Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(15):3407-3419. doi:10.7150/jca.30858 This issue Cite
Research Paper
MiR-HCC2 Up-regulates BAMBI and ELMO1 Expression to Facilitate the Proliferation and EMT of Hepatocellular Carcinoma Cells
1. Tianjin Life Science Research Center and Department of Pathogen Biology, Collaborative Innovation Center of Tianjin for Medical Epigenetics, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China
2. State Key Laboratory of Oncology in Southern China, Department of Hepatobiliary Oncology, Cancer Center, Sun Yat-sen University, 651 Dong-Feng Road East, Guangzhou 510060, China
*These authors contributed equally to this work
Received 2018-10-22; Accepted 2019-4-23; Published 2019-6-9
Abstract
MicroRNAs (miRNAs) are a class of gene expression regulators that participate in the occurrence and development of hepatocellular carcinoma (HCC), although the underlying mechanism by which they function in HCC has not been fully elucidated. Here, small RNA deep sequencing was used to identify aberrantly expressed miRNAs in HCC tissues, and a novel miRNA named miR-HCC2 was identified. RT-qPCR analysis demonstrated that miR-HCC2 displayed higher expression in HCC tissues than in adjacent non-tumor tissues. We documented that miR-HCC2 facilitated the growth, migration and invasion of HCC cells by accelerating cell cycle progression, incressing the expression of epithelial-to-mesenchymal transition (EMT)-associated marker vimentin but decreasing the expression of E-cadherin. MiR-HCC2 directly targeted the 3′ UTR of BAMBI and ELMO1 and up-regulated their expression. Both BAMBI and ELMO1 had the same patterns of expression with miR-HCC2 in HCC tissues. Additionally, blocking BAMBI or ELMO1 counteracted the phenotypic alterations elicited by miR-HCC2. Collectively, our investigation identified miR-HCC2 as a new positive modulator of HCC aggressiveness that may serve as a potential biomarker for the development of diagnostic and therapeutic approaches for HCC.
Keywords: BAMBI, ELMO1, HCC, miRNAs, migration, invasion
Introduction
Hepatocellular carcinoma (HCC) is an aggressive cancer; it is the sixth most prevalent cancer and the third leading cause of cancer-related deaths [1]. In recent decades, surgical resection, radiotherapy and chemotherapy have been remarkably improved, but the prognosis of HCC patients is far from desirable owing to the high risk of recurrence and distant metastasis [2]. Consequently, understanding the underlying mechanism of hepatocarcinogenesis appears to be crucial for managing HCC. Increasing evidence has shown that abnormal expression of many oncogenes and tumor suppressor genes, including the dysregulation of microRNAs involved in those gene pathways, is linked to the pathogenesis and development of various types of human cancer, such as T-cell acute lymphoblastic leukemia [3], breast cancer [4], lung cancer [5], cervical cancer [6] and hepatocellular carcinoma [7].
MicroRNAs (MiRNAs) are a group of small noncoding RNAs ~22 nt in length that serve as crucial post-transcriptional regulators of genes by targeting the 3′ untranslated regions (3' UTR) of their target mRNAs [8]. Typically, miRNAs negatively regulate multiple genes, leading to mRNA degradation or translation repression [9]. However, previous studies have shown that miRNAs can also positively regulate gene expression. For instance, miRNAs can up-regulate translation, switching from repression to activation [10]. For example, miR-346 up-regulates human telomerase reverse transcriptase (hTERT) [11], miR-346 positively regulates Argonaute 2 (AGO2) protein to augment the activity of other miRNAs [12], and miR-1 directly enhances mitochondrial gene translation during muscle differentiation [13]. It has been reported that miRNAs participate in diverse physiological processes, such as cell proliferation, cell death, differentiation and apoptosis [14-16]. Recent studies have documented the contribution of miRNAs to carcinogenesis by functioning as oncogenes or tumor suppressors. As reported, gain of miR-151 contributes to cell migration and invasion by down-regulating RhoGDIA in hepatocellular carcinoma [17]. MiR-193a-3p is down-regulated in ulcerative colitis neoplasia, which promotes carcinogenesis through up-regulation of IL17RD [18]. MiRNAs with an AAGUGC seed motif constitute an integral part of an oncogenic signaling network [19].
BMP and activin membrane-bound inhibitor (BAMBI), a non-signaling pseudo-receptor in the transforming growth factor (TGF)-β type I receptor family, negatively modulates the TGF-β kinase signaling that is activated by TGF-β family members [20]. Previous reports have suggested that BAMBI increases colorectal cancer metastasis by connecting the Wnt and beta-catenin pathways and inhibiting the TGF-β-signaling pathway [21], and its elevated expression, together with β-sitosterol expression, ameliorates NSCLC via autophagy inhibition and inactivation of the TGF-β/Smad2/3 pathway [22]. Additionally, BAMBI cooperates with Wnt to stimulate the expression of cyclin D1 and c-Myc and to promote cell cycle progression from G0/G1 to G2/M in the osteosarcoma cells [23]. Previous investigations showed that BAMBI expression, driven by Wnt/beta-catenin signaling, was elevated in HCC [24]. These observations suggested the importance of BAMBI in HCC and led us to investigate its dysregulation in HCC.
Engulfment and cell motility 1 (ELMO1), originally identified as Dock180-binding protein, promotes the invasion of human glioma cells and ovarian cancer cells in combination with Dock180, a two part Rac1 guanine nucleotide exchange factor [25, 26]. It was reported that ELMO1 is essential for Dock2-dependent lymphocyte migration [27] and the migration and invasion of lung cancer cells [28]. The phosphorylation of Y724 in ELMO1 may modulate Rac1 activation, leading to cancer cell adhesion to the extracellular matrix (ECM), invasion into surrounding tissues, and subsequent distant metastasis [29]. ELMO1 expression is positively correlated with the metastatic potential of HCC cells and was found to be an independent prognostic factor of survival [30]. Nonetheless, the mechanism by which ELMO1 expression is regulated in HCC cells has yet to be elucidated.
We previously have applied deep sequencing to obtain some new miRNA candidates in the HCC tissue. Here, we aimed to ascertain the impact of a novel miRNA, named miR-HCC2, on cell growth and EMT in HCC cells. miR-HCC2 was overexpressed in HCC tissues relative to adjacent non-tumor tissues. Ectopic expression of miR-HCC2 promoted cell proliferation by accelerating G1/S and S/G2/M phase transitions. Furthermore, we validated that miR-HCC2 enhanced migration, invasion and EMT in HCC cells. BAMBI and ELMO1 can be directly targeted by miR-HCC2, which results in an increase in mRNA and protein levels that ultimately exert the carcinogenic functions of miR-HCC2. The identification of miR-HCC2 may provide new insights into the molecular mechanisms of tumorigenesis.
