Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(21):5015-5021. doi:10.7150/jca.30828 This issue Cite
Review
Emerging roles of circRNA in formation and progression of cancer
Department of Pathology, Guangdong Medical University, Dongguan 523808, Guangdong Province, China
*Contributed equally
Received 2018-10-21; Accepted 2019-8-5; Published 2019-8-28
Abstract
Circular RNAs (circRNAs) are recently discovered as a special novel type of endogenous noncoding RNAs (ncRNAs), which form a covalently closed continuous loop and are highly represented in the eukaryotic transcriptome. Recent research revealed that circRNAs can function as microRNA (miRNA) sponges, regulators of splicing and transcription, as well as interact with RNA-binding proteins (RBPs). In this review, not only the function and mechanism, but also the experimental methods of circRNA are summarized. The summary of the current state of circRNA will help us in the discovery of novel biomarkers, the therapeutic targets and their potential significance in diagnosis and treatment of diseases. CircRNAs might play important roles in cancers especially in hepatocellular carcinoma, gastric carcinoma and colorectal cancer as well as serving as diagnostic or predictive biomarkers of some diseases and providing new treatments of diseases.
Keywords: circular RNA, microRNA, cancer, colorectal cancer
Introduction
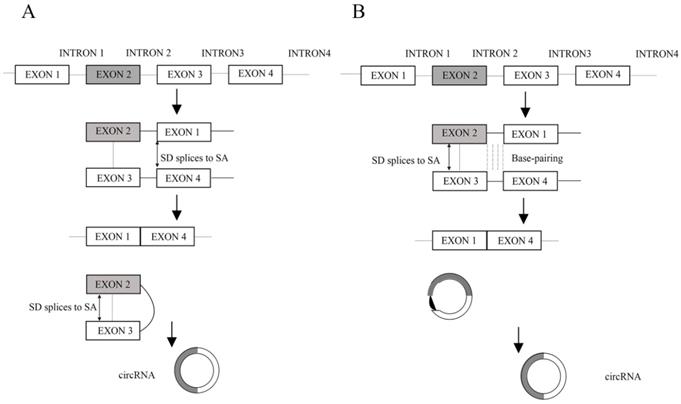
Unlike linear RNAs that are terminated with 5′ caps and 3′ tails, circRNAs form covalently closed loop structures with neither 5′-3′ polarities nor polyadenylated tails, which makes them much more stable than linear RNA and insusceptible to degradation by RNA exonuclease or RNase R [1]. CircRNA was first found in RNA viruses as early as the 1970s [2] and considered to be the result of the erroneously alternative splicing because of its low level expressions [3, 4]. With the development of technology and bioinformatics, Jeck et al. put forward two models of circRNA formation [5]. Model 1 is termed 'lariat-driven circularization' or 'exon skipping' (Figure 1A), and model 2 is termed 'intron-pairing-driven circularization' or 'direct back splicing' (Figure 1B) [5]. Later, Zhang et al. discovered a new type of circRNA which is derived from introns and is termed circular intronic RNAs (ciRNAs) [6]. Recently, Li et al. also revealed exons which are circularized with introns 'retained' between the exons and termed them EIciRNAs [7]. In addition, the muscle blind protein (MBL) can bind to circMbl flanking introns to provoke the formation of circRNAs. In this way, MBL can act as RBPs (RNA-binding proteins, proteins that bind to RNA molecules, are found in the cytoplasm and nucleus, and they are important in forming ribonucleoproteins (RNPs), generally target single-stranded regions within secondary structure domains where the functional groups of the bases may be easily available for sequence specific recognition) to bridge two flanking introns close together [8]. Furthermore, the interactions between RBPs form a bridge between the flanking introns, which bring the splice donor and splice acceptor into close proximity to promote circRNA biogenesis [9]. According to the advances in our understanding of circRNA biogenesis, these properties of circRNAs can be summarized as follows. Firstly, circRNAs are much more stable than linear RNAs [1]. Secondly, circRNAs are mainly composed of exons, which primarily reside in the cytoplasm and possibly have miRNA response elements (MREs) (miRNAs often bind their mRNA targets based on sequence complementarity in specific locations on the 3′ untranslated region (UTR) of the mRNA, termed MREs) [5, 10, 11]. Thirdly, the abundance of circular molecules exceeds those of the corresponding linear mRNAs by >10-fold in some cases [5]. Fourthly, circRNAs are evolutionarily conserved between different species [5, 10, 12]. Lastly, the vast majority of circRNAs are endogenous noncoding RNAs (ncRNAs) [13].
