3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(11):2520-2533. doi:10.7150/jca.29207 This issue Cite
Research Paper
Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma
1. Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Province, China;
2. Department of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450000, Henan Province, China;
3. Department of Medical Cosmetology, The Second Affiliated Hospital of Guangxi Medical University, Nanning 530000, Guangxi Province, China
Received 2018-8-14; Accepted 2019-4-27; Published 2019-6-2
Abstract
Objective: MicroRNAs (miRNAs) have been explored in malignancies. We investigated the functions of clustered miRNAs hsa-miR-221/222-3p in hepatocellular carcinoma (HCC).
Methods: Human miRNA tissue atlas website was determined expression levels in liver tissue. Four databases, TarBase, miRTarBase, miRecords and miRPathDB, were found experimentally validated target genes of clustered miRNAs. TargetScanHuman was predicted target genes. The STRING website was depicted protein-protein interaction (PPI) networks. The OncoLnc website analyzed prognostic values for hsa-miR-221/222-3p and their target genes. The MCODE plugin calculated modules of PPI networks. Receiver operating characteristic (ROC) curves were predicted 1, 3, and 5 years prognostic values.
Results: Expression of clustered miRNAs was high in liver tissues. A total of 1577 target genes were identified. Enrichment analysis showed that target genes were enriched mainly in cancer, Wnt signaling and ErbB signaling pathways. Two modules were calculated using PPI networks. Has-miR-221-3p was not associated with prognosis (P = 0.401). Has-miR-222-3p and target genes ESR1, TMED7, CBFB, ETS2, UBE2J1 and UBE2N of the clustered miRNAs were associated with HCC survival (all P < 0.05). Has-miR-222-3p, CBFB, and UBE2N showed good performance of ROC in prognosis prediction at 1, 3, and 5 years (all area under curves > 0.600).
Conclusion: Has-miR-222-3p and target genes, especially CBFB, UBE2N, may serve as prognostic predictors for HCC.
Keywords: microRNA, hsa-miR-221-3p, hsa-miR-222-3p, gene, hepatocellular carcinoma, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of most common malignancies worldwide, especially in China 1. HCC accounts for more than 60,000 deaths and roughly 750,000 newly diagnosis each year 2. Etiologically, both environmental factors including hepatitis B and C viral infection and genetic factors are suggested to be involved in HCC development 3,4. Treatments such as transcatherter chemoembolization, radial frequency ablation, percutaneous ethanol injection, liver transplantation and microwave ablation are used as beneficial medical procedures for HCC 5. However, due to the obscure symptoms and a lack of sensitive molecular biomarkers, early diagnosis of HCC is difficult and prognosis is poor. The 5-year survival rate is approximately 3-5% 6. This situation makes early diagnosis and treatment in clinics challenging and identifying potential molecular markers urgent. The mechanism by which microRNAs (miRNAs) regulate HCC development is often a topic of molecular biological studies 7. Increasing evidence shows that miRNAs are could be potential molecular biomarkers and therapeutic targets for HCC 7.
MiRNAs are small, single-stranded, endogenous noncoding RNA molecules of 19-25 nucleotides that are documented to have important functions. They regulate cell differentiation, proliferation, migration and apoptosis 8,9. By binding to the 3'-untranslated regions of target genes, miRNAs negatively modulate them at the post-transcription level 10,11. MiRNA deregulation is responsible for initiation and progression of HCC 12. Overexpression of the clustered mRNAs hsa-miR-221 and hsa-miR-222 13 is reported to directly lead to the downregulation of the tumor suppressor and cell cycle regulator p27 (Kip1) 14. Hsa-miR-222 is a potential biomarker and therapeutic target for Epstein-Barr virus+ diffuse large B-cell lymphoma in older people 15. Differential expression of miR-221 is documented in comparisons of pancreatic cancer and chronic pancreatitis tissues and miR-221 and miR-222 have been documented in comparisons of pancreatic cancer and normal tissues 16. Euu et al. 17 suggested that aberrant expression of miR-221 could provide insights into pancreatic tumorigenesis and may be a prognostic biomarker for pancreatic adenocarcinoma. Expression of miR-221 and miR-221 increase in response to cellular stress in cirrhotic and hepatitis-positive livers 18. MiR-221 potentiates the malignant progression of liver cancer by governing the tumor suppressor HDAC6 19. Induced by HBx protein and targeting estrogen receptor-α, upregulation of miRNA-221 facilitates aberrant proliferation of hepatitis B virus-related HCC 20. TarBase can provide for the first time hundreds of thousands of high quality manually curated experimentally validated miRNA: gene interactions, enhanced with detailed meta-data (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/index). miRTarBase is an experimentally validated microRNA-target interaction database. It possesses more than 360 miRNA-target interactions, which are validated by reporter assay, western blot, microarray, and next-generation sequencing experiments (http://mirtarbase.mbc.nctu.edu.tw/php/index.php). miRecords is an animal miRNA-target interactions database and is made up of two component: The Validated Targets component and The Predicted Targets component. The Validated Targets component, a large, high-quality database originated from meticulous literature curation, hosts 2705 interaction records between 644 miRNAs and 1901 target genes. The Predicted Targets component is an integration of predicted miRNA target: DINAN-microT, MicroInspector, miRanda, MirTarget2, miTarget, NBmiRTar, PicTar, PITA, RNA22, RNAhybrid, and TargetScan/TargetScanS (http://c1.accurascience.com/miRecords/). miRPathDB-miRNA Pathway Dictionary Database-is freely accessible for everyone at https://mpd.bioinf.uni-sb.de/. The database attempts to increase available target pathway by offering easy access to the information of miRNA regulated pathways. It contains 2599 human miRNAs, 14773 experimentally validated target genes and 19281 predicted target genes, 280 KEGG, 1300 Reactome pathways, 6169 biological processes, 1550 molecular functions, and 758 cellular components (https://mpd.bioinf.uni-sb.de/).
Many studies on miR-221 and/or miR-222 have focused on their target genes to determine their potential prognostic value for malignancies. Our study investigated clustered miRNAs miR-221/222 and their target genes and metabolic pathways and the biological processes that they were involved in. Our aim was to predict their prognostic value and provide insights into individualized therapy for HCC patients.
Materials and Methods
Analysis of stem-loop structure, sequences and tissue expression of hsa-miR-221/222
The stem-loop structure and sequences of hsa-miR-221/222 were determined using the miRBase database13, a searchable database of published miRNAs sequences and annotations, and the Vienna RNAfold WebServer website 21 for predicting minimum free energy structures and base-pair probabilities from single RNA and DNA sequences. The human miRNA tissue atlas website22, for identifying miRNA tissues of origin and providing insights into the specificity and heterogeneity of miRNAs in tissues, was used to obtain expression levels of hsa-miR-221/222 in multiple tissues.
