;58(23):5315-5320
35. Froehlich EV, Rinner B, Deutsch AJA, Meditz K, Knausz H, Troppan K, Scheipl S, Wibmer C, Leithner A, Liegl B. et al. Examination of Survivin Expression in 50 Chordoma Specimens-A Histological and In Vitro Study. Journal of Orthopaedic Research. 2015;33(5):771-778
36. Polier G, Ding J, Konkimalla BV, Eick D, Ribeiro N, Kohler R, Giaisi M, Efferth T, Desaubry L, Krammer PH. et al. Wogonin and related natural flavones are inhibitors of CDK9 that induce apoptosis in cancer cells by transcriptional suppression of Mcl-1. Cell Death Dis. 2011;2:e182
37. Bragelmann J, Dammert MA, Dietlein F, Heuckmann JM, Choidas A, Bohm S, Richters A, Basu D, Tischler V, Lorenz C. et al. Systematic Kinase Inhibitor Profiling Identifies CDK9 as a Synthetic Lethal Target in NUT Midline Carcinoma. Cell Rep. 2017;20(12):2833-2845
38. Yang C, Hornicek FJ, Wood KB, Schwab JH, Choy E, Iafrate J, Rosenberg A, Nielsen GP, Xavier RJ, Mankin H. et al. Characterization and analysis of human chordoma cell lines. Spine (Phila Pa 1976). 2010;35(13):1257-1264
Author contact
![]() Corresponding authors: Zhenfeng Duan, Sarcoma Biology Laboratory, Department of Orthopaedic Surgery, David Geffen School of Medicine at University of Los Angeles, 615 Charles E. Young Dr. S., Los Angeles, CA 90095, USA. Phone: +1-310-206-0029. Fax: +1-310-825-5409. E-mail address: zduanucla.edu; Quancheng Kan, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China. E-mail address: quanchengkannet
Corresponding authors: Zhenfeng Duan, Sarcoma Biology Laboratory, Department of Orthopaedic Surgery, David Geffen School of Medicine at University of Los Angeles, 615 Charles E. Young Dr. S., Los Angeles, CA 90095, USA. Phone: +1-310-206-0029. Fax: +1-310-825-5409. E-mail address: zduanucla.edu; Quancheng Kan, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450052, China. E-mail address: quanchengkannet

 Global reach, higher impact
Global reach, higher impact