Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(11):2501-2509. doi:10.7150/jca.29285 This issue Cite
Review
Role of Autophagy in Renal Cancer
1. Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China
2. Department of Cardiovascular Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China
*Contributed equally
Received 2018-8-16; Accepted 2019-4-26; Published 2019-6-2
Abstract
Autophagy is a highly conserved catabolic process with critical functions in maintenance of cellular homeostasis under normal growth conditions and in preservation of cell viability under stress. The role of autophagy in cancer is dual-sided. Autophagy-deficient cells are often more tumorigenic than their wild type counterparts in association with DNA damage accumulation, oxidative stress. At the same time, autophagy is a major cell survival mechanism. In recent years, it has been well demonstrated that autophagy may have relation with renal cell carcinoma (RCC). This review focuses on the research progress in relation between autophagy and RCC and the pharmacologic manipulation of autophagy for RCC treatment.
Keywords: autophagy, renal cell carcinoma, therapy
1. Introduction
Autophagy is a highly conserved metabolic process in eukaryotic cells and plays an important role in maintaining the viability of cells in a stable or stressed state [1, 2]. Substrate to be degraded in cells are packed with double membrane autophagosomes and transported to lysosomes for degradation and recycling [3]. Autophagy mainly occurs under the conditions of hypoxia, immune injury, stress and nutrient deficiency [4], which is considered as a defense mechanism of cells against adverse environmental stimuli [5]. Studies have shown that autophagy is involved in the pathologic process of various diseases, such as tumor [6], neurodegenerative disease [7], cardiovascular disease [8], infection and immune deficiency [9]. In recent years, many researches have studied the correlation between autophagy and renal cancer. However, the role of autophagy in the pathogenesis of renal cancer and the exact mechanism of its action are not clear.
Renal cell carcinoma (RCC) is the most common malignancy in renal neoplasia and clear cell renal cell carcinoma (ccRCC) is the most common subtype [10, 11]. Progress has been achieved with regard to the pathogenesis and therapy of RCC; however, its incidence continues to rise. Many suffered patients will experience metastasis or local recurrence. It has been reported that autophagy is a potential cell survival mechanism in metastatic RCC cells and autophagy inhibition could create synergistic cytotoxicity when combined with mTOR inhibitors in ccRCC [12, 13]. Autophagic gene polymorphisms are associated with progression-free survival (PFS) of ccRCC patients treated with pazopanib [14]. This led us to speculate that the regulation and function of autophagy is likely connected to maintenance of homeostasis of renal cancer cells, disease pathogenesis, and targeting therapy resistance. But the role of autophagy in renal cancer remains elusive. In this review, we have fully identified the studies in PubMed/MEDLINE and the Web of Science which were focusing on the effect and mechanism of autophagy on renal cancer, especially on the renal cell carcinoma. We have highlighted the autophagy related signaling pathways and autophagy related drugs in renal cancer in this review. Exploring the delicate mechanisms and regulation of autophagy in renal cancer may lead to optimization in therapeutic strategies.
2. Autophagy and its regulatory mechanisms
Autophagy has been observed by researchers for more than 40 years and is considered to be a non-specific process of degradation of large intracellular materials [15]. Subsequent studies have found a close relationship between autophagy and cellular stress response. It is now widely believed that autophagy plays a key role in many aspects, including cell quality control, tissue homeostasis and energy supply [16]. In recent years, a variety of key molecules have been found to be involved in the formation of autophagosomes. This process is highly evolutionary conservative in yeast and humans. A series of autophagy related genes (ATGs) in yeast were found and their mammalian homologues were also found [17].
At present, it is found that the following four functional units are involved in the regulation of autophagy process: a. The ATG1/unc-51-like kinase (ULK) complex contains ATG13 and FIP200 [18]; b. Vps34, III phosphatidyl inositol 3 kinase (PI3K) and ATG6/Beclin1 compounds [18]; c. Two ubiquitin-like proteins, microtubule-associated proteins 1 light chain 3 (LC3) and ATG12. LC3 is embedded in the inner and outer membranes of autophagosomes [19]. ATG12 and ATG5 are conjugated and interact with ATG16L, and participate in the lipidation of LC3 [20]; d. Transmembrane protein ATG9 and VMP1. The specific role of ATG9 in autophagy is unclear. The interaction between VMP1 and Beclin1 is necessary for autophagy. Overexpression of VMP1 can induce autophagy [21].
The canonical autophagy processes involves following three steps. a. Initiation of the isolation membrane. Under starvation, ATG1/ULK1 is localized on the initial membrane and forms a complex with ATG13 and FIP200 [22-24]. The ULK1 complex recruits the VPS34/Beclin1 complex and increases VPS34 activity, thereby promoting PI3P production [25]. PI3P aggregates on the endoplasmic reticulum membrane and promotes growth of autophagosome membranes [26]. b. Elongation and closure of the autophagosome membrane. The protein LC3 is cleaved by ATG4 protease to generate cytosolic LC3 (LC3-I) [27, 28]. LC3-I can bind to phosphatidylethanolamine to form LC3-II (LC3-PE) on the membrane of the autophagosome [28], which is regulated by the conjugation system ATG5-ATG12 [29] and the modification of ATG5 by ATG12 is essential for the elongation of the isolation membrane [30]. The abnormal intracellular proteins, excess or damaged organelles are surrounded by the initial membrane that will form a autophagosome with double layer membranes. c. Autolysosome formation. The autophagosome containing cytoplasmic components moves to the lysosome [31]. The outer autophagosome membrane fuses with the lysosomal membrane to form an autolysosome, resulting in transporting its cargo into the lysosomal cavity. This fusion process is mediated by the small GTPase Rab7 and SNARE [32, 33]. The newly formed autolysosomes eventually degrade the autophagosome-delivered contents and its inner membrane by lysosome's hydrolases [34].
