Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(11):2386-2396. doi:10.7150/jca.31088 This issue Cite
Research Paper
Prognostic Value of Long Noncoding RNA CRNDE as a Novel Biomarker in Solid Cancers: An Updated Systematic Review and Meta-Analysis
1. Organ Transplant Center, Hospital of University of Electronic Science and Technology of China and Sichuan Provincial People's Hospital, Chengdu 610072, China;
2. Sichuan Provincial Key Laboratory for Human Disease Gene Study, Institute of Laboratory Medicine, Hospital of University of Electronic Science and Technology of China and Sichuan Provincial People's Hospital, Chengdu 610072, China;
3. Renal Division, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
Received 2018-10-30; Accepted 2019-4-13; Published 2019-5-26
Abstract
Background: Long noncoding RNA colorectal neoplasia differentially expressed (CRNDE) has been reported to exhibit a potential oncogenic role in the development of human cancers. However, the clinical value of CRNDE expression in various cancers still remains unclear. Herein, we conducted a meta-analysis to investigate the association between CRNDE and clinical outcomes in solid cancers.
Methods: A systematic search was performed though the PubMed, EMBASE, Web of Science, Ovid, Cochrane library, CNKI and WanFang databases for eligible studies on clinical values of CRNDE in solid cancers. The pooled hazard ratios (HRs) or odd ratios (ORs) with 95% confidence intervals (CIs) were used to evaluate the link between CRNDE and clinical outcomes.
Results: A total of 3690 patients from 20 studies (including 2 studies have 2 cohorts, respectively) were included. The results suggested that elevated CRNDE expression predicted a poor overall survival (OS) for in 13 types of solid cancers (HR=1.46, 95% CI: 1.33-1.58, P<0.001) with no heterogeneity (I2=21.8%, P=0.19). Subgroup analysis indicated a significant association between high CRNDE expression and shorter OS in the studies with digestive system cancers (HR=1.42, 95% CI: 1.28-1.55, P<0.001), qRT-PCR method (HR=1.45, 95% CI: 1.30-1.59, P<0.001), sample size >100 (HR=1.44, 95% CI: 1.32-1.57, P<0.001), and NOS>7 (HR= 1.50, 95% CI: 1.23-1.78, P<0.001). Furthermore, the pooled results showed that CRNDE was an independent prognostic factor for OS in cancer patients (HR=1.37, 95% CI: 1.22-1.52, P<0.001). In addition, we also revealed that CRNDE was positively related to tumor size (OR=2.10, 95%CI: 1.68-2.63, P<0.001), TNM stage (OR=2.86, 95%CI: 2.29-3.56, P<0.001), lymph node metastasis (LNM) (OR=3.21, 95%CI: 2.01-5.13, P<0.001), and distant metastasis (OR=4.36, 95%CI: 2.36-8.07, P<0.001). Although the probable evidences of publication bias were found in the studies with OS, tumor size, TNM stage or LNM, the trim and fill analysis confirmed the reliability of these results was not affected.
Conclusion: Elevated CRNDE expression was associated with larger tumor size, advanced TNM stage, worse LNM and distant metastasis, and shorter OS, suggesting that CRNDE may act as an independent prognostic biomarker in solid cancers.
Keywords: long noncoding RNA, CRNDE, cancer, prognosis, meta-analysis
Introduction
Cancer has already become one of the major public health issues worldwide. Recently, cancer becomes the second leading cause of death in the United States [1]. Unfortunately, with increasing incidence and mortality in China, cancer has been the leading cause of death since 2010 [2]. Cancer is a complex human disease associated with a variety of chromosomal translocations, deletions and amplification, epigenetic alterations and genetic mutations. Despite recent advances in the diagnosis and treatment of human cancer, clinical prognosis still remains poor. However, one of the main reasons is the lack of effective biomarker to early diagnose cancer and predict clinical prognosis of cancer patients.
Long noncoding RNAs (lncRNAs), which are longer than 200 nucleotides in length, are an emerging class of evolutionarily conserved non-protein coding RNAs [3]. Although most lncRNAs cannot translate into any functional proteins, they are involved in most aspects of cellular and physiologic processes through acting as transcriptional modulators, posttranscriptional processors, enhancers, splicing regulators, chromatin remodelers, molecular decoys for miRNAs, or as guides for protein-DNA, protein-RNA and protein-protein interactions [4-8]. Notably, increasing evidences suggest that lncRNAs are aberrantly expressed in a broad spectrum of human cancers, and may play crucial roles in promoting and maintaining cancer initiation and progression, demonstrating their clinical potential as biomarkers and therapeutic targets [9-12]. Among these cancer-related lncRNAs, a new discovered lncRNA termed as 'colorectal neoplasia differentially expressed' (CRNDE) has drawn increasing attentions [13, 14].