Analysis of experimentally validated target genes of hsa-miR-221-3p/222-3p
Four databases, TarBase v7.0 23, miRTarBase 24, miRecords 25 and miRPathDB v1.0 26, were used to obtain experimentally validated target genes. A literature review was performed to avoid missing potential target genes in the four online databases.
Enrichment analysis of KEGG pathways and gene ontology of hsa-miR-221-3p/222-3p
Enrichment analysis of KEGG pathway and gene ontology (GO) terms were performed using the online Database for Annotation, Visualization and Integrated Discovery (DAVID v6.7) 27,28, which provides a comprehensive set of functional annotation tools for investigators to understand the biological meaning behind large list of genes. Plugins ClueGO and CluePedia from Cytoscape software were used to obtain KEGG pathway enrichment analysis from the miRTarBase database.
Analysis of hsa-miR-221-3p/222-3p target genes
The online database TargetScanHuman Release 7.1 29 was accessed to predict biological targets of miRNAs, for targets with conserved sites and predicted targets irrespective of site conservation. TargetScanHuman was used to predict consequential pairing of target regions and miRNAs. All remaining genes that were ruled out as experimentally validated target genes from all conserved genes were used for KEGG pathway and GO term enrichment analysis.
Analysis of genes related to HCC on MalaCards and differentially expressed genes in Gene Expression Omnibus
To acquire differentially expressed genes (DEGs) between normal and primary liver tissue, the online dataset GSE14520 30,31 (GPL3971 platform) in the Gene Expression Omnibus (GEO) database was used for analysis. DEGs were chosen by the criteria |FC| ≥ 2 and P ≤ 0.05. Genes reported to be related to HCC were accessed on the MalaCards website 32. Online resources Venn Diagrams and Venny 2.1 33 were used to calculate the intersections of genes.
Survival analysis of hsa-miR-221-3p/222-3p in HCC
Survival analysis of hsa-miR-221-3p/222-3p and target gene expression for HCC prognosis used the online database OncoLnc 34. Mutation analysis of target genes of hsa-miR-221-3p/222-3p used the online source cBioportal 35,36. The website miRTargetLink Human Release 6.0 37 was used to visualize experimentally validated target genes with strong evidence. Correlation analysis was used to visualize expression levels of hsa-miR-221-3p/222-3p and target genes. The MCODE plugin of Cytoscape v3.5.0 software was used to identify modules in numerous genes 38,39.
Statistical analysis
Pearson correlation analysis was calculated by SPSS v16.0 (IBM, Chicago, IL). Correlation result diagrams were made using R v3.2.0 (https://www.r-project.org/).
Results
Mature sequences, structure and expression analysis of hsa-miR-221/222
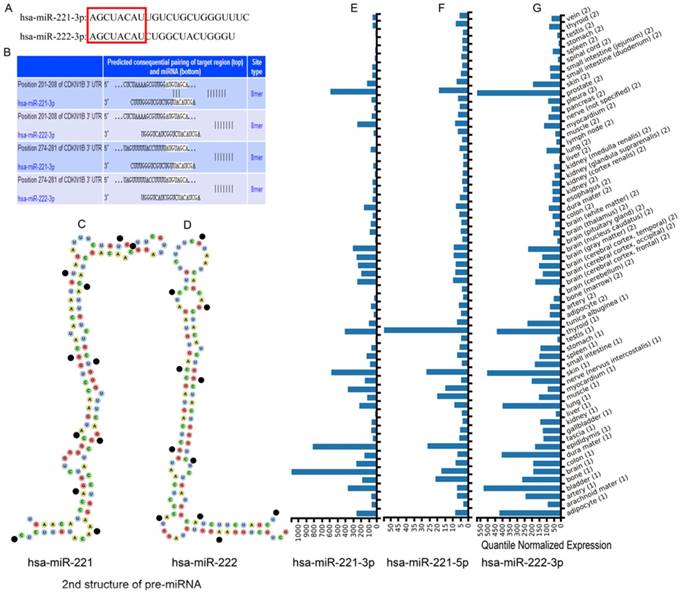
Sequences of mature clustered miRNAs hsa-miR-221-3p and hsa-miR-222-3p were obtained from miRBase databases. The first 8 bases were same while other bases were not (Figure 1A). The secondary structures of pre-miRNAs were from the Vienna RNAfold WebServer website. Hsa-miR-221 showed a reverse Z shape and 8 closed loops and hsa-miR-221 showed a natural L shape and 6 closed loops (Figure 1C-D). Expression of the clustered miRNAs hsa-miR-221-3p/5p and hsa-miR-222-3p in multiple organs is in Figure 1E-G. The miRNAs had relatively high expression levels in liver tissue. Hsa-miR-222-5p was not recognized in the website.
Experimentally validated target genes of hsa-miR-221-3p/222-3p
A total of 1577 target genes was determined from four websites. Specifically, 1092 experimentally validated genes for hsa-miR-221-3p/222-3p were determined from the TarBase database; 638 experimentally validated genes of hsa-miR-221-3p/222-3p were determined from the miRTarBase database; 20 experimentally validated genes of hsa-miR-221-3p/222-3p were determined from the miRecords database; and 417 experimentally validated genes of hsa-miR-221-3p/222-3p were determined from the miRPathDB database (Table 1). Detailed gene lists for the clustered miRNAs are in Supplementary Table 1.
Analysis of experimentally validated genes of hsa-miR-221-3p/222-3p
| Databases | miRNAs | Sum | Repetition | Remaining | Combination 1 | Combination 2 |
|---|---|---|---|---|---|---|
| TarBase | hsa-miR-221-3p | 900 | 112 | 788 | 1092 | 1577 |
| hsa-miR-222-3p | 800 | 88 | 712 | |||
| miRTarBase | hsa-miR-221-3p | 368 | 0 | 368 | 638 | |
| hsa-miR-222-3p | 394 | 0 | 394 | |||
| miRecords | hsa-miR-221-3p | 20 | 4 | 16 | 20 | |
| hsa-miR-222-3p | 16 | 2 | 14 | |||
| miRPathDB | hsa-miR-221-3p | 348 | 1 | 347 | 417 | |
| hsa-miR-222-3p | 374 | 2 | 372 |
Sequences, structures and expression in different tissues of hsa-miR-221/222. (A) Mature sequences of hsa-miR-221/222. (B) Predicted sequential pairing of target region and miRNA. (C-D) Secondary structure of hsa-miR-221 and hsa-miR-222 pre-miRNAs. (E-G) Expression of hsa-miR-221-3p, hsa-miR-221-5p and hsa-miR-222-3p in different tissues.