Under normal circumstances, the autophagy process in cells is at a low level, but it is necessary to maintain basic cellular activities, such as protein and organelle quality control. Certain stress states induce autophagy. Deficiency of nutrients is a typical activation factor of autophagy, which is mainly triggered by the mammalian target of rapamycin (mTOR), especially the signal pathway of mTOR complex 1 (mTORC1). When nutrients are sufficient, mTOR binds and phosphorylates the ULK1 complex, reducing its kinase activity, thereby inhibiting the autophagy initiation process [35]. Conversely, when nutrients are scarce, ULK1 is activated, promoting the initiation of autophagy. ULK1 can also be activated by AMP-activated protein kinase (AMPK) in the low-energy state (up-regulation of AMP/ATP) with the inhibition of mTORC1 and promotion of autophagy [36]. Initiation of autophagy is also activated by phosphatidylinositol 3-phosphate (PI3P), a product of the action of type III PI3K and Vps34. Downstream proteins of the Vps34 and ULK1 complexes, two pairs of conjugated complexes, ATG5-ATG12 and LC3-PE (LC3II), are involved in the autophagosomal membrane extension process. Other autophagy activation factors include anti-tumor therapy, reactive oxygen species (ROS), endoplasmic reticulum stress, and unfolded protein response (UPR) [21]. The detailed autophagy processes are depicted in Fig. 1.
3. Autophagy in tumors
The researchers found that the loss of the autophagy regulator Beclin1 (BECN1) was found in breast, ovarian, and prostate cancer cells, suggesting that autophagy has an inhibitory effect on tumor formation [37]. Liang et al [38] found that after the BECN1 protein was re-expressed in breast cancer cell lines, autophagy recovered and tumorigenesis was inhibited. Loss of other autophagy-related regulators also tends to promote tumorigenesis: ATG4C-/- mice exhibit high sensitivity to fibrosarcoma induced by chemical carcinogens [39]; The UVRAG-binding protein BIF1 is a positive regulator of autophagy that interacts with BECN1 and its complete deletion results in spontaneous tumorigenesis in mice [40]; ATG5-/- immortalized neonatal mouse kidney cells (iBMK) and BECN1+/- immortalized mouse mammary epithelial cells (iMMECs), which are deficient in autophagy, are more likely to form tumors in nude mice than autophagy-complete cells [41]; Systemic mosaic deletion of ATG5 and liver-specific deletion of ATG7 can lead to hepatic benign adenoma in autophagy-defective hepatocytes [42].
Studies on the genetic phenotype of autophagy in mice suggest that intact functional autophagy is essential for the maintenance of cell survival and cell homeostasis. Degenhardt et al [43] found that autophagic defects impair the viability of apoptosis-deficient mouse cells in the absence of growth factors and in metabolic stress state. This result has a great correlation with tumors because tumors often exhibit high metabolic demands when activated by oncogenes. The fact that the cells in the hypoxic region of the tumor show higher autophagy level also supports the above assertion [42]. Some researchers believe that autophagy plays an important role in the survival of tumor cells in the treatment of tumor radiotherapy and chemotherapy, and inhibition of autophagy improves the sensitivity of tumors to treatment [44].
The process of autophagy in eukaryotic cells.
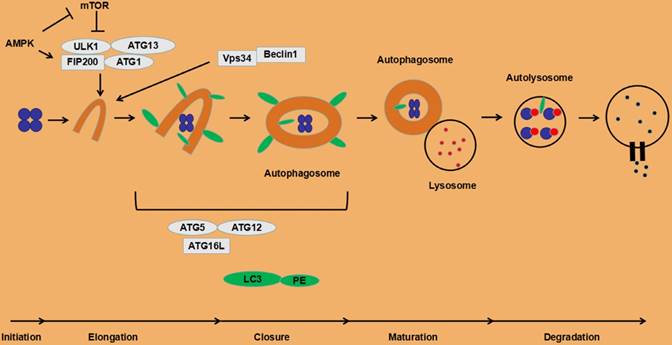
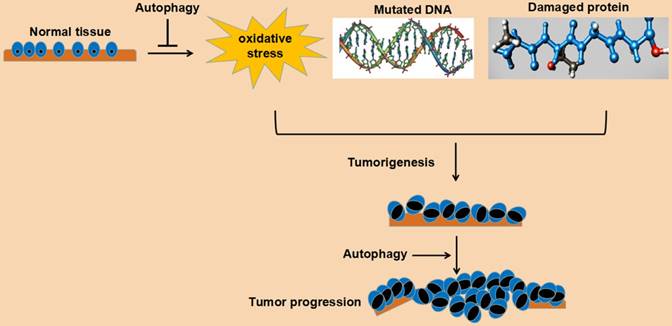
In a word, autophagy plays an important role in maintaining cell survival and homeostasis. Its role in tumorigenesis may be bidirectional: on the one hand, autophagy can reduce the pressure of oxidative stress, degrade mutated and damaged DNA and protein, and play a tumor suppressing role; on the other hand, autophagy can relieve various pressures, such as oxidative stress, damaged DNA and protein aggregation, and promote cell survival and play a cancer-promoting role. This bidirectional effect may be related to the following factors: a. tumor stage, such as the initial stage, advanced stage, metastatic stage, or gradual drug resistance stage; b. the tissue type of the tumor; c. genetic changes of the tumor. Understanding the role of autophagy in tumorigenesis undoubtedly benefits the establishment of a rational anti-tumor therapy program for autophagy. The detailed role of autophagy in cancer tumorigenesis and progression is depicted in Fig. 2.