1. The functins of circRNAs
1.1 CircRNAs function as miRNA sponges
The competitive endogenous RNAs (ceRNAs) can compete for miRNAs binding with their MREs [14]. MiRNAs can prevent the translation of target mRNA by complementary pairing with target mRNA 3′-UTR region, which affects the stability of target mRNA and regulates the expression in the nucleus by binding to promoters [15, 16]. Therefore, with its MREs, circRNAs, a new member of ceRNAs, plays an important role in the expression of RNAs by adsorbing miRNAs [17]. It was tested that ciRS-7/cerebellar degeneration related protein 1 antisense (CDR1as) is shown to bind miR-7 [18]. Murine Sex-determining region Y (Sry) is the gene responsible for mammalian sex determination and can produce a circRNA which has 16 binding sites for miR-138, indicating that the circular Sry RNA likely acts as a miR-138 sponge [17, 19, 20]. Additionally, Peng et al. found that cir-ZNF609 (ID: hsa_circ_0000615 in circBase) may act as a sponge for miR-150-5p to modulate the expression of AKT3 [21].
1.2 CircRNAs regulate transcription and alternative splicing
Some research revealed that the knockdown of ciRNA derived from intron of ANKRD52 (Ci-ankrd52) leads to reduced expression of their parent genes by combinding RNA Pol II, and lines of evidence suggest one possible function for circRNAs as positive regulators of RNA Pol II transcription [6]. Also, detailed studies discovered that circMbl is generated by the second exon of the splicing factor muscleblind, which competes with canonical premRNA splicing [8]. Well, circMbl flanking introns and circMbl itself have conserved MBL binding sites, suggesting that general splicing factors, such as MBL, may have effects on alternative splicing that modulate the balance between circRNA biogenesis and canonical splicing [8].
A. Lariat-driven circularization. Exon-skipping leads to a lariat whose restricted structure promotes circularization. B. Intron-pairing-driven circularization. Intron 1 and intron 3 are formed circular structure via base-pairing. Introns are removed or retained to form circRNA. (SD: splice donor, SA: splice acceptor).
1.3 CircRNAs interact with RBPs
Multiple evidence demonstrated noncoding RNA controls gene expression both at the transcriptional and post-transcriptional level through physical interaction with RBPs or other noncoding RNAs [22]. It was demonstrated that ectopic expression of circ-Foxo3 (a circular RNA generated from a member of the forkhead family of transcription factors, Foxo3) repressed cell cycle progression by binding to the cell cycle proteins cyclin-dependent kinase 2 (also known as cell division protein kinase 2, CDK2) and cyclin-dependent kinase inhibitor 1 (CDKN1 or p21) [23]. As a result, silencing endogenous circ-Foxo3 promotes cell proliferation [23]. RNA-binding motif protein 20 (RBM20) is critical for the formation of a subset of circRNAs originating from the titin gene, which is known to undergo complex alternative splicing in mammalian hearts [24].
1.4 CircRNAs regulate translation
As a member of ncRNAs, few circRNAs can be translated. Evidences are presented based on electron microscopy and electrophoretic behaviour that hepatitis delta virus (HDV) contains a single stranded circular RNA molecule [25]. This is the first animal virus identified with a circular RNA genome. Another interesting discovery is about covalently closed circular RNA (CCCRNA). It is the smallest one among all known viroids and virusoids and the only one that codes proteins [26]. Its sequence possesses an internal ribosome entry site (IRES) and is directly translated through two (or three) completely overlapping ORFs (shifting to a new reading frame at the end of each round) [26].
2. Experimental methods of circRNAs
2.1 CircRNAs chip
The circRNAs chip (Arraystar Human circRNAs chip, ArrayStar) containing 5396 probes specific for human circular RNAs splicing sites can be used to investigate different expressions of circRNAs between tumor tissues and normal tissues. After hybridization and washing with samples, samples (tumor tissues and matched non-tumor tissues) were analyzed on the circRNAs chips. These chosen circRNAs are screened for further analysis[27].