Materials and Methods
Human tissue samples and cell lines
Twenty pairs of fresh specimens containing human HCC and adjacent non-tumor tissues were collected from the Cancer Center of Sun Yat-Sen University of Medicine Science, and their histology was confirmed by pathologists (Table S1). Informed consent was obtained from each patient, and the study was approved by the ethical committee of the institution.
The human HCC cell lines Hep3B and Huh7 were originally purchased from ATCC and maintained in DMEM (GIBCO BRL, Grand Island, NY) and supplemented with 10% FBS (fetal bovine serum) and 1% PS (100 units/ml penicillin, 100 μg/ml streptomycin). These cell lines were incubated at 37°C in a humidified atmosphere with 5% CO2 and transfected with Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. All transfections were conducted in three independent experiments.
RNA extraction and RT-qPCR analysis
Extraction of total RNA from tissue samples and cells was performed using a mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer's instructions and then reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega, Madison, WI). RT-qPCR was performed with a SYBR Premix Ex TaqTM Kit (Promega, Madison, WI). Related primers were synthesized from AuGCT, Inc. (Beijing, China), and detailed sequences can be found in Table S2.
Western blot
To determine protein levels, Western blot was applied as described in a previous study [31]. Primary antibodies used in this study were purchased from Saier Biotech Co. (Tianjin, China), including antibodies against BAMBI (Catalog No: SRP07313), ELMO1 (Catalog No: SRP07468), E-cadherin (Catalog No: SRP05266), vimentin (Catalog No: SRP01327) and GAPDH (Catalog No: SRP00849). The secondary goat anti-rabbit antibody was obtained from Sigma. Endogenous GAPDH was used as a loading control to normalize the proteins of interest.
Plasmid construction
The plasmid pri-miR-HCC2, which included a precursor of the miR-HCC2 sequence, was inserted into the pcDNA3 vector at BamHI and EcoRI sites. For knockdown of miR-HCC2, we purchased the synthesized 2'-O-methyl-modified antisense oligonucleotides of miR-HCC2 (ASO-miR-HCC2) and the scramble control oligonucleotides (ASO-NC) from GenePharma (Shanghai, China). The wild-type 3′ UTRs of BAMBI and ELMO1 containing the target sites of miR-HCC2 or its respective mutant were constructed by annealing the oligos and cloning them into the pcDNA3-EGFP vector between the BamHI and EcoRI sites (downstream of EGFP). The wild-type and mutant 3′ UTRs of ELMO1 that were used for the enhanced green fluorescent protein reporter assay were constructed in the same manner.
A cDNA fragment containing the BAMBI or ELMO1 coding sequence was amplified by PCR and inserted into the pCD3×Flag/KBE vector between the BamHI and XhoI sites, resulting in overexpression plasmids of ELMO1 and BAMBI (Flag-BAMBI, Flag-ELMO1). To generate antisense oligonucleotides to knock down BAMBI and ELMO1, synthesized oligonucleotides were purchased from GenePharma (Shanghai, China), annealed and respectively inserted into the pSilencer2.1/neo vector (Ambion, Austin, TX) between the BamHI and HindIII sites. All constructed vectors were confirmed by sequencing, and the primers and oligonucleotides used are listed in Table S2.
Enhanced green fluorescent protein (EGFP) reporter assay
To validate the targeting of the 3′ UTRs of BAMBI and ELMO1 by miR-HCC2, pcDNA3, pri-miR-HCC2, ASO-NC or ASO-miR-HCC2 was co-transfected with BAMBI-3′ UTR, BAMBI-3′ UTR-mut, ELMO1-3′ UTR, or ELMO1-3′ UTR-mut in 48-well plates. The RFP (red fluorescent protein) expression vector pDsRed2-N1 (Clontech, Mountain View, CA) was co-transfected for normalization. At 48 h after transfection, the fluorescence intensities of EGFP and RFP were measured with an F-4500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan).
MTT and colony formation assays
For the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays, the transfected cells were counted after 24 h incubation and seeded in a 96-well-plate at a density of 4,000 cells/well (Hep3B) or 3,000 cells/well (Huh7).The subsequent protocol was reported before [32].
Following transfection and counting, 1,000 Hep3B and 300 Huh7 cells per well were seeded in 12-well plates. During the growth of colonies, the culture medium was replaced every 3 days. After 10 and 12 days, crystal violet was used to stain the cells. Only colonies containing more than 50 cells were quantified. The rate of colony formation was calculated with the following equation: colony formation rate = (number of colonies/number of seeded cells) × 100%.
Migration and invasion assays
Transwell chambers with 8 µm pores (Corning, Cambridge, MA) were used for the migration and invasion assays. To analyze the invasion abilities of the HCC cells, 2 mg/ml Matrigel (Clontech, Mountain View, CA) was added into the chambers. For migration and invasion assays, 80,000 and 120,000 Hep3B cells/well were seeded, respectively. For Huh7 cells, 60,000 and 80,000 cells/well were used for migration and invasion assays, respectively. The detailed procedure was described in a previous study [32].
Cell cycle analysis via flow cytometry
These analyses were performed in a previous study [12].
Statistical analysis
Each experiment was performed in triplicate. The data are presented as the means ± SD, and the comparisons between two groups were determined using ANOVA analysis. To clarify the association between two factors, Pearson's correlation analysis was performed. A p value of less than 0.05 was considered significant (*p<0.05; **p<0.01; ***p<0.001).
Results
MiR-HCC2 facilitates proliferation and accelerates cell cycle progression in human HCC cells
To globally identify novel miRNAs specifically altered in HCC tissues, a Solexa sequencing approach was performed using HCC tissue samples. Sequencing analysis showed approximately 3 million reads and their size distribution was enriched in 21-24 nt. Several novel miRNAs with high abundance were discovered. Herein we focused on a novel miRNA named miR-HCC2, which has the sequence “UCUGUUUGUCGUAGGCAGAUGG” and was located in Homo sapiens chromosome 5, GRCh38.p12 Primary Assembly (NCBI Reference Sequence: NC_000005). The pre-miR-HCC2 and its secondary structure were predicted (Figure 1A). To confirm the Solexa sequencing result, RT-qPCR was applied to detect the levels of miR-HCC2 in HCC tissues and the adjacent non-tumor tissues. As shown in Figure 1B, the expression levels of miR-HCC2 in HCC tissues were higher than those in adjacent non-tumor tissues (Figure 1B). As shown in the results, we wondered whether miR-HCC2 contributed to the malignant phenotypes of HCC cells.