4. Autophagy associated signaling pathways in renal cancer
4.1 Autophagy-related PI3K/AKT/mTOR pathway
Many factors (Table 1) are involved in the regulation of autophagy in renal cancer. The continuously activated PI3K/AKT/mTOR signaling axis is a typical survival mechanism for human tumor cells [45]. Many cases, such as tumor suppressor gene of phosphatase and tensin homolog (PTEN) and tuberous sclerosis complex (TSC) 1 and TSC2 deletions, type I PI3K mutations, AKT overexpression, sustained activation of tyrosine kinase growth factor receptors and so on, will lead to the abnormal activation of this signaling pathway and ultimately inhibit the autophagy process [46]. Activation of the PI3K/AKT/mTOR axis not only inhibits autophagy but also promotes protein translation and cell proliferation. Inhibiting the PI3K signaling axis will have an adverse effect on rapidly proliferating tumor cells, thereby inhibiting tumor growth. Sourbier et al [47] found that the increased phosphorylation of AKT at S473 and T308 increased the expression of AKT in 7 types of renal cancer cell lines (786-O, UOK-126, UOK-128, A498, ACHN, Caki-1, and Caki-2), whereas the expression of AKT was positively correlated with the expression of PI3K and inversely correlated with the expression of PTEN. To confirm whether the PI3K/AKT pathway is involved in renal tumor cell proliferation, the team treated the 786-O and Caki-1 cell lines with the specific PI3K inhibitor LY294002 and found that the number of cell deaths was significantly increased compared to the control group, and the difference was statistically significant (P<0.05). Seo et al [48] found that co-treatment with PP242 (inhibitor of mTORC1 and mTORC2) and curcumin induced the downregulation of the Rictor (an mTORC2 complex protein) and AKT protein levels, which led to lysosomal damage and induced autophagy in renal carcinoma cells. The authors believe that this results reveal that combined PP242 and curcumin treatment could induce autophagy-mediated cell death in renal cancer.
Signaling pathways of autophagy in renal cancer
| PI3K/AKT/mTOR pathway |
| tumor suppressor p53 |
| LC3B-dependent autophagy pathway |
| microtubule associated protein 1S (MAP1S) |
| transcription factor NF-E2-associated factor 2 (NRF2)/Kelch-like epichlorohydrin associated protein 1 (KEAP1) |
| Transient receptor potential melastatin 3 (TRPM3) |
| HOXA transcript at the distal tip (HOTTIP) |
4.2 Autophagy-associated p53 protein and renal cancer
The tumor suppressor p53 is an important checkpoint protein in mammalian cells [49]. It is activated under conditions of genetic stress such as DNA damage, hypoxia, and oncogene activation. In these cases, p53 can transactivate autophagy-inducing genes and inhibit mTOR through AMPK and TSC1/TSC2 dependent pathways to promote autophagy [50]. p53 can also act directly on the target of damage-regulated autophagy modulator (DRAM) to induce autophagy [51]. However, some studies have found that removal of p53 in the cytoplasm via gene or drug pathways can induce autophagy, indicating that extranuclear p53 is an effective inhibitor of autophagy [52]. However, it is still unclear in which environment p53 activates autophagy by which molecular pathway to inhibit tumor cell growth. In recent years, two large sample clinical studies have found the overexpression of p53 in renal cell carcinoma tissues (36%, n=97; 29.5%, n=297) [53, 54] suggesting that p53 is involved in the development of renal cell carcinoma (RCC). But a study found that RCC cells can survive and grow by inactivating p53 through TGase-2 mediated autophagy, which supplies recycled amino acids and bases under condition of starvation [55]. It is surprising that p53 levels are suppressed in RCC, although only 2.7% of RCC samples have p53 alterations in cBioPortal database. Warburton et al [56] showed that after UV irradiation of three renal cell carcinoma cell lines (ACHN, Caki-2, A498) to mediate DNA damage, the transcriptional activity of p53 was 1.4-fold, 2-fold, and 8-fold compared to control groups, respectively. The increase in transcriptional activity is positively correlated with the dose of UV, which suggests that p53 plays a role in repairing DNA damage and maintaining cell growth. The above studies provide some inspirations for us to improve the efficacy of certain drugs on renal tumors by inhibiting p53, and this effect may play a role in inhibiting the autophagic process induced by p53.
Schematic representation of function of autophagy in cancer cells.