Tissue expression, KEGG pathways and GO enrichment of hsa-miR-221-3p/222-3p target genes
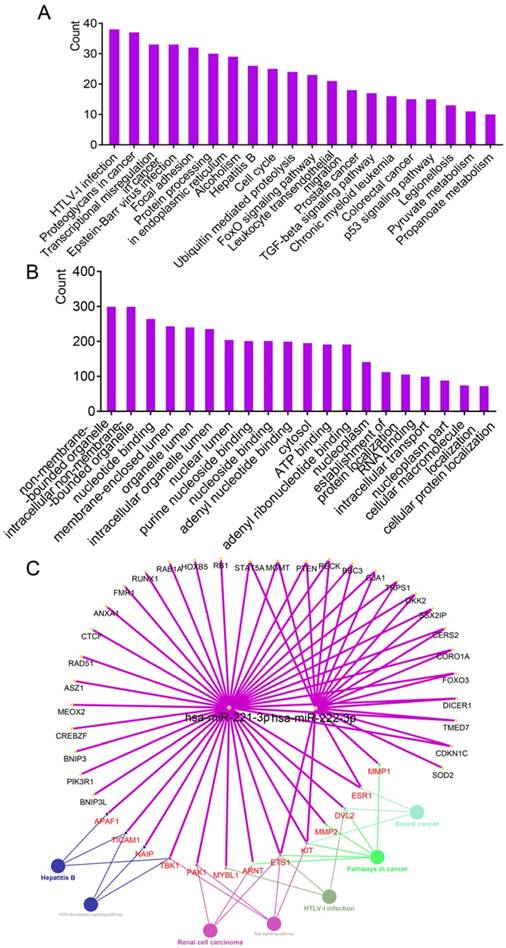
The top 20 significantly enriched tissues from DAVID website analysis are in Table 2. Many tissues, including liver, were in the results. Liver tissue had 216 enriched genes, ranking fourth of all enriched tissues, and accounted for 13.9987% that were significant (P < 0.001). Results of enrichment analysis are in Supplementary Table 2. Significant enrichment results for the top 20 KEGG pathways and top 20 GO terms are in Figure 2A. HTLV-1 infection, proteoglycans in cancer, and transcriptional misregulation in cancer were ranked with the top 3 counts in the KEGG pathway enrichment results. Enrichment results of GO terms, including biological process (BP), cellular component (CC) and molecular function (MF), are in Figure 2B. Nonmembrane-bounded organelles, intracellular nonmembrane-bounded organelles, and nucleotide binding, were ranked as the top 3 counts. Detailed enrichment results are in Supplementary Table 3.
Top 20 up-tissues of tissue expression of experimentally validated genes
| Category | Term | Count | % | P value | Benjamini | FDR |
|---|---|---|---|---|---|---|
| UP_TISSUE | Epithelium | 405 | 26.24757 | 2.95E-37 | 9.37E-35 | 3.97E-34 |
| UP_TISSUE | Brain | 785 | 50.87492 | 8.60E-13 | 1.37E-10 | 1.16E-09 |
| UP_TISSUE | Cajal-Retzius cell | 51 | 3.30525 | 5.13E-11 | 5.44E-09 | 6.91E-08 |
| UP_TISSUE | T-cell | 62 | 4.018146 | 8.82E-10 | 7.01E-08 | 1.19E-06 |
| UP_TISSUE | Uterus | 214 | 13.86909 | 2.72E-09 | 1.73E-07 | 3.66E-06 |
| UP_TISSUE | Placenta | 368 | 23.84964 | 6.42E-09 | 3.40E-07 | 8.65E-06 |
| UP_TISSUE | Platelet | 86 | 5.573558 | 1.82E-08 | 8.28E-07 | 2.46E-05 |
| UP_TISSUE | Fetal brain cortex | 48 | 3.110823 | 2.56E-07 | 1.02E-05 | 3.45E-04 |
| UP_TISSUE | Bone marrow | 108 | 6.999352 | 6.88E-07 | 2.43E-05 | 9.27E-04 |
| UP_TISSUE | Kidney | 167 | 10.82307 | 5.46E-05 | 0.001735 | 0.073588 |
| UP_TISSUE | Ovarian carcinoma | 30 | 1.944264 | 5.88E-05 | 0.001699 | 0.079251 |
| UP_TISSUE | Lung | 275 | 17.82242 | 1.14E-04 | 0.003026 | 0.154066 |
| UP_TISSUE | Cervix carcinoma | 56 | 3.629294 | 2.40E-04 | 0.005854 | 0.32305 |
| UP_TISSUE | Human endometrium | 7 | 0.453662 | 2.56E-04 | 0.005796 | 0.344413 |
| UP_TISSUE | Eye | 122 | 7.906675 | 5.03E-04 | 0.01061 | 0.676051 |
| UP_TISSUE | Fibroblast | 25 | 1.62022 | 6.41E-04 | 0.012672 | 0.861353 |
| UP_TISSUE | Peripheral Nervous System | 14 | 0.907323 | 6.92E-04 | 0.012862 | 0.928636 |
| UP_TISSUE | Liver | 216 | 13.9987 | 7.65E-04 | 0.013431 | 1.026589 |
| UP_TISSUE | Skin | 192 | 12.44329 | 0.001247 | 0.020671 | 1.668452 |
| UP_TISSUE | Uterus endothel | 8 | 0.518471 | 0.001512 | 0.023776 | 2.019658 |
Note: FDR: false discovery rate.
The results of connecting hsa-miR-221-3p/222-3p, target genes and metabolic pathways are in Figure 2C. A total of 43 genes and 7 metabolic pathways were enriched in this network. Of these, 18 genes, including STAT5A, MGMT, PTEN, RECK, BBC3, GJA1 and TRPS1 were connected to both miRNAs; 13 genes including APAF1, TICAM1, NAIP, TBK1, PAK1 and MYBL1, were involved in metabolic pathways. Seven metabolic pathways were NOD-like receptor signaling pathway, renal cell carcinoma, hepatitis B, ras signaling pathway, HTLV-1 infection, and pathways in cancer and breast cancer.
Enrichment analysis of GO terms and KEGG pathways by experimentally validated genes. (A-B) Enrichment results of KEGG pathways and GO terms by DAVID. (C) Enrichment results of KEGG pathways constructed by hsa-miR-221/222-3p and genes using ClueGO and CluePedia plugins.