MiR-HCC2 facilitates proliferation and accelerates the cell cycle progression of human HCC cells. (A) The sequence and predicted secondary structure of pre-miR-HCC2 and the mature sequence of miR-HCC2. (B) The relative expression levels of miR-HCC2 in 20 pairs of HCC tissues and adjacent non-tumor tissues were detected using RT-qPCR. U6 snRNA was used as an internal control. (C) MTT assays were performed to assess the influence of miR-HCC2 on cell viability. ASO: 2'-O-methyl-modified antisense oligonucleotides; ASO-NC: the scramble control oligonucleotides. (D) Cell clonogenicity was analyzed by colony formation assays. The colony formation rate = (number of colonies/number of seeded cells) × 100%. (E, F) The proportion of Hep3B (E) and Huh7 (F) cells in three phases (G1, S, or G2/M) of the cell cycle were measured by flow cytometric analysis. The proliferation index (PI) = (S+G2/M)/G1. All error bars indicate the means ± SD of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
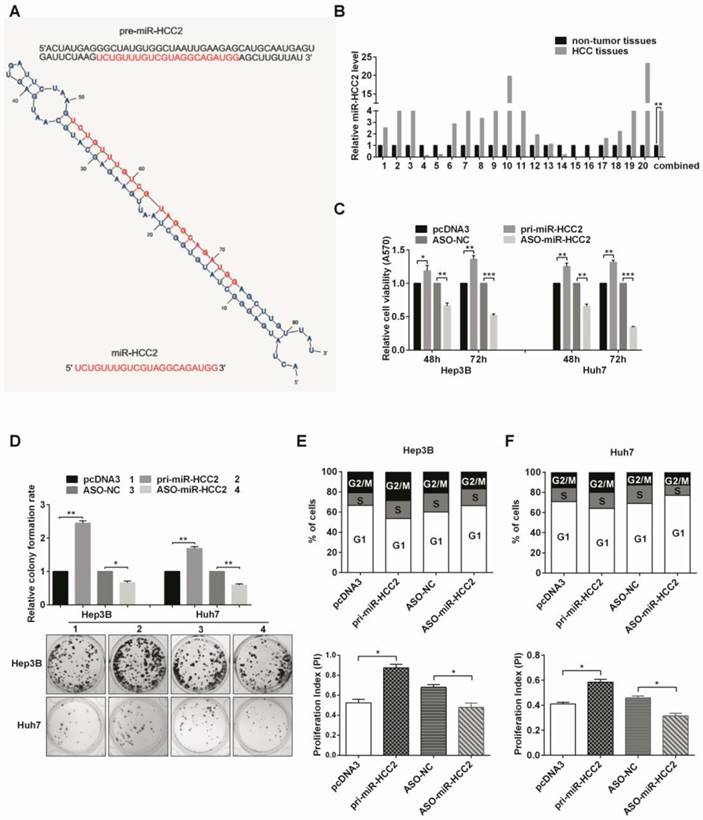
The levels of miR-HCC2 in the pri-miR-HCC2-transfected cells increased approximately 16-fold and 56-fold in Hep3B and Huh7 cells, respectively, compared with the control vector. Conversely, miR-HCC2 levels in those cells transfected with ASO-miR-HCC2 decreased by 50% (Hep3B) and 70% (Huh7) compared with the scrambled oligomer group (Figure S1). It was found that miR-HCC2 promoted cell viability, whereas ASO-miR-HCC2 inhibited cell viability in both Hep3B and Huh7 cells (Figure 1C). As shown in Figure 1D, overexpression of miR-HCC2 increased the colony formation rates of Hep3B and Huh7 cells, whereas suppressing miR-HCC2 expression decreased the rates. These results indicated that miR-HCC2 facilitates the proliferation of Hep3B and Huh7 cells.
We further explored the mechanism responsible for the modulation of cell growth in HCC cells. An increased percentage of cells in the S and G2/M phases and a decreased percentage of cells in the G1 phase were observed in pri-miR-HCC2-transfected cells compared with the control in both Hep3B and Huh7 cells. The proliferation index was also markedly higher than that of the control. In contrast, ASO-miR- HCC2-transfected cells had a markedly increased percentage of cells in the G1 phase and a reduction in the percentage of cells in the S and G2/M phases; a lower proliferation index was obtained compared with the ASO control (Figure 1E and 1F). Taken together, these observations suggested that miR-HCC2 is capable of facilitating cell proliferation by accelerating cell cycle progression in human HCC cells.
MiR-HCC2 enhances cell migration and invasion and modulates EMT-associated molecules in human HCC cells
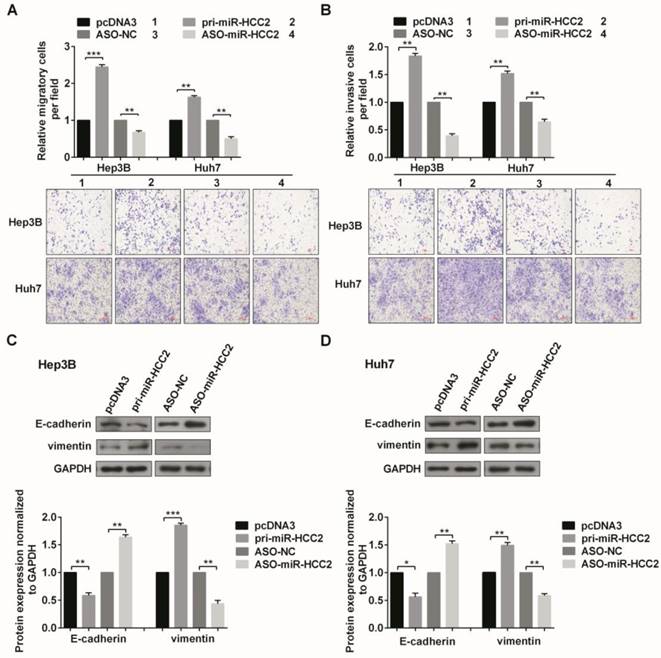
Then we evaluated the effect of miR-HCC2 on migration and invasion in human HCC cells. In Hep3B cells, the overexpression of miR-HCC2 increased migration and invasion capacities. In contrast, the opposite effect was observed in the ASO-miR-HCC2-transfected cells. Consistent with the results in Hep3B cells, miR-HCC2-transfection enhanced the migration and invasion capacities of Huh7 cells, whereas the inhibition of miR-HCC2 decreased these capacities (Figure 2A and 2B). These results revealed that miR-HCC2 enhances both the migration and invasion capacities of human HCC cells.
Next, we checked the expression of key molecular markers in EMT (E-cadherin and vimentin) to explore the role of miR-HCC2 in this process. In Hep3B and Huh7 cells, miR-HCC2 overexpression reduced E-cadherin protein levels and increased vimentin protein levels, while ASO-miR-HCC2- transfection increased E-cadherin but decreased vimentin protein levels (Figure 2C and 2D). Collectively, these results showed that miR-HCC2 promotes the migration, invasion and EMT of human HCC cells.