2.2 RNA extraction and microarray analysis
After being extracted from each sample using a homogenizer and TRIzol regent, total RNA is digested with RNase R to remove linear RNA and enrich circRNA. Subsequently, the enriched circRNA is amplified and transcribed into fluorescent cRNA (complementary RNA) utilizing a random priming method. After hybridizing, hybridized arrays are supposed to be scanned. When comparing two groups of profile differences, the fold change (i.e. the ratio of the group averages) between the groups for each circRNA is computed [28].
2.3 Quantitative real-time (qRT-PCR)
cDNA is synthesized with the Reverse Transcription System using random primers. qRT-PCR analysis is carried out to detect circRNAs expressions [29]. After silencing and overexpression of circRNAs, the results of qRT-PCR are necessary to detect the expression of them.
2.4 Fluorescence in situ hybridization (FISH)
FISH can be used to investigate the expression and intracellular location of circRNAs in tissue and cell lines [29].
2.5 Cell Counting Kit-8 (CCK-8) and 5-ethynyl-2'-deoxyuridine (EDU)
Cell proliferation rates are detected with CCK-8 [29] and EDU [30].
2.6 Gene ontology (GO) analysis
GO analysis (geneontology.org) can be used to construct meaningful annotation of gene products. GO contains three domains, including biological process (BP), cellular components (CC) and molecular function (MF) [28]. The Gene Ontology, which provides the logical structure of the biological functions ('terms') and their relationships to one another, manifested as a directed acyclic graph the corpus of GO annotations, evidence-based statements relating a specific gene product (a protein, non-coding RNA, or macromolecular complex, which we often refer to as 'genes' for simplicity) to a specific ontology term.
2.7 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
KEGG is utilized to harvest pathway clusters covering the knowledge of the molecular interaction and reaction networks in genes producing differentially expressed circRNA [28]. The higher order functional information is stored in the PATHWAY database, which contains graphical representations of cellular processes, such as metabolism, membrane transport, signal transduction and cell cycle. The KEGG databases are daily updated and made freely available (http://www. genome.ad.jp/kegg/).
2.8 The University of California Santa Cruz (UCSC) genome browser
Launched in 2001 to showcase the draft human genome assembly, the UCSC Genome Browser database (http://genome.ucsc.edu) and associated tools continue to grow, providing a comprehensive resource of genome assemblies and annotations to scientists and students worldwide [31]. The UCSC Genome Browser database hosts a large repository of genomes with 166 assemblies from GenBank that represent over 93 different organisms across the tree of life [32].
2.9 CircNet Database
CircNet database (http://circnet.mbc.nctu.edu.tw/) provides the following resources: (i) novel circRNAs, (ii) integrated miRNA-target networks, (iii) expression profiles of circRNA isoforms, (iv) genomic annotations of circRNA isoforms (e.g. 282 948 exon positions), and (v) sequences of circRNA isoforms [33].
2.10 CircRNA Identifier (CIRI)
CIRI is able to unbiasedly and accurately detect circRNAs from transcriptome data by employing multiple filtration strategies. By applying CIRI to encode RNA-seq data, the prevalence of intronic/intergenic circRNAs as well as fragments specific to them in the human transcriptome can be identified and experimentally validated [34].
2.11 CircBank
The circular RNA database circBank was officially launched on July 7th, 2018. A total of 140,790 human circRNA records are recorded in the circBank database, which also develops a dedicated ID number, based on the name of its' Host gene and the corresponding location.