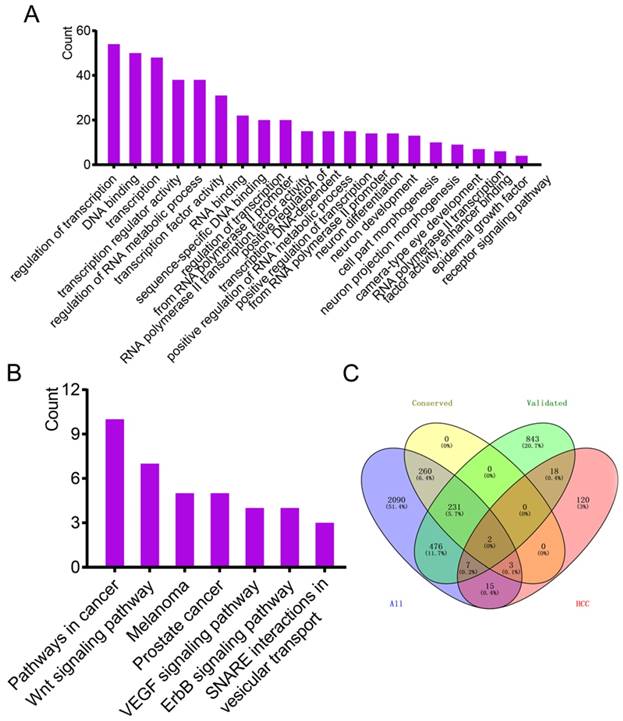
Enrichment analysis of GO terms and KEGG pathways by predicted target genes and Venn diagram analysis. (A-B) Enrichment analysis of GO terms and KEGG pathways of predicted target genes (ruled out of experimentally validated genes) by TargetScanHuman. (C) Venn diagram of intersection analysis using four datasets.
Prediction, enrichment analysis of target genes and intersection analysis from four data sources
The website TargetScan was used to predict target genes of hsa-miR-221-3p/222-3p irrespective of site conservation to find target genes with potential research value. A total of 496 target genes with conserved sites and 3084 target genes irrespective of site conservation were identified by the website. CDKN1B, ranked first of all the predicted genes, was predicted to have consequential paring of a target region at position 201-208 in the 3'-untranslated region with hsa-miR-221-3p (Figure 1B). Hsa-miR-222-3p was not recognized. Removing 238 common target genes that were experimentally validated left a predicted 258 target genes enriched by DAVID, generating KEGG pathways and GO terms. The top 20 enriched GO terms and KEGG pathways are in Figure 3A-B. A protein-protein interaction (PPI) network of 258 target genes is depicted in Supplementary Figure 1. Detailed lists of 496 target genes (238 common and 258 other predicted genes 258) and 3084 target genes are in Supplementary Table 5.
Online source MalaCards was used to identify genes validated with HCC. A total of 165 genes were identified (Supplementary Table 5). Four data sources, 1577 validated genes, 496 predicted genes with conserved sites, 3084 predicted genes irrespective of site conservation and 165 HCC-related genes were used for intersection analysis using the Venny online tool. Only two genes were common to the four datasets (Figure 3C).
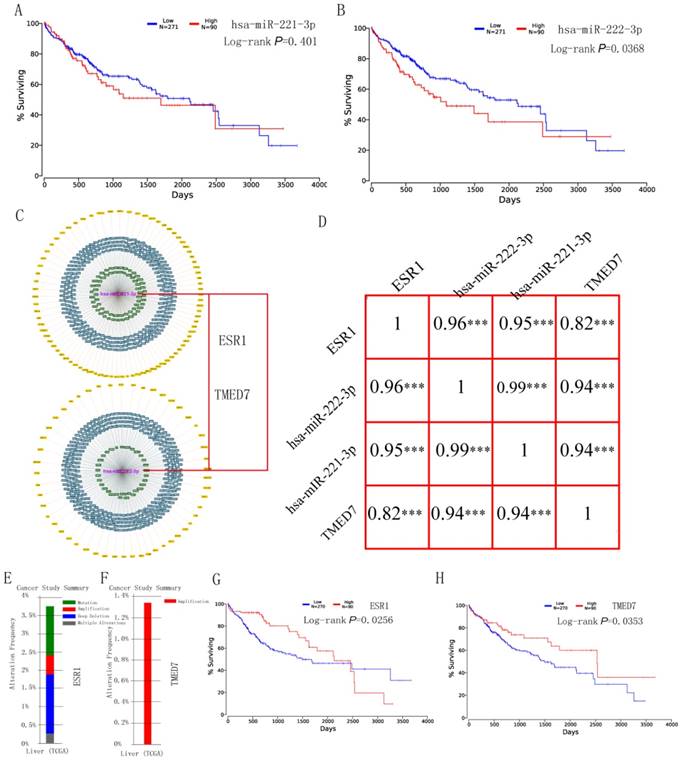
Survival analysis, Pearson correlation analysis and mutation analysis of hsa-miR-221/222-3p and target genes. (A-B) Survival analysis of hsa-miR-221-3p and hsa-miR-222-3p. (C) Target genes of hsa-miR-221/222-3p by miRTargetLink and two target genes of them. (D) Pearson correlation diagram of hsa-miR-221/222-3p and two target genes. (E-F) Mutation analysis of ESR1 and TMED7. (G-H) Survival analysis of ESR1 and TMED7.
Survival analysis of hsa-miR-221-3p/222-3p and target genes in HCC
Survival analysis of hsa-miR-221-3p/222-3p was performed using the OncoLnc website at quatile in Figure 4A-B. Hsa-miR-222-3p was significant (P = 0.0368); while hsa-miR-222-3p was not (P = 0.401). The MiRTargetLink website was used to determine experimentally validated genes of hsa-miR-221-3p/222-3p. Target genes with strong evidence are in Figure 4C. Detailed target gene lists are in Supplementary Table 4. Of the target genes, ESR1 and TMED7 were chosen for further analysis. Pearson correlation results for hsa-miR-221-3p, hsa-miR-222-3p, ESR1 and TMED7 are in Figure 4D. Gene expression was highly and positively correlated with the miRNAs (all R > 0.9, P < 0.001). Mutation analysis of ESR1 and TMED7 was plotted in the Cbioportal website (Figure 4E-F) and survival analysis of these two genes was depicted in the OncoLnc website (Figure 4G-H). ESR1 showed mutation analysis in mutation, amplification, deep deletion and multiple alterations, TMED7 showed mutation analysis only in amplification. Both were significant in survival analysis (P = 0.0256 for ESR1; P = 0.0253 for TMED7).