BAMBI and ELMO1 are directly up-regulated by miR-HCC2
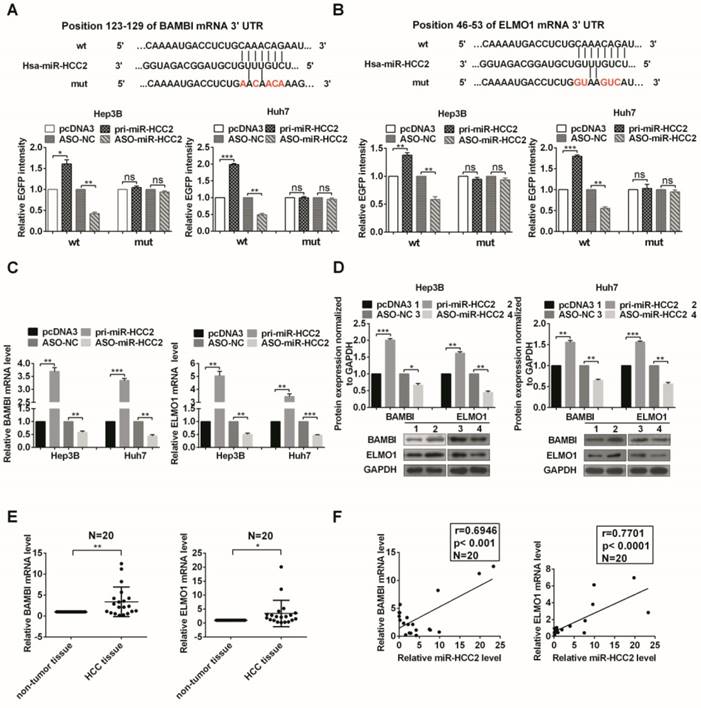
We performed bioinformatics analysis using the prediction algorithms of TargetScan, PicTar, and miRanda to identify possible candidates that mediate the malignant effects of miR-HCC2 in HCC cells. Interestingly, we found that both BAMBI and ELMO1 transcripts have a putative miR-HCC2 binding site in their 3′ UTRs. BAMBI was previously reported to expedite cell cycle progression [23], and ELMO1 is a related protein that promotes cell phagocytosis and migration [33, 34]. Combined with our previous observations, we focused on validating BAMBI and ELMO1 as targets of miR-HCC2.
To determine whether miR-HCC2 directly targets BAMBI or ELMO1, we performed EGFP reporter analyses by co-transfecting the reporter vectors (containing wild-type or mutated putative binding sites in the 3′ UTR) and the miR-HCC2 expression vector or ASO-miR-HCC2. Compared with their respective controls, pri-miR-HCC2 significantly enhanced the fluorescent intensity of the wild- type reporter vector (pcDNA3/EGFP-BAMBI-wt), while ASO-miR-HCC2 exhibited the opposite effects in both Hep3B and Huh7 cells. However, modulations of miR-HCC2 expression did not affect the fluorescence intensity of the mutant form of the reporter vector (pcDNA3/EGFP-BAMBI-mut). Similar results were obtained for the reporter vectors of ELMO1 (Figure 3A and 3B).
Then we addressed whether miR-HCC2 impacts the expression of endogenous BAMBI and ELMO1. The results showed that overexpression of miR-HCC2 increased their expression at both mRNA and protein levels, whereas suppression of miR-HCC2 decreased their expression (Figure 3C and 3D). Taken all together, miR-HCC2 directly targeted the 3′ UTR of BAMBI and ELMO1 and up-regulated their expression.
We further evaluated the correlations between BAMBI/ELMO1 and miR-HCC2 in HCC tissues by examining the expression of BAMBI and ELMO1 in the 20 paired clinical specimens mentioned earlier. Both BAMBI and ELMO1 had higher expression in HCC tissues compared to that in the adjacent non-tumor tissues (Figure 3E). The correlation analysis indicated that the expression of both BAMBI and ELMO1 was positively correlated with miR-HCC2 expression (Figure 3F).
MiR-HCC2 enhances the migration and invasion of human HCC cells and modulates EMT-associated markers. (A, B) Transwell migration assays and invasion assays were performed to detect the impact of miR-HCC2 on the migration (A) and invasion (B) abilities of Hep3B and Huh7 cells. Five random fields were counted under a light microscope, and representative fields of the migratory and invasive cells are presented. (C, D) Western blot was used to confirm the expression of key molecular markers associated with EMT (E-cadherin and vimentin) in Hep3B (C) and Huh7 (D) cells. All images were photographed at 100× magnification, and the data quantification is represented in bar graphs. All error bars indicate the means ± SD of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
BAMBI and ELMO1 contribute to the malignant phenotypes of human HCC cells