4.3 LC3B-dependent autophagy pathway
LC3B is a yeast autophagy-related protein ATG8 homolog in mammalian cells [57]. Its C-terminal glycine is bound to phosphatidylethanolamine (PE) to form lipidated LC3 (LC3-II). LC3-II is embedded in the autophagosome membrane and participates in the elongation of the autophagosome membrane [58]. Mikhaylova et al [59] found that LC3B-dependent autophagy is essential for the growth of renal cell carcinoma. They injected LC3B shRNA lentiviral particles into subcutaneous renal cancer cell 786-O tumors in nude mice and found that the tumor volume was significantly smaller at 9 days than in the control group (P=0.0007). Then 786-O cells stably expressing LC3B shRNA were injected into the renal capsule of the nude mice. After 4 weeks, the mass of the tumor was also significantly lower than that of the control group (P<0.05). Similar results were also found in the other kidney cancer cell line, A498, indicating that LC3B-mediated autophagy is essential for the growth of renal cell carcinoma in nude mice. The team used quantitative immunoblotting to measure the expression level of LC3B in human clear cell renal cell carcinoma (ccRCC) tissue and normal kidney tissue and found that its expression level was positively correlated with tumor stage (P<0.05). But another group found that both mRNA and protein levels of LC3 were significantly decreased in ccRCC compared with paired adjacent tissue [60]. They also found that a low level of LC3-II was associated with poor prognosis in ccRCC, indicating that autophagy might be suppressed and associated with progression in ccRCC.
4.4 MAP1S activated autophagy pathway
MAP1S is a member of the cell microtubule-associated protein family 1, which interacts with LC3 and is a positive regulator of autophagy [61]. Loss of MAP1S leads to autophagy defects, which can cause mitochondrial dysfunction and affect cell growth. At the same time, MAP1S was found to be an important survival-related gene in cancer patients [62]. Hepatocellular carcinoma in MAP1S-deficient mice has a greater tendency to metastasize [63]. Low expression of MAP1S in human prostate cancer will reduce the average survival time of patients [64]. Based on this, we believe that MAP1S-mediated autophagy may be associated with tumor metastasis and patient prognosis. ccRCC is the most common type of human renal cell carcinoma. Xu et al [65] found that the expression level of MAP1S in the four ccRCC cell lines (786-O, RCC4, A498, Caki-1) was significantly lower than that of the human normal renal cell line (HK-2), whereas the expression level of MAP1S in the tumor specimens of 76 ccRCC patients was also significantly lower than that in the normal tissue adjacent to the cancer. The cumulative survival time of patients with high expression of MAP1S was significantly higher than that of patients with low expression (P<0.01) by plotting Kaplan-Meier curves of ccRCC patients. The above studies indicate that MAP1S-mediated autophagy was associated with the development and prognosis of ccRCC. High levels of MAP1S activate autophagy, reduce cellular genome instability, attenuate the invasiveness of ccRCC, and increase patients' survival time.
4.5 Autophagy related KEAP1/NRF2 pathway
The transcription factor NF-E2-associated factor 2 (NRF2) activates the transcription of many antioxidant target genes, and Kelch-like epichlorohydrin-associated protein 1 (KEAP1) is its inhibitor. KEAP1 can "lock" NRF2 in the cytoplasm and promote its degradation [66]. This pathway plays an important role in both acute and chronic renal injury as well as in renal tumors [67, 68]. Studies have shown that the succinylation of KEAP1 was increased with decreased NRF2 degradation, which activated HMOX1 and other stress response genes to promote tumor cell survival in the fumarate hydratase (FH) deficient type II papillary renal cell carcinoma [69]. Fabrizio et al [70] found that the expression level of KEAP1 gene was decreased after promoter methylation, which increased the expression of NRF2 and played an important role in ccRCC. P62, a substrate protein of autophagy, is a key agonist of NRF2 [71]. In autophagy-defective cells, P62 degradation was decreased. When P62 competitively bound KEAP1, NRF2 was released and entered into the nucleus to activate transcription of downstream target genes and promote tumor cell survival [72].
4.6 TRPM3-dependent autophagy pathway
Transient receptor potential melastatin 3 (TRPM3) is specifically present in ccRCC cells and regulates calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) and autophagy by modulating calcium flux [73]. This pathway is associated with known autophagy regulatory networks such as AMP-activated protein kinase (AMPK) and unc-51 like autophagy activating kinase 1 (ULK1). In addition, TRPM3 regulates autophagy through the action of zinc ion flux and miR-204 on autophagosome ligand LC3.
4.7 HOTTIP-dependent autophagy pathway
HOXA transcript at the distal tip (HOTTIP) is a long non-coding RNA that is upregulated in several human cancers [74]. A group found that HOTTIP expression was elevated in the RCC tissues and cell lines, and it was closely associated with prognosis of patients. HOTTIP can induce autophagy and affect proliferation, migration and invasion of RCC cell via autophagy dependent manner. And they further found that HOTTIP regulate autophagy through the PI3K/Akt/Atg13 signaling pathway [75].
5. Application of autophagy-related drugs in renal carcinoma
5.1 Autophagy-promoting drugs in renal cancer
Several pharmaceutical agents targeting autophagy in renal cancer have been described (Table 2). Sunitinib can effectively prolong the tumor-specific survival and overall survival of patients with advanced renal cell carcinoma [76]. Sunitinib is an oral oxindol, multitargeted tyrosine kinase inhibitor, which selectively inhibits vascular endothelial growth factor receptor 1 (VEGFR1), VEGFR2, VEGFR3, platelet-derived growth factor receptor (PDGFRα), PDGFRβ, stem-cell growth factor receptor, fms-related tyrosine kinase 3 (FLT3), RET and CSF1 receptor (CSF1R). Sunitinib can inhibit AKT/mTOR signaling pathway and cause autophagy of renal cancer cells, and its induced autophagy is associated with apoptosis [77].