Hsa-miR-221-3p/222-3p may facilitate HCC carcinogenesis
The results indicated that hsa-miR-221-3p/222-3p and target genes of ESR1 and TMED7 had verified prognostic value in HCC. Other validated genes, for example APAF1, NAIP, TICAM1, TBK1, PAK1, ETS1, KIT, ARNT, MMP2, DVL2, MMP1, were identified as involved in the tumor-related pathways NOD-like receptor signaling pathway, hepatitis B, Ras signaling pathway and pathways in cancer. Their target genes were enriched in BP, CC, and MF such as RNA binding, cellular protein localization, and epidermal growth factor receptor signaling pathway, which are relevant to tumorigenesis. These results suggested that hsa-miR-221-3p/222-3p played key role in HCC carcinogenesis.
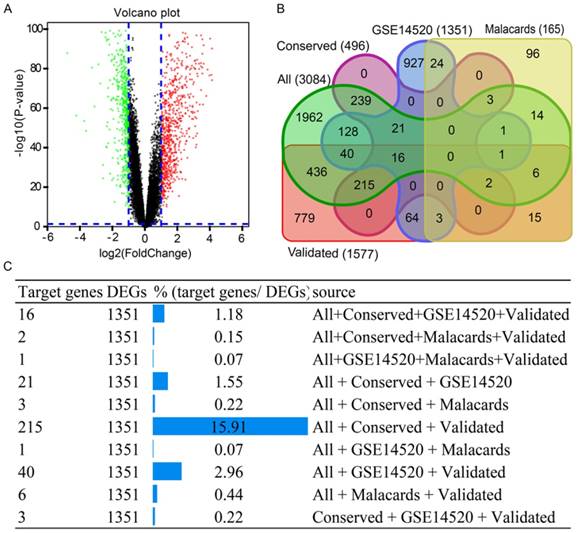
To investigate this hypothesis, GEO datasets and the GSE14520 GPL3921 platform, were used for analysis. Using the criterion described above, 1351 DEGs were identified (Supplementary Table 6). A volcano plot of the GSE14520 dataset is in Figure 5A. A Venn diagram was generated using the datasets of 1571 validated genes, 3084 genes, conservative sites 496 genes, 1351 genes and 165 validated genes in Figure 5B. A total of 215 target genes/DEGs, accounting for 15.91%, ranked first of all of the more than three dataset intersection groups (Figure 5C). Detailed results on 215 target genes and 215 DEGs are in Supplementary Table 7.
Results of volcano plot and Venn diagram analysis. (A) Volcano plot of genes in GSE14520 dataset. (B) Venn diagram analysis of five datasets. (C) Detailed results of Venn diagram by source origin (at least 3 datasets).
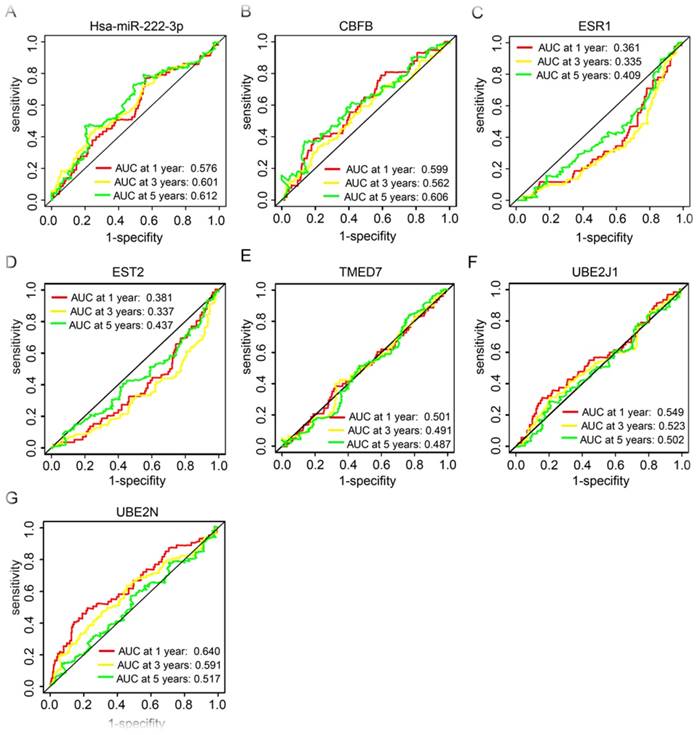
Receiver operating characteristic curve (ROC) analysis of HCC prognostic related miRNA and target genes
In our further analysis, HCC prognosis related miRNA and target genes, has-miR-222-3p, CBFB, ESR1, EST2, TMED7, UBE2J1, and UBE2N, were performed for prognostic ROC analysis in 1, 3, and 5 years, respectively. Has-miR-222-3p showed a good performance in clinical outcome prediction in both 3 and 5 years (P = 0.601 and 0.612 respectively, Figure 6A). CBFB showed a good performance in 5 years prediction (P = 0.606, Figure 6B). UBE2N presented a good performance in 1 year prediction (P = 0.640, Figure 6G).
Molecular mechanism of hsa-miR-221-3p/222-3p implicated in HCC tumorigenesis
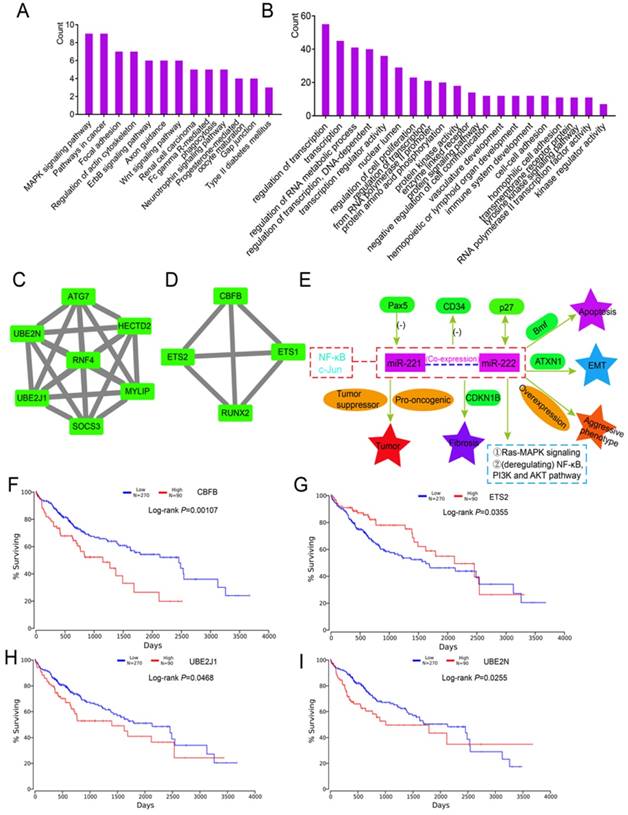
To determine the potential functions of 215 DEGs, enrichment analysis of KEGG pathways and GO terms was performed using DAVID. Enriched were 13 metabolic pathways including the MAPK signaling pathway, pathways in cancer, and the Wnt signaling pathway (Figure 7A). The top 20 enriched GO terms are in Figure 7B. Detailed enrichment results are in Supplementary Table 8. A PPI network of 215 DEGs is in Supplementary Figure 2. Two modules were identified by MCODE, which included 7 and 4 genes (Figure 7C-D). In the 11 enriched genes, CBFB (P = 0.00107), ETS2 (P = 0.0355), UBE2J1 (P = 0.0468) and UBE2N (P = 0.0255) were associated with HCC survival (Figure 7F-I). Specific mechanisms of involvement of these miRNAs are in Figure 7E.