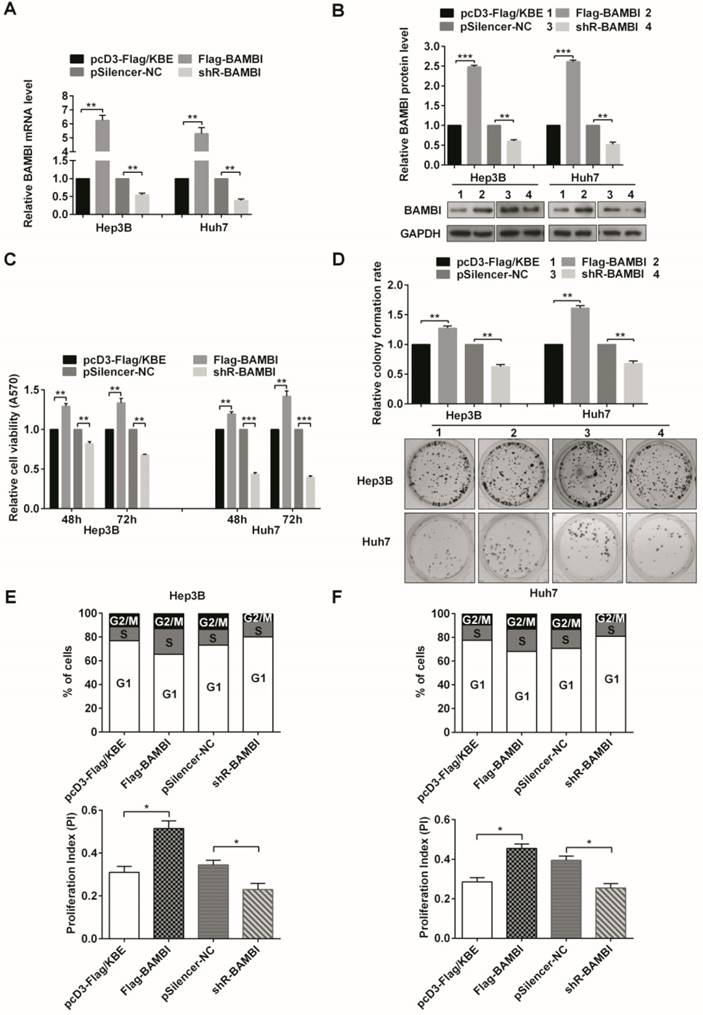
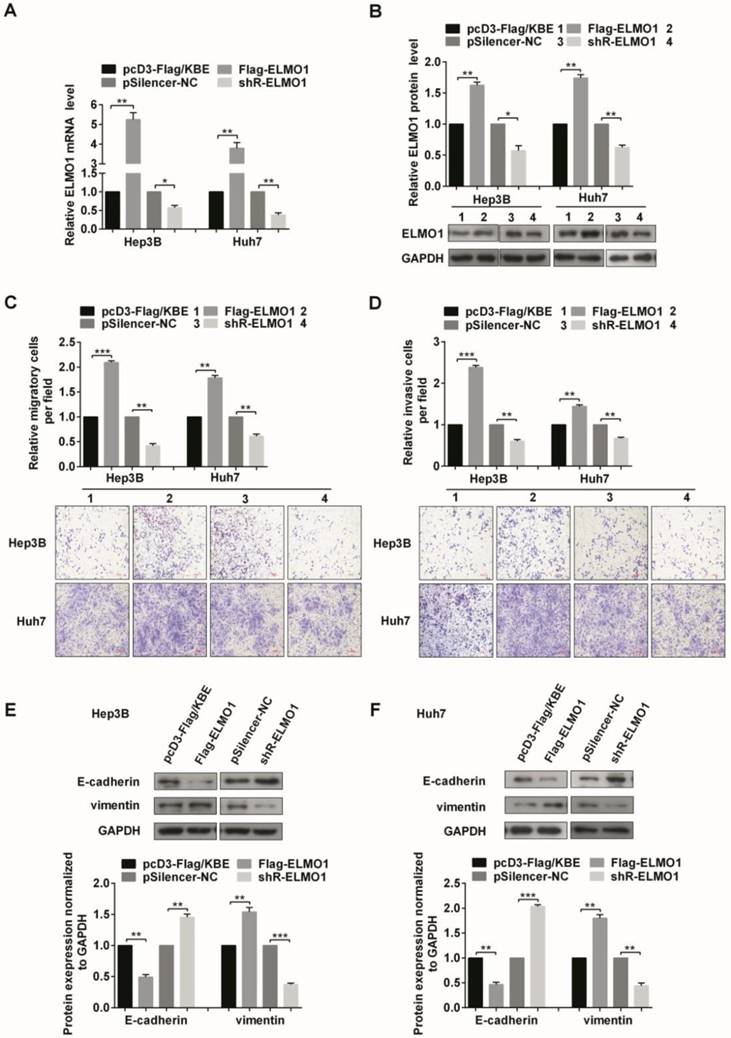
To further investigate the biological roles of BAMBI and ELMO1 in HCC cells, we constructed overexpression (Flag-BAMBI, Flag-ELMO1) and knockdown vectors (shR-BAMBI, shR-ELMO1) for each and determined the effectiveness of these constructs (Figure 4A, 4B and Figure 5A, 5B).
MTT assays indicated that increasing the level of BAMBI enhanced cell viability in Hep3B and Huh7 cells, whereas the inhibition of BAMBI decreased the viability (Figure 4C). The colony formation assays showed that Flag-BAMBI-transfection promoted the colony formation rate, while shR-BAMBI-transfection decreased the rate (Figure 4D). BAMBI accelerated the G1 to S and the S to G2/M transitions (Figure 4E and 4F). However, after overexpression or knockdown of ELMO1, no significant effects on cell viability, colony formation rate, and cell cycle progression were observed (Figure S2).
ELMO1 enhanced both migration and invasion capacities in the two cell lines (Figure 5C and 5D). We also explored the effect of ELMO1 on EMT-associated markers. The overexpression of ELMO1 decreased E-cadherin but increased vimentin levels, which was consistent with its role in facilitating migration and invasion. In contrast, the knockdown of ELMO1 increased E-cadherin and decreased vimentin levels (Figure 5E and 5F). Whereas the migration and invasion capacities changes after BAMBI overexpression or knockdown is not so impressive, similar results were obtained in EMT-associated markers (Figure S3).
In summary, BAMBI and ELMO1 may play oncogenic roles in human HCC cells and contribute to the malignant phenotypes of human HCC cells.
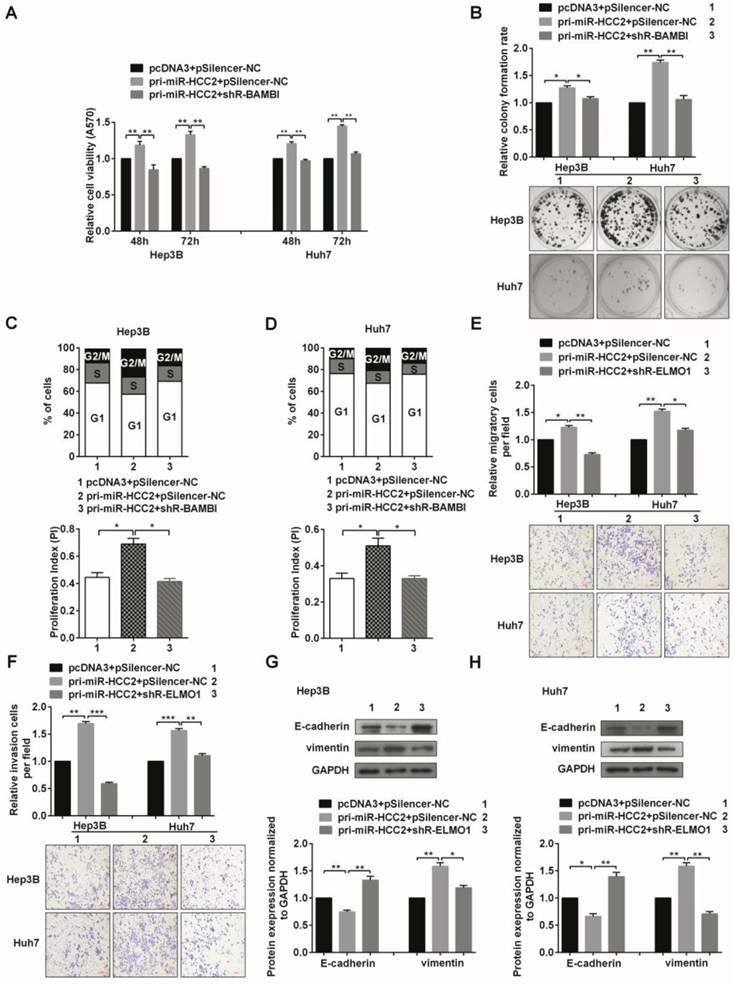
Knockdown of BAMBI and ELMO1 abolishes the effects induced by miR-HCC2 in human HCC cells
Given that BAMBI, ELMO1 and miR-HCC2 had similar effects on malignant processes in HCC cells and that the expression of BAMBI and ELMO1 was positively regulated by miR-HCC2 at both mRNA and protein levels, we asked whether the effects of miR-HCC2 in HCC cells were achieved via its up-regulation of BAMBI and ELMO1 expression. To address this question, pri-miR-HCC2 along with shR-BAMBI or shR-ELMO1 were co-transfected into Hep3B and Huh7 cells, and a series of functional rescue experiments were conducted. As anticipated, the knockdown of BAMBI mostly blocked the effects of miR-HCC2 overexpression on cell viability (Figure 6A). The miR-HCC2-mediated increases in the colony formation rate (Figure 6B) and cell cycle progression were significantly counteracted when BAMBI was knocked down (Figure 6C and 6D). Additionally, the migration and invasion capacities that were promoted by miR-HCC2 expression were impaired by ELMO1 knockdown (Figure 6E and 6F), and the EMT-associated molecular makers were restored to normal levels (Figure 6G and 6H). Overall, the knockdown of BAMBI or ELMO1 abolished the phenotypic alterations elicited by miR-HCC2, demonstrating that BAMBI and ELMO1 serve as direct functional targets of miR-HCC2.