CRNDE was originally discovered to be upregulated in colorectal cancer (CRC), and located on the long arm of chromosome 16 on the strand opposite to the adjacent IRX5 gene [15, 16]. CRNDE contains a total of six exons, five of which are core exons (E1, E2, E4, E5 and E6) and one of which is less mentioned exon (E3). Till now, 14 splice variants of CRNDE have been identified and named as CRNDE 201-214 [15, 16]. Recent studies demonstrated that CRNDE is not only overexpressed in CRC tissues [17-19], but also abnormally expressed in different types of cancers, including hepatocellular carcinoma (HCC) [20, 21], non-small cell lung cancer (NSCLC) [22, 23], glioma [24, 25], renal cell carcinoma (RCC) [26], breast cancer [27, 28], gastric cancer (GC) [29], intrahepatic cholangiocarcinoma (IHCC) [30], pancreatic cancer (PC) [31], papillary thyroid cancer (PTC) [32], bladder cancer [33], ovarian cancer (OC) [34], and cervical cancer (CC) [35]. It has been demonstrated that CRNDE may play an important role in tumorigenesis. CRNDE could function as a crucial molecular switch for diverse cancer-related pathways including cell proliferation, differentiation, apoptosis, cytoskeletal dynamism, angiogenesis, invasion and metastasis [19, 36-40].
As a cancer-related lncRNA, elevated CRNDE expression is closely related to poor prognosis as well as aggressive clinicopathologic features for cancer patients. Increasing evidences demonstrated that CRNDE may server as a negative prognostic biomarker in various cancers, and high CRNDE expression markedly correlated with larger tumor size, advanced TNM stage, worse lymph node metastasis and distant metastasis [29]. However, most studies reported about the prognostic role of CRNDE expression are limited by sample size and discrete clinical outcomes. Therefore, we conducted a systematic review and quantitative meta-analysis to investigate the clinicopathological and prognostic value of CRNDE as a potential biomarker in human solid cancers.
Materials and Methods
Search strategy and study selection
We searched though the PubMed, EMBASE, Web of Science, Ovid, Cochrane library, CNKI and WanFang databases for potentially eligible studies on clinical values of CRNDE expression in human solid cancers from inception up to October 24, 2018. The search terms were included: “CRNDE”, “colorectal neoplasia differentially expressed”, “lncRNA”, “long noncoding RNA”, “cancer”, “carcinoma”, “neoplasm”, “prognosis” and “survival”. The reference lists of the retrieved articles were searched manually, and the literature search was performed by two independent researchers (Yu Zhou and Tian Xu).
The studies were considered eligible if they met the following criteria: any type of human cancer was studied; the studies investigated the prognostic value of CRNDE expression in cancers; the levels of CRNDE expression in cancerous tissues were detected; patients were grouped according to the levels of CRNDE expression; the studies included an association between CRNDE and clinicopathologic parameters; the studies provided sufficient data to estimate the HRs with corresponding 95% CI for OS; and the studies were published in English. The exclusion criteria were as follows: letters, editorials, expert opinions, case reports and reviews; the studies only investigated the molecular structure and functions of CRNDE; the studies did not include the usable data for further analysis; and duplicate publications.
Data extraction and quality assessment
Two investigators (Yu Zhou and Tian Xu) independently evaluated and extracted the data from each eligible study, and a consensus was achieved for disagreements by a third investigator (Shikai Zhu). According to the above inclusion and exclusion criteria, the following elements were extracted: first author, publication date, country, tumor type, TNM stage, sample size, cut-off value, follow-up period, detection method, adjuvant therapy before the surgery, survival analysis methodology, HRs with corresponding 95% CIs for OS, disease-free survival (DFS) and progression‑free survival (PFS), and other clinicopathologic parameters including age, gender, tumor size, tumor differentiation, TNM stage, lymph node metastasis and distant metastasis. HRs with corresponding 95% CIs were preferentially extracted from univariate or multivariate analyses. If these were not available, the HR estimates were calculated from Kaplan-Meier survival curves using Engauge Digitizer V4.1 software according to the method of Tierney et al [41].
The quality of included studies was assessed based on the Newcastle-Ottawa scale (NOS) criteria. The NOS criteria apply a “star” rating system ranges from 0 to 9 stars for the judgment of methodological quality, which was based on selection (0-4 stars), comparability (0-2 stars) and outcome (0-3 stars) [42]. Based on previous recommendations, studies with 5 points were considered to be of high quality. The two aforementioned investigators independently evaluated the quality of each study. Conflicting evaluations or inconsistent data from the included studies were resolved through discussion with a third investigator (Shikai Zhu).