3. CircRNAs and cancers
3.1 CircRNAs and hepatocellular carcinoma (HCC)
Growing evidence indicates that circRNA expression alterations have a broad impact in biological characteristics of HCC. CircRNAs act as oncogenes or tumor suppressors in HCC. Furthermore, circRNAs interfere with hepatitis virus infection [35]. Therefore, circRNAs can serve as potential diagnostic biomarkers for HCC [35]. For instance, the oncogenic circRNA, CDR1as, is shown to be deregulated in a variety of cancers including HCC by acting as a sponge of miR-7 that sequesters and competitively inhibits the activity of miR-7 [36]. Liu et al. demonstrated that miR-7 can inhibit the growth of cancer cells and promote apoptosis[37]. It is known to all that target genes of miR-7 mainly including epidermal growth factor receptor (EGFR), AKT and so on, which are oncogenes or tumor suppressor genes in cancers[38, 39]. As for EGFR, it is highly expressed in pancreatic cancer, oral cancer, cervical cancer and so on[38-40]. The inhibition the expression of EGFR can strengthen the curative effect of chemoradiotherapy based on cisplatin[41]. In a word, ciRS-7 may act as a ceRNA of miR-7, competitively inhibiting the activity of miR-7 and promotes the expression of oncogenes. As a result, it can promote the initiation and development of cancer. We consider ciRS-7 as the target of the early diagnosis and therapy in cancer because the inhibition expression of ciRS-7 may affect the activities of multiple oncogenes. The expression of hsa_circ_0005986 is lower in HCC tissues compared with adjacent normal tissues as a tumor suppressor in HCC carcinogenesis [42]. Especially, a total of 99 dysregulated circRNAs are identified to be associated with chronic hepatitis B (CHB) by circRNA/miRNA regulatory axes [43] [44]. Among them, miR-122 is one of the most abundant miRNAs in the liver and plays a central role in the HCV life cycle [44]. Recent studies indicate that circRNAs play a crucial role in controlling antiviral immune responses [45]. In addition, Hsa_circ_0001649 is significantly downregulated in HCC, indicating that it may serve as a novel potential biomarker for HCC and may function in tumorigenesis and metastasis of HCC [46]. Hsa_circ_0005075 can also act as a potential HCC biomarker because the expression of hsa_circ_0005075 correlates with HCC tumor size [47] (Table 1).
Overview of deregulated circRNAs in HCC
| CircRNA | Expression Change | Relative miRNA | Signal Path | Reference |
|---|---|---|---|---|
| ciRS-7 | up | miR-7 | —— | [36] |
| Has_circ_0005986 | down | —— | —— | [42] |
| Hsa_circ_0001649 | down | —— | —— | [46] |
| Hsa_circ_0079299 | down | —— | PI3K/AKT/mTOR | [59] |
| Circ_0067934 | up | miR-1324 | Wnt/β-catenin | [60] |
| Has_circ_0000567 | down | miR-421 | —— | [61] |
| Circ_101368 | up | miR-200a | HMGB1/RAGE | [62] |
| cSMARCA5 | down | miR-17-3p/miR-181b-5p | —— | [63] |
| Circ_0016788 | up | miR-486 | —— | [64] |
| CircADAMTS13 | down | miR-484 | —— | [65] |
| Circ-ZEB1.33 | up | miR-200a-3p | —— | [66] |
| Circ_100338 | up | miR-141-3p | —— | [67] |
| Circ_0008450 | up | miR-548p | —— | [68] |
| Has_circ_101280 | up | miR-375 | —— | [69] |
| Has_circ_0103809 | up | miR-490-5p | —— | [70] |
| CircADAMTS14 | up | miR-572 | —— | [71] |
3.2 CircRNAs and gastric carcinoma (GC)
Present study finds that different expressions of circRNAs and the corresponding miRNAs interact through circRNA binding sites to regulate the expression of target genes [48]. Results showed that a decrease in the circPVRL3 (Has_circ_0066779 is in gene symbol PVRL3 and it is named as circPVRL3) expression level is associated with the presence of GC and also with higher TNM stage and lower overall survival rates compared with that in adjacent noncancerous tissues [49]. The receiver operating characteristic (ROC) curve can be used to investigate the diagnostic value of circPVRL3 in distinguishing GC tissues from adjacent nontumorous tissues and different TNM stages. It deserves to be mentioned that Kaplan-Meier overall survival curve shows that the survival time of patients with low expression is shortened. We are convinced that circPVRL3 may play a protection role in GC and can be applied as a powerful independent prognostic factor even a treatment target. In this study, a total of 9 miRNAs (miR-203, miR-1272, miR-1283, miR-31, miR-638, miR-496, miR-485-3p, miR-766, and miR-876-3p) and corresponding target mRNAs are predicted to have an interaction with circPVRL3. In addition, Zheng et al have reported that miR-203 can suppress invasion of GC cells by targeting ERK1/2/Slug/E-cadherin signaling[50]. Evidence showed that miR-31 can function as a suppressor regulated by epigenetic mechanisms and target integrin α5 suppressing tumor cell invasion and metastasis by indirectly regulating phosphatidylinositol 3-kinase (PI3K)/AKT pathway in human GC SGC7901 cells[51, 52]. Chen et al. characterized circPVT1 may promote cell proliferation by acting as a sponge for members of the miR-125 family. The level of circPVT1 is observed as an independent prognostic marker for overall survival and disease-free survival of patients with GC as well as a novel proliferative factor and prognostic marker in GC [53] (Table 2).