Discussion
In this study, we conducted a comprehensive investigation of the clustered miRNAs hsa-miR-221 and hsa-miR-222 and HCC. We found that the miRNAs were highly expressed in liver tissue and their secondary pre-miRNA structures showed specific characteristics. By determining their experimentally validated target genes, we identified their characterized biological processes and metabolic pathways. We found that these clustered miRNAs and their target genes had potential prognostic value for HCC, especially hsa-miR-222-3p, which functioned as a tumor promotor in hepatic tumorigenesis. DEGs were validated for metabolic processes and pathways. Many metabolic pathways and functions in biological processes were detailed, depicted and clarified.
Receiver operating characteristic curves in 1, 3 and 5 years of prognostic related miRNA and genes. (A-G): receiver operating characteristic curves in 1, 3 and 5 years of has-miR-222-3p, CBFB, ESR1, EST2, TMED7, UBE2J1, and UBE2N, respectively.
Enrichment, modules and survival analysis of differentially expressed genes (DEGs) and mechanism analysis by literature review. (A-B) Enrichment analysis of KEGG pathways and GO terms for 215 DEGs. (C-D) Two modules of 215 DEGs by MCODE plugin. (E) Mechanisms of miR-221/222 in tumorigenesis by literature review. (F-I) Survival analysis of genes in the above modules.
Generally, an miRNA originates from a primary transcript pri-miRNA, which is transcribed by RNA polymerase II 40. An miRNA can extend to more than 100 kilobases and contain more than one precursor miRNA hairpin (pre-miRNA) 40. Several studies show that more than half of miRNAs are intragenic and located mainly in the regions of protein-coding genes 41,42. Only one-third of intronic miRNAs are transcribed from promotors. The majority are co-expressed and co-regulated with host genes in which they are reside 40. Thus, host miRNAs and miRNAs may result from a common primary transcript 40. Intergenic pri-miRNAs are poorly characterized and lack well-annotated databases 40. However, similar to transcriptional regulation of protein-coding genes, intergenic pri-miRNA expression is predominantly regulated by transcription factors through promotors and enhancers 40. In addition, protein-coding genes and promotors and enhancers of intergenic pri-miRNAs share common epigenetic characteristics, including histone modification marks 40. Produced from the same pri-miRNA, expression alterations of them are observed synchronously 40.
Hsa-miR-221 and hsa-miR-222 (miR-221/222), two highly homologous miRNAs, are clustered on the short arm of chromosome X 40. Patterns of miRNA expression are generally accepted to be associated with cancer types, stages and other clinical variables43. Analysis of miRNA expression indicates that miRNAs function both oncogenically and as tumor suppressors 43. They are acknowledged to be overexpressed in most epithelial malignancies including breast, liver, pancreatic and lung cancer and to have oncogenic functions in these cancers 40,44-47. Expression of miRNAs 221/222 is reported to regulate acute myeloid leukemia at a post-transcriptional level and may be a novel potential biomarker and putative oncogene in this disease 48. Overexpression of hsa-miR-221/222 is identified in glioblastoma multiforme-initiating cells, which are pivotal regulators in dynamic expression 49. Increased expression of miR-221 is reported in pancreatic cancer 17, glioblastoma 50 and thyroid cancer 45.
Tathiana et al.15 reported different functions of hsa-miR-222 by targeting multiple target genes: hsa-miR-222 functions in tumor suppression by targeting PTEN, in cell cycle regulation by targeting CDKN1B (p27) and CDKN1C, in transcription by targeting STAT5A, as a oncogene by targeting FOS and KIT, as an adhesion molecule by targeting ICAM1, and in oxidative stress by targeting SOD2. Roehle et al.51 reported hsa-miR-222 is involved in immune regulation and B cell-related tumors. Mediated by Rbm24 protein, miRNA-222 alters myogenic differentiation in an alternative splicing process 52.
Upregulation of clustered miR-221/222 is controversial and particularly relevant to astrocytic glioblastoma multiforme subtype. 53 MiR-221/222 were observed in glioblastoma multiforme-initiating cell differentiation and these clustered miRNAs are overexpressed in glioblastoma multiforme compared to nontransformed tissues 49,54. In addition, these miRNAs demonstrate a inhibitory effect on proliferation in an erythroleukemic cell line and decreased stem cell repopulating activity in cord blood CD34+ cells by inhibiting KIT 55. Therefore, the miRNAs exert both oncogenic and tumor-suppressive effects, depending on the cellular context 49.
Our research found that miRNA-221/222 are involved in the NOD-like receptor signaling pathway by targeting TICAM1, NAIP and TBK1 genes. The NOD-like receptor signaling pathway is composed of regulating factors and receptors including NLRC and NLRX family gene members. This result parallels our previous report that NLRC and NLRX gene subfamily members are potential biomarkers for HCC 56. Hepatitis B was enriched in the results by targeting APAF1, TICAM1 and TBK1. The Ras signaling pathway and pathways in cancer were present in results. In the next enrichment analysis, pathways in cancer, the Wnt signaling pathway and the ErbB signaling pathway were shown in different results. Both miR-222 and some target genes had significant values for prognosis. Given these results, we postulated that the clustered miRNAs may be involved in these pathways and the target genes may be involved in metabolic pathways.