BAMBI and ELMO1 are directly up-regulated by miR-HCC2. (A, B) The predicted binding sites of miR-HCC2 (middle) with the wild-type (top) and mutated (bottom) 3′ UTR of BAMBI (A) and ELMO1 (B). The red letters represent the mutant nucleotides. The EGFP fluorescence intensities of Hep3B and Huh7 cells co-transfected with wild-type or mutant pcDNA3/EGFP- BAMBI (or pcDNA3/EGFP- ELMO1) 3′ UTR with pri-miR-HCC2 or ASO-miR-HCC2 were detected. The effects of miR-HCC2 on BAMBI and ELMO1 mRNA (C) and protein (D) levels were determined using RT-qPCR and Western blot, respectively. (E) The relative expression levels of BAMBI and ELMO1 mRNA in 20 pairs of HCC tissues and adjacent non-tumor tissues were detected by RT-qPCR. β-actin was used as an internal control. (F) Pearson′s correlation analysis indicated that BAMBI and ELMO1 expression was positively correlated with the expression of miR-HCC2. All experiments were repeated at least three times. *p<0.05; **p<0.01; ***p<0.001, ns: no significance.
BAMBI accelerates the proliferation and the G1/S and S/G2/M phase transitions of human HCC cells. (A, B) The efficiencies of BAMBI expression plasmid Flag-BAMBI and knockdown plasmid shR-BAMBI were determined by measurement of mRNA (A) and protein (B) levels. (C) MTT assays of cells that were transiently transfected with Flag-BAMBI, shR-BAMBI, or a control vector. (D) Colony formation assays tested the impact of BAMBI on the proliferation of Hep3B and Huh7 cells. (E, F) Flow cytometric analysis was utilized to assess cell cycle progression in Hep3B cells (E) and Huh7 cells (F). The proliferation index of cells transfected with Flag-BAMBI or shR-BAMBI is presented. All error bars indicate the means ± SD of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
ELMO1 contributes to the cell migration, invasion, and drives the EMT process in human HCC cells. (A, B) The efficiencies of ELMO1 expression plasmid Flag-ELMO1 and knockdown plasmid shR-ELMO1 were determined by measurement of mRNA (A) and protein (B) levels. (C, D) The migratory (C) and invasive (D) capabilities of Hep3B and Huh7 cells transfected with either Flag-ELMO1 or shR-ELMO1 were measured. (E, F) Protein expression levels of EMT-associated markers were detected by Western blot. All error bars indicate the means ± SD of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
Knockdown of BAMBI and ELMO1 abolishes the effects induced by miR-HCC2 in human HCC cells. (A) MTT assays of Hep3B and Huh7 cells co-transfected with pri-miR-HCC2 and shR-BAMBI. (B) The miR-HCC2-mediated increase in the colony formation rate was counteracted when BAMBI was suppressed. (C, D) Cell cycle progression of the co-transfected cells was obtained via flow cytometry. The populations of cells in different phases and the proliferation indexes are shown. (E, F) The migration (E) and invasion (F) capacities promoted by miR-HCC2 were impaired by suppression of ELMO1. (G, H) Western blot was performed to determine the expression of EMT-associated markers (E-cadherin and vimentin) after shR-ELMO1 transfection in the presence of miR-HCC2. All error bars indicate the means ± SD of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
In conclusion, our results showed that miR-HCC2 up-regulates the expression of BAMBI and ELMO1 to facilitate the proliferation, migration, and invasion of human HCC cells.
Discussion
Recently, much evidence has indicated the relevance of a high level of tumor invasiveness, increased intrahepatic spread, and increased extrahepatic metastasis in HCC aggressiveness. MiRNAs influence many cell behaviors and are well established as fundamental regulators of migration and invasion in HCC cells. MiR-7 functions as a tumor suppressor and plays a substantial role in the inhibition of tumor growth and metastasis in HCC through the PI3K/Akt pathway [35]. MiR-188-5p suppresses tumor cell proliferation and metastasis via targeting of FGF5 in hepatocellular carcinoma [36]. MiR-429 was identified as a key inducer of HCC metastasis via regulation of the classic Wnt pathway rather than via regulation of epithelial-mesenchymal transition [37]. Here, we focus on a novel miRNA named miR-HCC2 identified from HCC tissues by deep sequencing, which emphasizes its potential roles in HCC. We are the first to report the up-regulation of miR-HCC2 in HCC tissues compared with adjacent non-tumor tissues. Functionally, miR-HCC2 promoted the proliferation of HCC cells. In addition, the number of migratory and invasive cells was markedly increased when miR-HCC2 was overexpressed, whereas the inhibition of miR-HCC2 decreased their number, suggesting that miR-HCC2 may act as an oncogene via facilitation of the proliferation, migration, and invasion of HCC cells.
Currently, there is a considerable body of literature available indicating that EMT is a pivotal event associated with tumor cell invasion and metastasis in HCC progression [38]. EMT is a complex process that allows epithelial cells to lose their differentiated features and acquire a mesenchymal phenotype during metastasis. EMT-induced changes are associated with decreased expression of epithelial markers, such as E-cadherin, occludin, plakophilin, and cytokeratin, and the increased expression of mesenchymal markers, including vimentin, N-cadherin, and fibronectin. EMT can be influenced by several distinct pathways and thus can be regulated by miRNAs [39]. The observation that up-regulation of miR-HCC2 can markedly enhance the expression of vimentin but decrease that of E-cadherin suggests a positive role of miR-HCC2 in the EMT process.