Overview of deregulated circRNAs in GC
| CircRNA | Expression Change | Relative miRNA | Signal Path | Reference |
|---|---|---|---|---|
| CircFAT1(e2) | down | miR-548g | —— | [72] |
| CircPDSS1 | up | miR-186-5p | —— | [73] |
| Circ_100269 | down | miR-630 | —— | [74] |
| Circ-DONSON | up | —— | —— | [75] |
| CircNRIP1 | up | miR-149-5p | AKT1/mTOR | [76] |
| Has_circ_0001368 | down | miR-6506-5p | —— | [77] |
| Circ_LARP4 | down | miR-424 | —— | [78] |
3.3 CircRNAs and colorectal cancer (CRC)
Bachmayr-Heyda A et al. were the first to report a global reduction of circular RNA abundance in CRC cell lines and tumor tissues compared to normal tissues, and they discovered a negative correlation of global circular RNA abundance and proliferation [54]. Hsa_circ_0000069 knockdown can notably inhibit cell proliferation, migration, invasion, and induce G0/G1 phase arrest of cell cycle in vitro. It is demonstrated that hsa_circ_0000069, an important regulator in cancer progression, can be a promising target in the diagnosis and therapy in CRC [55]. The expression of hsa_circ_001988 is significantly correlated with differentiation and perineural invasion, and hsa_circ_001988 may become a novel potential biomarker in the diagnosis of CRC and a potential novel target for the treatment of CRC [56]. Zhu et al. conducted circular RNA profiles to identify circ-BANP as being enhance the growth of CRC cell by PI3K/Akt pathway. The biological function of circ-BANP (validated one circRNA generated from Exon 5-11 of BANP gene, termed circ-BANP) is convinced to be related with cell proliferation [29]. What's more, the expression of hsa_circ_0007534 is significantly up-regulated in CRC tumor tissues compared with adjacent non-tumor tissues, and hsa_circ_0007534 expression is correlated with tumor stage and lymph node metastasis [57]. Furthermore, the silence of hsa_circ_0007534 by siRNA significantly inhibited proliferation and induced apoptosis of CRC cells [57]. Hsa_circ_0126897_CBC1 is thought to be a potential biomarker for CRC, and the cell cycle is closely associated with the occurrence and development of CRC using CapitalBio microarray technology [58]. According to the study, 431 circRNAs are found differentially expressed in CRC tissues from patients with pulmonary metastasis compared with tissues without metastasis [28]. Another study revealed that hsa_circRNA_105055 (upregulated), hsa_circRNA_086376 (downregulated) or hsa_circRNA_102761 (downregulated) may regulate the pulmonary metastasis of CRC through binding with miR-7 to regulate protein kinase C beta (PRKCB) that is involved in the NF-κB or Wnt signaling pathway [28] (Table 3).
Overview of deregulated circRNAs in CRC
| CircRNA | Expression Change | Relative miRNA | Signal Path | Reference |
|---|---|---|---|---|
| Circ-BANP | down | —— | PI3 K/Akt | [29] |
| CircHIPK3 | up | miR-7 | c-Myb | [30] |
| Hsa_circ_0000069 | up | —— | —— | [55] |
| Hsa_circ_001988 | down | —— | —— | [56] |
| Hsa_circ_007534 | up | —— | —— | [57] |
| CircITGA7 | down | miR-370-3p | Ras | [79] |
| Circ_0026344 | down | miR-21,miR-31 | —— | [80] |
4. Discussion and Conclusion
During the last decade, studies have convincingly documented that ncRNAs participate in regulating of cellular structure, function, and physiological development, and they may contribute to the pathogenesis and development of cancer. Among them, circRNAs are considered a new star in the field of ncRNAs research and extensively investigated. Considering the stability and cytoplasmic localization circRNAs, engineered circRNAs could be exploited for a range of molecular tools or therapies [9]. Circular RNA constructs have been engineered both in vitro and in Vivo which could be applied to effectively sequester not only microRNAs or other RNAs of choice, but any RNA-binding protein with known sequence or structure specificity [9]. In this review, we briefly summarize the characteristics and functions of circRNAs with emphasis on their functional role in biological processes associated with cancer. Firstly, circRNAs function as miRNA sponges. Secondly, circRNAs regulate transcription and alternative splicing. Thirdly, circRNAs interact with RBPs. Fourthly, circRNAs regulate translation. Taken together, these functions indicate that circRNAs have the potential to play important roles in transcription and post-transcription and to become ideal biomarkers in the diagnosis of diseases especially in cancer.