In a literature review, we found that Pax5 reduces miR-221 and miR-222 expression when co-expressed with miR-221 by inducing differentiation from CD19- common lymphoid progenitors to CD19+ pre-B-I cells 57,58. By targeting Bmf, miR-221 inhibits apoptosis and overexpression is associated with a more aggressive phenotype of tumor multifocality 59. By targeting CDKN1B, miR-221/222 expression is associated with liver fibrosis and upregulated in disease propagation 60. NF-κB and c-Jun, two transcription factors, are involved in cancer onset and progression, contributing to tumorigenesis by inducing miR-221/222 transcription 61. In addition, p27 hsa both positive and negative functions in the process of regulating cell proliferation, cell motility and apotosis 62.
Our investigation demonstrated that high expression of hsa-miR-222-3p produced a better prognosis than low expression. Thus, we conclude from our study that hsa-miR-222 may be a tumor promotor in liver tumorigenicity, consistent with previous research 63. Beatriz et al.49 found that these miRNAs have pro-oncogenic or tumor-suppressor functions, depending on the cellular context. Our study found that overexpression of miR-221 did not induce an undesirable prognosis for HCC patients. Of note, this opinion was not consistent with Pascal et al. 64 who found that overexpression of miR-221 contributes risk of hepatic tumorigenesis. Thus, we concluded that the potential functions of miR-221-3p need to be further explored and miR-222 may be oncogenic in HCC. Several target genes of miR-221/222-3p and miR-222 may participate in pathways in cancer, the Wnt signaling pathway and the ErbB signaling pathway. Several target genes of miR-221/222-3p and miR-222-3p itself may be potential prognostic biomarkers for HCC. Identification of these biomarkers provides new insight for HCC treatment, which means early detection of their expressions and drug target determination can give each HCC patient individualized therapy based on each patient characteristic.
The study has several limitations that need to be recognized. First, more highly specific miRNA or clustered miRNAs need to be recognized in liver tissue for analysis. Second, clinical data should be included to further validate these findings.
Third, experiments on potential biomarkers are necessary to determine their prognostic value and clarify concrete mechanisms of the clustered miRNAs. Therefore, further investigations are still warranted to address these issues.
Conclusions
In conclusion, although our study had some deficiencies, we found target genes of miR-221/222-3p, both experimentally validated and predicted, in different databases. We analyzed potential KEGG pathways and biological processes, cellular components, and molecular functions using the target genes. We analyzed the potential prognostic abilities of the miRNAs and their target genes and performed mutation analysis of target genes. Another dataset validated these findings and specific mechanisms were reviewed. Potentially important genes were enriched into two modules and prognostic predictions were performed to identify the genes in modules of molecular biomarker values.
Abbreviations
miRNAs: microRNAs; HCC: hepatocellular carcinoma; ROC: receiver operating characteristic curve; PPI: protein-protein interaction; GEO: Gene Expression Omnibus; BP: biological process; CC: cellular component; MF: molecular function; GO: gene ontology; FDR: false discovery rate.
Supplementary Material
Supplementary figures.
Supplementary table 1.
Supplementary table 2.
Supplementary table 3.
Supplementary table 4.
Supplementary table 5.
Supplementary table 6.
Supplementary table 7.
Supplementary table 8.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (No.: 81560535, 81802874, 81072321, 30760243, 30460143 and 30560133), Natural Science Foundation of Guangxi Province of China (Grant No.2017JJB140189y), 2009 Program for New Century Excellent Talents in University (NCET), Guangxi Natural Sciences Foundation (No.: GuiKeGong 1104003A-7), and Guangxi Health Ministry Medicine Grant (Key-Scientific Research-Grant Z201018). The present study is also partly supported by Scientific Research Fund of the Health and Family Planning Commission of Guangxi Zhuang Autonomous Region (Z2016318), Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (GKE2018-01), The Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (2018KY0110), Guangxi Key Laboratory for the Prevention and Control of Viral Hepatitis (No.GXCDCKL201902), 2018 Innovation Project of Guangxi Graduate Education (JGY2018037), and 2018 Innovation Project of Guangxi Graduate Education (YCBZ2018036). As well as, the present study is also partly supported by Research Institute of Innovative Think-tank in Guangxi Medical University (The gene-environment interaction in hepatocarcinogenesis in Guangxi HCCs and its translational applications in the HCC prevention). We would also acknowledge the supported by the Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education.
Authors' Contributions
Xiangkun Wang and Tao Peng designed this manuscript; Xiwen Liao, Ketuan Huang, Xianmin Zeng, Zhengqian Liu, Xin Zhou, Tingdong Yu, Chengkun Yang, Long Yu, Qiaoqi Wang, Chuangye Han, Guangzhi Zhu, Xinping Ye and Tao Peng conducted the study and analyzed the data. Xiangkun Wang wrote the manuscript, and Tao Peng guided the writing.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Xiong G, Wang Y, Ding Q, Yang L. Hsa-mir-1269 genetic variant contributes to hepatocellular carcinoma susceptibility through affecting SOX6. Am J Transl Res. 2015;7(10):2091-2098
2. Liu Y, Zhao L, Ma W. et al. The Blockage of KCa3.1 Channel Inhibited Proliferation, Migration and Promoted Apoptosis of Human Hepatocellular Carcinoma Cells. Journal of Cancer. 2015;6(7):643
3. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nature Reviews Gastroenterology & Hepatology. 2010;7(8):448
4. Jiang DK, Sun J, Cao G. et al. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus–related hepatocellular carcinoma. Nature Genetics. 2013;45(1):72
5. Julius B, Iii VD, Asham EH. et al. Hepatocellular carcinoma: a review. Journal of Hepatocellular Carcinoma. 2016;3:41-53
6. Li H, Liu F, Zhu H. et al. Interaction Between Polymorphisms of IFN-gamma and MICA Correlated with Hepatocellular Carcinoma. Med Sci Monit. 2016;22:549-553
7. Chu R, Mo G, Duan Z. et al. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun Signal. 2014;12:45
8. Plasterk RHA. Micro RNAs in Animal Development. Cell. 2006;124(5):877
9. Tang W, Tang J, He J. et al. SLIT2/ROBO1-miR-218-1-RET/PLAG1: a new disease pathway involved in Hirschsprung's disease. Journal of Cellular & Molecular Medicine. 2015;19(6):1197-1207
10. S V, F R, N B. et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2015;137(6):1032-1046
11. Hyun S, Lee JH, Hua J. et al. Conserved MicroRNA miR-8/miR-200 and Its Target USH/FOG2 Control Growth by Regulating PI3K. Cell. 2009;139(6):1096-1108
12. Yin W, Zhao Y, Ji YJ. et al. Serum/plasma microRNAs as biomarkers for HBV-related hepatocellular carcinoma in China. Biomed Research International. 2015;2015(52):965185