Pharmaceutical agents targeting autophagy in renal cancer
| Drugs that promote autophagy |
| Sunitinib |
| AZD-2014 |
| Rasfonin |
| Silybin |
| Sinomenine |
| Drugs that inhibit autophagy |
| Chloroquine (CQ) and hydroxychloroquine (HCQ) |
| ROC-325 |
| 3-Methyladenine (3-MA) |
The dual mTORC1/2 inhibitor AZD-2014 inhibits the survival and growth of renal cancer cells more significantly than rapamycin and everolimus. AZD-2014 disrupts the accumulation and activation of mTORC1/2 by down-regulating the expression of HIF-1α/2α and cyclinD1 in renal cancer cells, leading to autophagy-dependent apoptosis of RCC [78].
Rasfonin is a alpha-pyrone metabolite that is isolated from fungi and has anti-cancer effects. Rasfonin-induced autophagy is associated with upregulation of AKT phosphorylation. Inhibition of AKT by small molecule inhibitors or genetic modifications can reduce rasfonin-dependent autophagic flux and PARP-1 cleavage. AKT promotes rasfonin-enhanced autophagy and caspase-dependent apoptosis by affecting the glycolytic pathway [79].
Silybin is a flavonoid prophylactic anticancer drug that has anti-metastasis effects in the treatment of renal cancer. Silybin can increase the expression of LC3-II in RCC cells, induce intracellular autophagic flow, and increase the formation of intracellular autophagic vacuoles. It is also possible to induce autophagy by AMPK/mTOR pathway and to inhibit the migration and invasion of RCC cells by activating autophagy [80]. In the same time, autophagy has a positive role in silybin induced anti-metastatic effects. The activation of autophagy enhances the inhibition of migration and invasion of RCC cells induced by silybin, and its inhibitory effect is reduced when autophagy is inhibited [81].
Sinomenine is extracted from Chinese medicinal plant Sinomenium acutum and can suppress several cancer cell growth [82-85]. Sinomenine significantly regulated the level of autophagy-related proteins such as p62 protein and Beclin1. Furthermore, sinomenine enhanced autophagy through PI3K/AKT/mTOR pathway in RCC [86].
5.2 Drugs that inhibit autophagy in renal cancer
Everolimus is a PI3K family protein kinase inhibitor for second-line treatment of RCC after sunitinib treatment failure. And it can inhibits mTOR, blocking a critical downstream effector of growth factor signaling. Although everolimus is safe and well tolerated, emerging drug resistance has been found [87]. Since inhibition of mTOR could induce autophagy, activation of autophagy may be a key mechanism for everolimus resistance. The in vitro studies demonstrated that everolimus and chloroquine synergistically inhibit the activity of RCC cells. The use of chloroquine and everolimus can effectively inhibit the autophagic flux and promote apoptosis, suggesting that combined use of targeted therapeutics can improve the therapeutic effect of renal cancer [88]. And an phase I/II trial of everolimus further validated that combining mTOR and autophagy inhibition could have >40% 6 month progression free survival (PFS) rate [13].
CQ/HCQ disrupts the degradation of autophagic proteins and prevents the conversion of LC3B-I to LC3B-II and inhibits the formation of autophagosomes. A series of phase I and phase I/II trials examined the safety and initial efficacy in an HCQ-based anticancer therapy [90-95]. Paflomeromycin A1 prevents the maturation of autophagosomes by inhibiting the fusion of autophagosomes and lysosomes. These inhibitors against different stages of autophagy can enhance the anti-renal cancer activity of sorafenib, suggesting that sorafenib-activated autophagy is a cancer-promoting factor that causes chemotherapy resistance. The use of chloroquine or hydroxychloroquine in combination with autophagy inhibitors has been used for the treatment of RCC [96].
ROC-325 is an orally available novel inhibitor of autophagic degradation. Based on the RCC cell model, a research group found that ROC-325 treatment could lead to accumulation of autophagosomes and inhibit autophagic flux. The in vivo study revealed that ROC-325 treatment could significantly and dose-dependently reduce the RCC xenografts growth and the inhibitory effect was better than HCQ [97].
Paclitaxel is a mitotic inhibitor and inducer of apoptosis, and its killing effect on FLCN-defective renal cancer cells is dependent on enhancing cell autophagy. Inhibition of autophagy with 3-Methyladenine (3-MA) can increase paclitaxel-induced apoptosis of FLCN-defective renal cancer cells, suggesting that paclitaxel combined with autophagy inhibitors can improve the efficacy of chemotherapy for FLCN-defective renal tumors [98].
6. Perspectives
The incidence of renal cancer is increasing year by year. Although surgical treatment is preferred, its long-term recurrence and metastasis rate is still high, and it is not sensitive to radiotherapy and chemotherapy. Therefore, postoperative adjuvant therapy with targeted drugs is necessary and can prevent the recurrence and metastasis of tumors, improve the postoperative survival rate and improve the quality of life. Autophagy is a hot issue in medical research in recent years. Its related studies have found a variety of new tumor pathogenesis mechanisms and promote the development of diagnosis and treatment of renal cancer. However, the theory of the intersection of renal cancer and autophagy needs more in-depth research and improvement.