To elucidate possible mechanisms by which miR-HCC2 exerts its function, we predicted its putative targets using the prediction algorithms of TargetScan, PicTar, and miRanda. Of the targets listed, BAMBI and ELMO1 were chosen for further investigation. EGFP reporter assays were performed and the results showed that miR-HCC2 directly targeted the 3′ UTR of BAMBI and ELMO1 mRNAs, thus up-regulating their expression. Furthermore, miR-HCC2 increased endogenous BAMBI and ELMO1 at both the mRNA and protein levels. RT-qPCR and correlation analysis validated that BAMBI and ELMO1 had the same patterns of expression as miR-HCC2 in HCC tissues, indicating that the positive regulation of BAMBI and ELMO1 by miR-HCC2 was clinically significant. Typically, miRNAs bind to the 3' UTR of target mRNAs, leading to mRNA cleavage or translation repression. However, documented evidence has revealed that miRNAs can also enhance gene expression by interacting with promoter or 3′ UTR of target gene. For example, miR-205 induced the expression of the interleukin (IL) tumor suppressor genes IL24 and IL32 by targeting specific sites in their promoters in prostate cancer [40]. AGO2 and Fragile-X mental retardation related protein 1(FXR1) associated with TNFα AU-Rich-Element and relieved the inhibition of TNFα gene expression by miR-369-3p in response to serum starvation [41]. Recently, our lab has found that miRNA-346 binds the 3′ UTR of hTERT in GRSF1 dependent manner [11]. Herein, we found miR-HCC2 targeted and enhanced the expression of BAMBI and ELMO1. Nevertheless, the underlying mechanism remains unclear and needs to be further clarified.
BAMBI is aberrantly elevated in most gastric cancer [42], colorectal cancer and hepatocellular carcinomas [24] but is absent in breast cancer [43] and silenced in a subset of high-grade bladder cancer [22]. Overexpression of BAMBI was observed in HCC tissues, which is consistent with previous reports [24]. In HepG2, transcription of BAMBI is modulated via a feedback loop that promotes the binding of Smad3 and Smad4 to Smad-responsive elements in the BAMBI promoter [44], and a previous study validated that Wnt/β-catenin signaling drove elevated BAMBI expression [24]. Here, we showed that miR-HCC2 can also directly up-regulate the expression of BAMBI. Overall, BAMBI is capable of increasing cellular growth through the enhancement of Wnt signaling and the inhibition of TGF-β-SMAD suppressive signaling in colorectal cancer [45]. Our data suggested that both miR-HCC2 and BAMBI facilitate the proliferation of HCC cells via accelerating the G1 to S and S to G2/M phase transitions. Furthermore, the phenotypic alterations elicited by miR-HCC2 were counteracted when BAMBI was knocked down, demonstrating that miR-HCC2 modulates HCC cells proliferation by increasing BAMBI expression.
Mammalian ELMO1 and Dock180 proteins and their homologs in Drosophila melanogaster and Caenorhabditis elegans function as crucial upstream regulators of Rac during development and cell migration. Dock180 and ELMO1 mediate the GTP loading of Rac via a domain within Dock180 that recognizes nucleotide-free Rac [46]. The association of Nck-1 with ELMO1 facilitated the activation of RhoG, thus promoting Rac1 activation and cell motility [47]. A previous study indicated that overexpression of ELMO1 promotes cell invasion and migration in hepatocellular carcinoma [25]. Similarly, our investigation validated that ELMO1 augmented the migration, invasion, and EMT of HCC cells. Suppression of ELMO1 antagonized the migration and invasion capacities induced by miR-HCC2 and reversed the expression of EMT-associated markers. In brief, miR-HCC2 up-regulated the expression of ELMO1, thereby promoting the migration and invasion of HCC cells by mediating the EMT process. To our knowledge, this study is the first demonstration that miR-HCC2 contributes to the high level of ELMO1 in HCC tissues. However, whether the effects of ELMO1 depend on Rac1 activation or other pathways is yet to be determined.
Collectively, our findings indicate that miR-HCC2 increased HCC cells proliferation, metastatic features, migration and invasion by maintaining high levels of BAMBI and ELMO1. Therefore, miR-HCC2 may function as an oncogene, thus adding complexity to the existing miRNAs regulatory network and presenting potential diagnosis and treatment strategies for the management of HCC.
Abbreviations
MiRNAs: MicroRNAs; HCC: hepatocellular carcinoma; EMT: epithelial-to-mesenchymal transition; 3' UTR: 3' untranslated regions; hTERT : human telomerase reverse transcriptase; BAMBI: BMP and activin membrane-bound inhibitor; TGF: transforming growth factor; ELMO1: Engulfment and cell motility 1; ECM: extracellular matrix; IL : interleukin; FXR1: Fragile-X mental retardation related protein 1.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos: 91629302; 81602410; 81572790; 31270818), the Natural Science Foundation of Tianjin (12JCZDJC25100; 17JCQNJC11300), and the Postgraduate Innovation Fund of '13th Five-Year comprehensive investment', Tianjin Medical University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-55
2. Wang C, Yao B, Xu M, Zheng X. RIP1 upregulation promoted tumor progression by activating AKT/Bcl-2/BAX signaling and predicted poor postsurgical prognosis in HCC. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:15305-13
3. Kramer AC, Kothari A, Wilson WC, Celik H, Nikitas J, Mallaney C. et al. Dnmt3a regulates T-cell development and suppresses T-ALL transformation. Leukemia. 2017;31:2479-90
4. Chen S, Wang H, Huang YF, Li ML, Cheng JH, Hu P. et al. WW domain-binding protein 2: an adaptor protein closely linked to the development of breast cancer. Molecular cancer. 2017;16:128
5. Adams MN, Burgess JT, He Y, Gately K, Snell C, Zhang SD. et al. Expression of CDCA3 Is a Prognostic Biomarker and Potential Therapeutic Target in Non-Small Cell Lung Cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2017;12:1071-84
6. Zhang X, Chen H, Wang X, Zhao W, Chen JJ. Expression and transcriptional profiling of the LKB1 tumor suppressor in cervical cancer cells. Gynecologic oncology. 2014;134:372-8
7. Tang B, Tang F, Wang Z, Qi G, Liang X, Li B. et al. Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/beta-catenin signaling. Journal of experimental & clinical cancer research: CR. 2016;35:82
8. Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW. et al. The microRNAs of Caenorhabditis elegans. Genes & development. 2003;17:991-1008
9. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77-94
10. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931-4
11. Song G, Wang R, Guo J, Liu X, Wang F, Qi Y. et al. miR-346 and miR-138 competitively regulate hTERT in GRSF1- and AGO2-dependent manners, respectively. Scientific reports. 2015;5:15793