We also discuss the experimental methods of circRNAs for the further research. A better understanding of circRNAs in diseases may contribute to the development of novel detection methods, resulting more reliable diagnosis and treatment in cancer.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81472275, 81702399), Natural Science Foundation of Guangdong Province (2014A030313542).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs[J]. Int J Mol Sci. 2014;15(6):9331-9342
2. Sanger H L, Klotz G, Riesner D. et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures[J]. Proc Natl Acad Sci U S A. 1976;73(11):3852-3856
3. Cocquerelle C, Mascrez B, Hetuin D. et al. Mis-splicing yields circular RNA molecules[J]. FASEB J. 1993;7(1):155-160
4. Danan M, Schwartz S, Edelheit S. et al. Transcriptome-wide discovery of circular RNAs in Archaea[J]. Nucleic Acids Res. 2012;40(7):3131-3142
5. Jeck W R, Sorrentino J A, Wang K. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA. 2013;19(2):141-157
6. Zhang Y, Zhang X O, Chen T. et al. Circular intronic long noncoding RNAs[J]. Mol Cell. 2013;51(6):792-806
7. Li Z, Huang C, Bao C. et al. Exon-intron circular RNAs regulate transcription in the nucleus[J]. Nat Struct Mol Biol. 2015;22(3):256-264
8. Ashwal-Fluss R, Meyer M, Pamudurti N R. et al. circRNA biogenesis competes with pre-mRNA splicing[J]. Mol Cell. 2014;56(1):55-66
9. Lasda E, Parker R. Circular RNAs: diversity of form and function[J]. RNA. 2014;20(12):1829-1842
10. Salzman J, Chen R E, Olsen M N. et al. Cell-type specific features of circular RNA expression[J]. PLoS Genet. 2013;9(9):e1003777
11. Memczak S, Jens M, Elefsinioti A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature. 2013;495(7441):333-338
12. Wang P L, Bao Y, Yee M C. et al. Circular RNA is expressed across the eukaryotic tree of life[J]. PLoS One. 2014;9(6):e90859
13. Qu S, Yang X, Li X. et al. Circular RNA: A new star of noncoding RNAs[J]. Cancer Lett. 2015;365(2):141-148
14. Shi X, Sun M, Liu H. et al. Long non-coding RNAs: a new frontier in the study of human diseases[J]. Cancer Lett. 2013;339(2):159-166
15. Salmena L, Poliseno L, Tay Y. et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?[J]. Cell. 2011;146(3):353-358
16. Salmanidis M, Pillman K, Goodall G. et al. Direct transcriptional regulation by nuclear microRNAs[J]. Int J Biochem Cell Biol. 2014;54:304-311
17. Li J Q, Yang J, Zhou P. et al. [The biological functions and regulations of competing endogenous RNA][J]. Yi Chuan. 2015;37(8):756-764
18. Jeck W R, Sharpless N E. Detecting and characterizing circular RNAs[J]. Nat Biotechnol. 2014;32(5):453-461
19. Capel B, Swain A, Nicolis S. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis[J]. Cell. 1993;73(5):1019-1030
20. Hansen T B, Jensen T I, Clausen B H. et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature. 2013;495(7441):384-388
21. Peng L, Chen G, Zhu Z. et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease[M]. Oncotarget. 2017:8 808-818
22. Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system[J]. Nat Immunol. 2014;15(6):484-491
23. Du WW, Yang W, Liu E. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2[J]. Nucleic Acids Res. 2016;44(6):2846-2858
24. Khan M A, Reckman Y J, Aufiero S. et al. RBM20 Regulates Circular RNA Production From the Titin Gene[J]. Circ Res. 2016;119(9):996-1003
25. Kos A, Dijkema R, Arnberg A C. et al. The hepatitis delta (delta) virus possesses a circular RNA[J]. Nature. 1986;323(6088):558-560
26. Abouhaidar M G, Venkataraman S, Golshani A. et al. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt[J]. Proc Natl Acad Sci U S A. 2014;111(40):14542-14547
27. Han D, Li J, Wang H. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression[J]. Hepatology. 2017;66(4):1151-1164
28. Zeng Y, Xu Y, Shu R. et al. Altered expression profiles of circular RNA in colorectal cancer tissues from patients with lung metastasis[M]. 2017. 1818 -1828