Acknowledgements
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QC designed the study. QC and PB carried out data acquisition and performed the research. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mizushima N. Autophagy: Process and function[J]. Genes Dev. 2007;21(22):2861-2873
2. Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy[J]. NAT CELL BIOL. 2010;12(9):814-822
3. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation[J]. SCIENCE. 2000;290(5497):1717-1721
4. Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells[J]. CELL STRUCT FUNCT. 2002;27(6):421-429
5. Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: Cell survival in the land of plenty[J]. Nat Rev Mol Cell Biol. 2005;6(6):439-448
6. Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy[J]. CANCER RES. 2010;70(9):3431-3434
7. Schaeffer V, Lavenir I, Ozcelik S. et al. Stimulation of autophagy reduces neurodegeneration in a mouse model of human tauopathy[J]. BRAIN. 2012;135(Pt 7):2169-2177
8. Liao X, Sluimer JC, Wang Y. et al. Macrophage autophagy plays a protective role in advanced atherosclerosis[J]. CELL METAB. 2012;15(4):545-553
9. Kim JJ, Lee HM, Shin DM. et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action[J]. CELL HOST MICROBE. 2012;11(5):457-468
10. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018[J]. CA Cancer J Clin. 2018;68(1):7-30
11. Moch H, Cubilla AL, Humphrey PA. et al. The 2016 WHO classification of tumours of the urinary system and male genital Organs-Part a: Renal, penile, and testicular tumours[J]. EUR UROL. 2016;70(1):93-105
12. Singla M, Bhattacharyya S. Autophagy as a potential therapeutic target during epithelial to mesenchymal transition in renal cell carcinoma: An in vitro study[J]. BIOMED PHARMACOTHER. 2017;94:332-340
13. Haas NB, Appleman LJ, Stein M. et al. Autophagy inhibition to augment mTOR inhibition: A phase I/II trial of everolimus and hydroxychloroquine in patients with previously treated renal cell carcinoma[J]. CLIN CANCER RES. 2019
14. Santoni M, Piva F, De Giorgi U. et al. Autophagic gene polymorphisms in liquid biopsies and outcome of patients with metastatic clear cell renal cell carcinoma[J]. ANTICANCER RES. 2018;38(10):5773-5782
15. De Duve C, Wattiaux R. Functions of lysosomes[J]. ANNU REV PHYSIOL. 1966;28:435-492
16. Mizushima N, Levine B, Cuervo AM. et al. Autophagy fights disease through cellular self-digestion[J]. NATURE. 2008;451(7182):1069-1075
17. Chen N, Karantza-Wadsworth V. Role and regulation of autophagy in cancer[J]. Biochim Biophys Acta. 2009;1793(9):1516-1523
18. Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes[J]. J CELL BIOL. 2009;186(6):773-782
19. Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: Beyond the usual suspects' review series[J]. EMBO REP. 2008;9(9):859-864
20. Hanada T, Noda NN, Satomi Y. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy[J]. J BIOL CHEM. 2007;282(52):37298-37302
21. Kimmelman AC. The dynamic nature of autophagy in cancer[J]. Genes Dev. 2011;25(19):1999-2010
22. Ganley IG, Lam DH, Wang J. et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy[J]. J BIOL CHEM. 2009;284(18):12297-12305
23. Hara T, Takamura A, Kishi C. et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells[J]. J CELL BIOL. 2008;181(3):497-510
24. Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy[J]. AUTOPHAGY. 2009;5(5):649-662
25. Russell RC, Tian Y, Yuan H. et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase[J]. NAT CELL BIOL. 2013;15(7):741-750
26. Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate[J]. FEBS LETT. 2010;584(7):1302-1312
27. Kabeya Y, Mizushima N, Ueno T. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing[J]. EMBO J. 2000;19(21):5720-5728
28. Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes[J]. J BIOL CHEM. 2004;279(46):47704-47710
29. Otomo C, Metlagel Z, Takaesu G. et al. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy[J]. NAT STRUCT MOL BIOL. 2013;20(1):59-66
30. Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: Variations on a common theme of self-eating?[J]. Nat Rev Mol Cell Biol. 2011;13(1):7-12
31. Fass E, Shvets E, Degani I. et al. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes[J]. J BIOL CHEM. 2006;281(47):36303-36316
32. Gutierrez MG, Munafo DB, Beron W. et al. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells[J]. J CELL SCI. 2004;117(Pt 13):2687-2697
33. Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes[J]. CELL. 2012;151(6):1256-1269
34. Mizushima N, Ohsumi Y, Yoshimori T. Autophagosome formation in mammalian cells[J]. CELL STRUCT FUNCT. 2002;27(6):421-429
35. Jung CH, Ro SH, Cao J. et al. MTOR regulation of autophagy[J]. FEBS LETT. 2010;584(7):1287-1295
36. Gwinn DM, Shackelford DB, Egan DF. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint[J]. MOL CELL. 2008;30(2):214-226
37. Aita VM, Liang XH, Murty VV. et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21[J]. GENOMICS. 1999;59(1):59-65
38. Liang XH, Jackson S, Seaman M. et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1[J]. NATURE. 1999;402(6762):672-676
39. Marino G, Salvador-Montoliu N, Fueyo A. et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3[J]. J BIOL CHEM. 2007;282(25):18573-18583