12. Guo J, Lv J, Liu M, Tang H. miR-346 Up-regulates Argonaute 2 (AGO2) Protein Expression to Augment the Activity of Other MicroRNAs (miRNAs) and Contributes to Cervical Cancer Cell Malignancy. The Journal of biological chemistry. 2015;290:30342-50
13. Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y. et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607-19
14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
15. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-5
16. Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biological reviews of the Cambridge Philosophical Society. 2009;84:55-71
17. Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M. et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nature cell biology. 2010;12:390-9
18. Pekow J, Meckel K, Dougherty U, Huang Y, Chen X, Almoghrabi A. et al. miR-193a-3p is a Key Tumor Suppressor in Ulcerative Colitis-Associated Colon Cancer and Promotes Carcinogenesis through Upregulation of IL17RD. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:5281-91
19. Zhou Y, Frings O, Branca RM, Boekel J, le Sage C, Fredlund E. et al. microRNAs with AAGUGC seed motif constitute an integral part of an oncogenic signaling network. Oncogene. 2017;36:731-45
20. Fan Y, Li X, Xiao W, Fu J, Harris RC, Lindenmeyer M. et al. BAMBI elimination enhances alternative TGF-beta signaling and glomerular dysfunction in diabetic mice. Diabetes. 2015;64:2220-33
21. Fritzmann J, Morkel M, Besser D, Budczies J, Kosel F, Brembeck FH. et al. A colorectal cancer expression profile that includes transforming growth factor beta inhibitor BAMBI predicts metastatic potential. Gastroenterology. 2009;137:165-75
22. Wang X, Li M, Hu M, Wei P, Zhu W. BAMBI overexpression together with beta-sitosterol ameliorates NSCLC via inhibiting autophagy and inactivating TGF-beta/Smad2/3 pathway. Oncology reports. 2017;37:3046-54
23. Zhou L, Park J, Jang KY, Park HS, Wagle S, Yang KH. et al. The overexpression of BAMBI and its involvement in the growth and invasion of human osteosarcoma cells. Oncology reports. 2013;30:1315-22
24. Sekiya T, Adachi S, Kohu K, Yamada T, Higuchi O, Furukawa Y. et al. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. The Journal of biological chemistry. 2004;279:6840-6
25. Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T. et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer research. 2007;67:7203-11
26. Wang H, Linghu H, Wang J, Che YL, Xiang TX, Tang WX. et al. The role of Crk/Dock180/Rac1 pathway in the malignant behavior of human ovarian cancer cell SKOV3. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2010;31:59-67
27. Stevenson C, de la Rosa G, Anderson CS, Murphy PS, Capece T, Kim M. et al. Essential role of Elmo1 in Dock2-dependent lymphocyte migration. Journal of immunology. 2014;192:6062-70
28. Shi L, Zhang B, Sun X, Zhang X, Lv S, Li H. et al. CC chemokine ligand 18(CCL18) promotes migration and invasion of lung cancer cells by binding to Nir1 through Nir1-ELMO1/DOC180 signaling pathway. Molecular carcinogenesis. 2016;55:2051-62
29. Makino Y, Tsuda M, Ohba Y, Nishihara H, Sawa H, Nagashima K. et al. Tyr724 phosphorylation of ELMO1 by Src is involved in cell spreading and migration via Rac1 activation. Cell communication and signaling: CCS. 2015;13:35
30. Jiang J, Liu G, Miao X, Hua S, Zhong D. Overexpression of engulfment and cell motility 1 promotes cell invasion and migration of hepatocellular carcinoma. Experimental and therapeutic medicine. 2011;2:505-11
31. Peng RQ, Wan HY, Li HF, Liu M, Li X, Tang H. MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7. The Journal of biological chemistry. 2012;287:14301-9
32. Ren ZJ, Nong XY, Lv YR, Sun HH, An PP, Wang F. et al. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell death & disease. 2014;5:e1387
33. Huang NN, Becker S, Boularan C, Kamenyeva O, Vural A, Hwang IY. et al. Canonical and noncanonical g-protein signaling helps coordinate actin dynamics to promote macrophage phagocytosis of zymosan. Molecular and cellular biology. 2014;34:4186-99
34. Li H, Yang L, Fu H, Yan J, Wang Y, Guo H. et al. Association between Galphai2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nature communications. 2013;4:1706
35. Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852-62
36. Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H. et al. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. Journal of hepatology. 2015;63:874-85
37. Tang J, Li L, Huang W, Sui C, Yang Y, Lin X. et al. MiR-429 increases the metastatic capability of HCC via regulating classic Wnt pathway rather than epithelial-mesenchymal transition. Cancer letters. 2015;364:33-43
38. van Zijl F, Zulehner G, Petz M, Schneller D, Kornauth C, Hau M. et al. Epithelial-mesenchymal transition in hepatocellular carcinoma. Future oncology. 2009;5:1169-79
39. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. Journal of hepatology. 2016;65:798-808
40. Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y. et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637-49
41. Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105-18
42. Zhang Y, Yu Z, Xiao Q, Sun X, Zhu Z, Zhang J. et al. Expression of BAMBI and its combination with Smad7 correlates with tumor invasion and poor prognosis in gastric cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:7047-56
43. Lang DS, Marwitz S, Heilenkotter U, Schumm W, Behrens O, Simon R. et al. Transforming growth factor-beta signaling leads to uPA/PAI-1 activation and metastasis: a study on human breast cancer tissues. Pathology oncology research: POR. 2014;20:727-32
44. Sekiya T, Oda T, Matsuura K, Akiyama T. Transcriptional regulation of the TGF-beta pseudoreceptor BAMBI by TGF-beta signaling. Biochemical and biophysical research communications. 2004;320:680-4
45. Carethers JM. Intersection of transforming growth factor-beta and Wnt signaling pathways in colorectal cancer and metastasis. Gastroenterology. 2009;137:33-6
46. Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC. et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nature cell biology. 2002;4:574-82
47. Zhang G, Chen X, Qiu F, Zhu F, Lei W, Nie J. A novel interaction between the SH2 domain of signaling adaptor protein Nck-1 and the upstream regulator of the Rho family GTPase Rac1 engulfment and cell motility 1 (ELMO1) promotes Rac1 activation and cell motility. The Journal of biological chemistry. 2014;289:23112-22
Author contact
![]() Corresponding author: Dr. Hua Tang, No. 22 Qi-Xiang-Tai Road, Tianjin 300070, China, Tel: +86 22 23542603; Fax: +86 22 23542503; E-mail: tanghedu.cn; htang2002com
Corresponding author: Dr. Hua Tang, No. 22 Qi-Xiang-Tai Road, Tianjin 300070, China, Tel: +86 22 23542603; Fax: +86 22 23542503; E-mail: tanghedu.cn; htang2002com

 Global reach, higher impact
Global reach, higher impact