29. Zhu M, Xu Y, Chen Y. et al. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer[J]. Biomed Pharmacother. 2017;88:138-144
30. Zeng K, Chen X, Xu M. et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7[J]. Cell Death & Disease. 2018:9 (4)
31. Rosenbloom K R, Armstrong J, Barber G P. et al. The UCSC Genome Browser database: 2015 update[J]. Nucleic Acids Res. 2015;43(Database issue):D670-D681
32. Speir M L, Zweig A S, Rosenbloom K R. et al. The UCSC Genome Browser database: 2016 update[J]. Nucleic Acids Res. 2016;44(D1):D717-D725
33. Liu Y C, Li J R, Sun C H. et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data[J]. Nucleic Acids Res. 2016;44(D1):D209-D215
34. Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification[J]. Genome Biol. 2015;16:4
35. Wang M, Yu F, Li P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma[J]. Cancers (Basel). 2018:10 (8)
36. Peng L, Yuan X Q, Li G C. The emerging landscape of circular RNA ciRS-7 in cancer (Review)[J]. Oncol Rep. 2015;33(6):2669-2674
37. Liu S, Zhang P, Chen Z. et al. MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells[J]. FEBS Lett. 2013;587(14):2247-2253
38. Ribeiro F A, Noguti J, Oshima C T. et al. Effective targeting of the epidermal growth factor receptor (EGFR) for treating oral cancer: a promising approach[J]. Anticancer Res. 2014;34(4):1547-1552
39. Tomao F, Di Tucci C, Imperiale L. et al. Cervical cancer: are there potential new targets? An update on preclinical and clinical results[J]. Curr Drug Targets. 2014;15(12):1107-1120
40. Stock A M, Hahn S A, Troost G. et al. Induction of pancreatic cancer cell migration by an autocrine epidermal growth factor receptor activation[J]. Exp Cell Res. 2014;326(2):307-314
41. Nogueira-Rodrigues A, Moralez G, Grazziotin R. et al. Phase 2 trial of erlotinib combined with cisplatin and radiotherapy in patients with locally advanced cervical cancer[J]. Cancer. 2014;120(8):1187-1193
42. Fu L, Chen Q, Yao T. et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma[J]. Oncotarget. 2017;8(27):43878-43888
43. Zhou T C, Li X, Chen L J. et al. Differential expression profile of hepatic circular RNAs in chronic hepatitis B[J]. J Viral Hepat. 2018
44. Bandiera S, Pfeffer S, Baumert T F. et al. miR-122-a key factor and therapeutic target in liver disease[J]. J Hepatol. 2015;62(2):448-457
45. Li X, Liu C X, Xue W. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection[J]. Mol Cell. 2017;67(2):214-227
46. Qin M, Liu G, Huo X. et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma[J]. Cancer Biomark. 2016;16(1):161-169
47. Shang X, Li G, Liu H. et al. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development[J]. Medicine (Baltimore). 2016;95(22):e3811
48. Sui W, Shi Z, Xue W. et al. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology[J]. Oncol Rep. 2017;37(3):1804-1814
49. Sun H D, Xu Z P, Sun Z Q. et al. Down-regulation of circPVRL3 promotes the proliferation and migration of gastric cancer cells[J]. Sci Rep. 2018;8(1):10111
50. Zheng Y, Liu W, Guo L. et al. The expression level of miR-203 in patients with gastric cancer and its clinical significance[J]. Pathol Res Pract. 2017;213(12):1515-1518
51. Wei J, Wang Z, Wang Z. et al. MicroRNA-31 Function as a Suppressor Was Regulated by Epigenetic Mechanisms in Gastric Cancer[J]. Biomed Res Int. 2017;2017:5348490
52. Zhang J, Bian Z, Zhou J. et al. MicroRNA-638 inhibits cell proliferation by targeting phospholipase D1 in human gastric carcinoma[J]. Protein Cell. 2015;6(9):680-688
53. Chen J, Li Y, Zheng Q. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer[J]. Cancer Lett. 2017;388:208-219
54. Bachmayr-Heyda A, Reiner A T, Auer K. et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues[J]. Sci Rep. 2015;5:8057