40. Takahashi Y, Coppola D, Matsushita N. et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis[J]. NAT CELL BIOL. 2007;9(10):1142-1151
41. Karantza-Wadsworth V, Patel S, Kravchuk O. et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis[J]. Genes Dev. 2007;21(13):1621-1635
42. Takamura A, Komatsu M, Hara T. et al. Autophagy-deficient mice develop multiple liver tumors[J]. Genes Dev. 2011;25(8):795-800
43. Degenhardt K, Mathew R, Beaudoin B. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis[J]. CANCER CELL. 2006;10(1):51-64
44. Wang K, Liu R, Li J. et al. Quercetin induces protective autophagy in gastric cancer cells: Involvement of Akt-mTOR- and hypoxia-induced factor 1alpha-mediated signaling[J]. AUTOPHAGY. 2011;7(9):966-978
45. LoPiccolo J, Blumenthal GM, Bernstein WB. et al. Targeting the PI3K/Akt/mTOR pathway: Effective combinations and clinical considerations[J]. Drug Resist Updat. 2008;11(1-2):32-50
46. Arico S, Petiot A, Bauvy C. et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway[J]. J BIOL CHEM. 2001;276(38):35243-35246
47. Sourbier C, Lindner V, Lang H. et al. The phosphoinositide 3-kinase/Akt pathway: A new target in human renal cell carcinoma therapy[J]. CANCER RES. 2006;66(10):5130-5142
48. Seo SU, Woo SM, Lee HS. et al. MTORC1/2 inhibitor and curcumin induce apoptosis through lysosomal membrane permeabilization-mediated autophagy[J]. ONCOGENE. 2018;37(38):5205-5220
49. Levine AJ. P53, the cellular gatekeeper for growth and division[J]. CELL. 1997;88(3):323-331
50. Feng Z, Hu W, de Stanchina E. et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: Stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways[J]. CANCER RES. 2007;67(7):3043-3053
51. Crighton D, Wilkinson S, O'Prey J. et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis[J]. CELL. 2006;126(1):121-134
52. Tasdemir E, Maiuri MC, Galluzzi L. et al. Regulation of autophagy by cytoplasmic p53[J]. NAT CELL BIOL. 2008;10(6):676-687
53. Haitel A, Wiener HG, Baethge U. et al. Mdm2 expression as a prognostic indicator in clear cell renal cell carcinoma: Comparison with p53 overexpression and clinicopathological parameters[J]. CLIN CANCER RES. 2000;6(5):1840-1844
54. Zigeuner R, Ratschek M, Rehak P. et al. Value of p53 as a prognostic marker in histologic subtypes of renal cell carcinoma: A systematic analysis of primary and metastatic tumor tissue[J]. UROLOGY. 2004;63(4):651-655
55. Kang JH, Lee JS, Hong D. et al. Renal cell carcinoma escapes death by p53 depletion through transglutaminase 2-chaperoned autophagy[J]. CELL DEATH DIS. 2016;7:e2163
56. Warburton HE, Brady M, Vlatkovic N. et al. P53 regulation and function in renal cell carcinoma[J]. CANCER RES. 2005;65(15):6498-6503
57. Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy[J]. Int J Biochem Cell Biol. 2004;36(12):2503-2518
58. Weidberg H, Shvets E, Shpilka T. et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis[J]. EMBO J. 2010;29(11):1792-1802
59. Mikhaylova O, Stratton Y, Hall D. et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma[J]. CANCER CELL. 2012;21(4):532-546
60. Deng Q, Wang Z, Wang L. et al. Lower mRNA and protein expression levels of LC3 and Beclin1, markers of autophagy, were correlated with progression of renal clear cell carcinoma[J]. JPN J CLIN ONCOL. 2013;43(12):1261-1268
61. Xie R, Nguyen S, McKeehan K. et al. Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation[J]. J BIOL CHEM. 2011;286(12):10367-10377
62. Vandin F, Clay P, Upfal E. et al. Discovery of mutated subnetworks associated with clinical data in cancer[J]. Pac Symp Biocomput. 2012:55-66
63. Xie R, Wang F, McKeehan WL. et al. Autophagy enhanced by microtubule- and mitochondrion-associated MAP1S suppresses genome instability and hepatocarcinogenesis[J]. CANCER RES. 2011;71(24):7537-7546
64. Jiang X, Zhong W, Huang H. et al. Autophagy defects suggested by low levels of autophagy activator MAP1S and high levels of autophagy inhibitor LRPPRC predict poor prognosis of prostate cancer patients[J]. Mol Carcinog. 2015;54(10):1194-1204
65. Xu G, Jiang Y, Xiao Y. et al. Fast clearance of lipid droplets through MAP1S-activated autophagy suppresses clear cell renal cell carcinomas and promotes patient survival[J]. Oncotarget. 2016;7(5):6255-6265
66. Tong KI, Padmanabhan B, Kobayashi A. et al. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response[J]. MOL CELL BIOL. 2007;27(21):7511-7521
67. Lee DF, Kuo HP, Liu M. et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta[J]. MOL CELL. 2009;36(1):131-140
68. Sporn MB, Liby KT. NRF2 and cancer: The good, the bad and the importance of context[J]. NAT REV CANCER. 2012;12(8):564-571
69. Kinch L, Grishin NV, Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2[J]. CANCER CELL. 2011;20(4):418-420
70. Fabrizio FP, Costantini M, Copetti M. et al. Keap1/Nrf2 pathway in kidney cancer: Frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma[J]. Oncotarget. 2017;8(7):11187-11198
71. Lau A, Wang XJ, Zhao F. et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62[J]. MOL CELL BIOL. 2010;30(13):3275-3285
72. Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: An insight into cullin-ring ubiquitin ligases[J]. Antioxid Redox Signal. 2010;13(11):1699-1712
73. Hall DP, Cost NG, Hegde S. et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma[J]. CANCER CELL. 2014;26(5):738-753
74. Lian Y, Cai Z, Gong H. et al. HOTTIP: A critical oncogenic long non-coding RNA in human cancers[J]. MOL BIOSYST. 2016;12(11):3247-3253
75. Su Y, Lu J, Chen X. et al. Long non-coding RNA HOTTIP affects renal cell carcinoma progression by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway[J]. J Cancer Res Clin Oncol. 2018
76. Faivre S, Demetri G, Sargent W. et al. Molecular basis for sunitinib efficacy and future clinical development[J]. NAT REV DRUG DISCOV. 2007;6(9):734-745
77. Cao P, Jiang XJ, Xi ZJ. [Sunitinib induces autophagy via suppressing Akt/mTOR pathway in renal cell carcinoma][J]. Beijing Da Xue Xue Bao Yi Xue Ban. 2016;48(1):584-589
78. Zheng B, Mao JH, Qian L. et al. Pre-clinical evaluation of AZD-2014, a novel mTORC1/2 dual inhibitor, against renal cell carcinoma[J]. CANCER LETT. 2015;357(2):468-475
79. Lu Q, Yan S, Sun H. et al. Akt inhibition attenuates rasfonin-induced autophagy and apoptosis through the glycolytic pathway in renal cancer cells[J]. CELL DEATH DIS. 2015;6:e2005
80. Zeng J, Liu W, Li F. et al. Mp92-15 suppression of chaperone-mediated autophagy: A novel mechanism of action of silibinin against bladder and renal cancer[J]. J UROLOGY. 2016;195(4):e1167
81. Li F, Ma Z, Guan Z. et al. Autophagy induction by silibinin positively contributes to its anti-metastatic capacity via AMPK/mTOR pathway in renal cell carcinoma[J]. INT J MOL SCI. 2015;16(4):8415-8429
82. Li X, Li P, Liu C. et al. Sinomenine hydrochloride inhibits breast cancer metastasis by attenuating inflammation-related epithelial-mesenchymal transition and cancer stemness[J]. Oncotarget. 2017;8(8):13560-13574
83. Jiang S, Gao Y, Hou W. et al. Sinomenine inhibits A549 human lung cancer cell invasion by mediating the STAT3 signaling pathway[J]. ONCOL LETT. 2016;12(2):1380-1386
84. Xie T, Ren HY, Lin HQ. et al. Sinomenine prevents metastasis of human osteosarcoma cells via S phase arrest and suppression of tumor-related neovascularization and osteolysis through the CXCR4-STAT3 pathway[J]. INT J ONCOL. 2016;48(5):2098-2112
85. Yang H, Yin P, Shi Z. et al. Sinomenine, a COX-2 inhibitor, induces cell cycle arrest and inhibits growth of human colon carcinoma cells in vitro and in vivo[J]. ONCOL LETT. 2016;11(1):411-418
86. Deng F, Ma YX, Liang L. et al. The pro-apoptosis effect of sinomenine in renal carcinoma via inducing autophagy through inactivating PI3K/AKT/mTOR pathway[J]. BIOMED PHARMACOTHER. 2018;97:1269-1274
87. Hagiwara N, Watanabe M, Iizuka-Ohashi M. et al. Mevalonate pathway blockage enhances the efficacy of mTOR inhibitors with the activation of retinoblastoma protein in renal cell carcinoma[J]. CANCER LETT. 2018;431:182-189
88. Grimaldi A, Santini D, Zappavigna S. et al. Antagonistic effects of chloroquine on autophagy occurrence potentiate the anticancer effects of everolimus on renal cancer cells[J]. CANCER BIOL THER. 2015;16(4):567-579
89. Carew JS, Kelly KR, Nawrocki ST. Autophagy as a target for cancer therapy: New developments[J]. CANCER MANAG RES. 2012;4:357-365
90. Rangwala R, Chang YC, Hu J. et al. Combined MTOR and autophagy inhibition: Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma[J]. AUTOPHAGY. 2014;10(8):1391-1402
91. Vogl DT, Stadtmauer EA, Tan KS. et al. Combined autophagy and proteasome inhibition: A phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma[J]. AUTOPHAGY. 2014;10(8):1380-1390
92. Rangwala R, Leone R, Chang YC. et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma[J]. AUTOPHAGY. 2014;10(8):1369-1379
93. Mahalingam D, Mita M, Sarantopoulos J. et al. Combined autophagy and HDAC inhibition: A phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors[J]. AUTOPHAGY. 2014;10(8):1403-1414
94. Rosenfeld MR, Ye X, Supko JG. et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme[J]. AUTOPHAGY. 2014;10(8):1359-1368
95. Barnard RA, Wittenburg LA, Amaravadi RK. et al. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma[J]. AUTOPHAGY. 2014;10(8):1415-1425
96. Lotze MT, Maranchie J, Appleman L. Inhibiting autophagy: A novel approach for the treatment of renal cell carcinoma[J]. CANCER J. 2013;19(4):341-347
97. Carew JS, Espitia CM, Zhao W. et al. Disruption of autophagic degradation with ROC-325 antagonizes renal cell carcinoma pathogenesis[J]. CLIN CANCER RES. 2017;23(11):2869-2879
98. Zhang Q, Si S, Schoen S. et al. Suppression of autophagy enhances preferential toxicity of paclitaxel to folliculin-deficient renal cancer cells[J]. J Exp Clin Cancer Res. 2013;32:99
Author contact
![]() Corresponding author: Qi Cao, Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (HUST), 1277 Jiefang Avenue, Wuhan, Hubei Province, China, 430022; E-mail: curkeycom.
Corresponding author: Qi Cao, Department of Urology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (HUST), 1277 Jiefang Avenue, Wuhan, Hubei Province, China, 430022; E-mail: curkeycom.

 Global reach, higher impact
Global reach, higher impact