13. Kozomara A, Griffithsjones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42(Database issue):D68
14. Galardi S, Mercatelli N, Giorda E. et al. miR-221 and miR-222 Expression Affects the Proliferation Potential of Human Prostate Carcinoma Cell Lines by Targeting p27Kip1. Journal of Biological Chemistry. 2007;282(32):23716-23724
15. Andrade TA, Evangelista AF, Campos AH. et al. A microRNA signature profile in EBV+ diffuse large B-cell lymphoma of the elderly. Oncotarget. 2014;5(23):11813-11826
16. Bloomston M, Frankel WL, Petrocca F. et al. MicroRNA Expression Patterns to Differentiate Pancreatic Adenocarcinoma From Normal Pancreas and Chronic Pancreatitis. Jama. 2007;297(17):1901-1908
17. Lee EJ, Gusev Y, Jiang J. et al. Expression profiling identifies microRNA signature in pancreatic cancer. International Journal of Cancer. 2007;120(5):1046
18. Jiang J, Gusev Y, Aderca I. et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clinical Cancer Research An Official Journal of the American Association for Cancer Research. 2008;14(2):419-427
19. Bae HJ, Jung KH, Eun JW. et al. MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J Hepatol. 2015;63(2):408-419
20. Chen JJ, Tang YS, Huang SF, Ai JG, Wang HX, Zhang LP. HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBVrelated hepatocellular carcinoma by targeting estrogen receptor-alpha. Oncol Rep. 2015;33(2):792-798
21. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Research. 2008;36(Web Server issue):70-74
22. Ludwig N, Leidinger P, Becker K. et al. Distribution of miRNA expression across human tissues. Nucleic Acids Research. 2016;44(8):3865
23. Vlachos IS, Paraskevopoulou MD, Karagkouni D. et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Research. 2015;43(Database issue):D153
24. Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW. et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Research. 2017:46 (Database issue)
25. Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database issue):D105-110
26. Backes C, Kehl T, Stöckel D. et al. miRPathDB: a new dictionary on microRNAs and target pathways. Nucleic Acids Research. 2017;45(D1):D90
27. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44
28. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37(1):1
29. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife Sciences. 2015:4 (e05005)
30. Roessler S, Jia HL, Budhu A. et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Research. 2010;70(24):10202-10212
31. Roessler S, Long EL, Budhu A. et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142(4):957-966
32. Rappaport N, Twik M, Plaschkes I. et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Research. 2016;45(Database issue):D877-D887
33. Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007
34. Anaya J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peerj Computer Science. 2016;2(2):e67
35. Gao J, Aksoy BA, Dogrusoz U. et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science Signaling. 2013;6(269):pl1
36. Cerami E, Gao J, Dogrusoz U. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012;2(5):401
37. Gaul U. MicroRNA targets in Drosophila. Genome Biology. 2003;5(1):R1
38. Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498-2504
39. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics. 2003;4:2
40. Gui B, Hsieh CL, Kantoff PW, Kibel AS, Jia L. Androgen receptor-mediated downregulation of microRNA-221 and -222 in castration-resistant prostate cancer. PLoS One. 2017;12(9):e0184166
41. Monteys AM, Wan RMS, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. Rna-a Publication of the Rna Society. 2010;16(3):495
42. Hinske LC, Galante PA, Kuo WP, Ohnomachado L. A potential role for intragenic miRNAs on their hosts' interactome. Bmc Genomics. 2010;11(1):533
43. Bandopadhyay M, Banerjee A, Sarkar N. et al. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer. 2014;14:721
44. Felicetti F, Errico MC, Bottero L. et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Research. 2008;68(8):2745
45. He H, Jazdzewski K, Li W. et al. The role of microRNA genes in papillary thyroid carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(52):19075-19080
46. Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Current Molecular Medicine. 2012;12(1):27-33
47. Kawaguchi T, Komatsu S, Ichikawa D. et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. British Journal of Cancer. 2013;108(2):361
48. Rommer A, Steinleitner K, Hackl H. et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer. 2013;13:364
49. Aldaz B, Sagardoy A, Nogueira L. et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PLoS One. 2013;8(10):e77098
50. Ciafrè SA, Galardi S, Mangiola A. et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical & Biophysical Research Communications. 2005;334(4):1351
51. Roehle A, Hoefig KP, Repsilber D. et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. British Journal of Haematology. 2008;142(5):732-744
52. Cardinali B, Cappella M, Provenzano C. et al. MicroRNA-222 regulates muscle alternative splicing through Rbm24 during differentiation of skeletal muscle cells. Cell Death Dis. 2016;7:e2086
53. Kim TM, Huang W, Park R, Park PJ, Johnson MD. A Developmental Taxonomy of Glioblastoma Defined and Maintained by MicroRNAs. Cancer Research. 2011;71(9):3387-3399
54. Zhang J, Han L, Ge Y. et al. miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. International Journal of Oncology. 2010;36(4):913
55. Felli N, Fontana L, Pelosi E. et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(50):18081-18086
56. Wang X, Yang C, Liao X, Han C, Yu T, Huang K. et al. NLRC and NLRX gene family mRNA expression and prognostic value in hepatocellular carcinoma. Cancer Med. 2017;6(11):2660-72
57. Knoll M, Simmons S, Bouquet C, Grun JR, Melchers F. miR-221 redirects precursor B cells to the BM and regulates their residence. Eur J Immunol. 2013;43(9):2497-2506
58. Schebesta A, Mcmanus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27(1):49
59. Gramantieri L, Fornari F, Ferracin M. et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15(16):5073-5081
60. Almas I, Afzal S, Idrees M. et al. Role of circulatory microRNAs in the pathogenesis of hepatitis C virus. Virusdisease. 2017;28(4):360-367
61. Galardi S, Mercatelli N, Farace MG, Ciafrè SA. NF-kB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Research. 2011;39(9):3892-3902
62. Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nature Reviews Cancer. 2008;8(4):253
63. Callegari E, Elamin BK, Giannone F. et al. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;56(3):1025-1033
64. Pineau P, Volinia S, Mcjunkin K. et al. miR-221 overexpression contributes to liver tumorigenesis. Journal of Hepatology. 2010;52(1):264-269
Author contact
![]() Corresponding author: Professor Tao Peng, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Province, China. Tel: (+86)-771-5350190. Fax: (+86)-771-5350031. E-mail: pengtaogmucom.
Corresponding author: Professor Tao Peng, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi Province, China. Tel: (+86)-771-5350190. Fax: (+86)-771-5350031. E-mail: pengtaogmucom.

 Global reach, higher impact
Global reach, higher impact