55. Guo J N, Li J, Zhu C L. et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer[J]. Onco Targets Ther. 2016;9:7451-7458
56. Wang X, Zhang Y, Huang L. et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances[J]. Int J Clin Exp Pathol. 2015;8(12):16020-16025
57. Zhang R, Xu J, Zhao J. et al. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells[J]. Eur Rev Med Pharmacol Sci. 2018;22(1):118-126
58. Chen S, Zhang L, Su Y. et al. Screening potential biomarkers for colorectal cancer based on circular RNA chips[J]. Oncol Rep. 2018;39(6):2499-2512
59. Zheng H, Chen T, Li C. et al. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma[J]. Cancer Manag Res. 2019;11:443-454
60. Zhu Q, Lu G, Luo Z. et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis[J]. Biochem Biophys Res Commun. 2018;497(2):626-632
61. Xu L, Feng X, Hao X. et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth[J]. J Exp Clin Cancer Res. 2019;38(1):98
62. Li S, Gu H, Huang Y. et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling[J]. Cell Cycle. 2018;17(19-20):2349-2359
63. Yu J, Xu Q G, Wang Z G. et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma[J]. J Hepatol. 2018;68(6):1214-1227
64. Guan Z, Tan J, Gao W. et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway[J]. J Cell Physiol. 2018;234(1):500-508
65. Qiu L, Huang Y, Li Z. et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma[J]. Mol Oncol. 2019;13(2):441-455
66. Gong Y, Mao J, Wu D. et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6[J]. Cancer Cell Int. 2018;18:116
67. Huang X Y, Huang Z L, Xu Y H. et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma[J]. Sci Rep. 2017;7(1):5428
68. Zhang J, Chang Y, Xu L. et al. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p[J]. J Cell Biochem. 2019;120(6):9487-9494
69. Cao S, Wang G, Wang J. et al. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2[J]. Immunol Cell Biol. 2019;97(2):218-228
70. Cai H, Hu B, Ji L. et al. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway[J]. Am J Transl Res. 2018;10(6):1690-1702
71. Song C, Li D, Liu H. et al. The competing endogenous circular RNA ADAMTS14 suppressed hepatocellular carcinoma progression through regulating microRNA-572/regulator of calcineurin 1[J]. J Cell Physiol. 2019;234(3):2460-2470
72. Fang J, Hong H, Xue X. et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus[J]. Cancer Lett. 2019;442:222-232
73. Ouyang Y, Li Y, Huang Y. et al. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2[J]. J Cell Physiol. 2019;234(7):10458-10469
74. Zhang Y, Liu H, Li W. et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630[J]. Aging (Albany NY). 2017;9(6):1585-1594
75. Ding L, Zhao Y, Dang S. et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4[J]. Mol Cancer. 2019;18(1):45
76. Zhang X, Wang S, Wang H. et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway[J]. Mol Cancer. 2019;18(1):20
77. Lu J, Zhang P Y, Li P. et al. Circular RNA hsa_circ_0001368 suppresses the progression of gastric cancer by regulating miR-6506-5p/FOXO3 axis[J]. Biochem Biophys Res Commun. 2019;512(1):29-33
78. Zhang J, Liu H, Hou L. et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression[J]. Mol Cancer. 2017;16(1):151
79. Li X, Wang J, Zhang C. et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7[J]. J Pathol. 2018
80. Yuan Y, Liu W, Zhang Y. et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31[J]. Biochem Biophys Res Commun. 2018;503(2):870-875
Author contact
![]() Corresponding author: Wei Zhu, MD, Department of Pathology, Guangdong Medical University, No.1 Xincheng Road, Dongguan 523808, Guangdong Province, China. E-mail: zhuweiedu.cn.
Corresponding author: Wei Zhu, MD, Department of Pathology, Guangdong Medical University, No.1 Xincheng Road, Dongguan 523808, Guangdong Province, China. E-mail: zhuweiedu.cn.

 Global reach, higher impact
Global reach, higher impact