The flow diagram of this meta-analysis.
Statistical analyses
All data analyses were performed with STATA statistical software version 14.0 (Stata Corporation, College Station, TX, USA). Pooled HRs with 95% CIs were used to estimate the prognostic value of CRNDE on OS, DFS and PFS in solid cancers. Pooled ORs with 95% CIs were used to estimate the association between CRNDE and clinicopathological characteristics such as age, gender, tumor size, tumor differentiation, TNM stage, lymph node metastasis and distant metastasis. X2-based Cochran Q test and Higgins I2 statistic were utilized to determine the heterogeneity among those studies. P-value <0.05 in combination with I2-value >50% was considered significant heterogeneity. Random-effects models were applied in cases with significant heterogeneity. Subgroup analysis and sensitivity analysis were performed to dissect the heterogeneity. In addition, publication bias was determined using Begg's funnel plots and Egger's linear regression tests. If there were the probable evidence of publication bias, the trim and fill analysis was performed. P-value <0.05 was considered statistically significant.
Results
Study Selection and Characteristics
A total of 325 potentially relevant articles were identified in this meta-analysis. To yield relevant articles, we evaluated the titles, abstract and author information of all collected articles, and 204 duplicate articles were excluded. After screening the titles and abstract carefully, 92 irrelevant articles were excluded such as letters, editorials, expert opinions, case reports, reviews and other type uninvolved publications for the analysis and full text review. For evaluated the eligibility of full-text articles, nine articles without the sufficient data or dividing into high and low expression groups were excluded. Finally, 20 eligible studies were included in this meta-analysis (Figure 1).
A total of 3690 patients from 20 studies (2 studies included 2 cohorts, respectively) between 2015 and 2018 were included (Table 1). A total of 17 studies were published in English and 3 in Chinese. The regions of those studies were from China (n=19), Poland (n=1) and USA (n=2). Among these studies, the sample size ranged between 54 and 512 patients, and 14 studies enrolled more than 100 patients. 14 types of solid cancers were recorded including CRC (n=4), glioma (n=3), RCC (n=2), breast cancer (n=2), GC (n=1), HCC (n=2), IHCC (n=1), PC (n=1), NSCLC (n=2), PTC (n=1), bladder cancer(n=1), OC (n=1) and CC (n=1). The levels of CRNDE expression were detected by quantitative real-time polymerase chain reaction (qRT-PCR) (n= 16), microarray (n=3) and RNA-seq (n=3). None of patients received adjuvant therapy before the surgery were conformed in 12 studies. Multivariate analyze was included in 10 studies. Clinical outcomes were recoded including 19 studies for OS, 2 for DFS, and 2 for PFS. HRs with the corresponding 95% CIs for OS were extracted from the original data in 10 studies, and calculated from Kaplan-Meier Curves in other 10 studies (Supplement Table 1). Clinicopathologic parameters were also recoded in 15 studies including age, gender, tumor size, tumor differentiation, TNM stages, lymph node metastasis (LNM) and distant metastasis (Supplement Table 2). Additionally, all studies were more than 6 according to the NOS score criteria and thus were considered high quality (Supplement Table 3).
Prognostic value of CRNDE expression in solid cancers
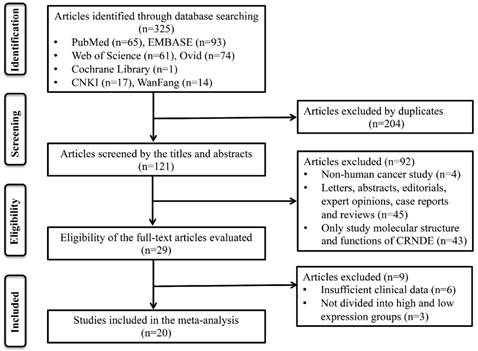
A total of 19 studies including 3496 patients were recruited to assess the effects of CRNDE expression on OS in solid cancers. The pooled results suggested that elevated CRNDE expression predicted a poor OS for in 13 types of solid cancers (HR=1.46, 95% CI: 1.33-1.58, P<0.001) with no heterogeneity (I2=21.8%, P=0.190) (Figure 2A). Furthermore, subgroup analysis was also conducted to investigate the association between HRs and these variables including cancer type, detection method, sample size and NOS. Stratified analysis indicated that there was a negatively relationship between CRNDE and OS in the studies with digestive system cancers (HR=1.42, 95% CI: 1.28-1.55, P<0.001), gynecologic cancers (HR=4.52, 95% CI: 1.49-7.56, P<0.001), and glioma (HR=1.83, 95% CI: 1.23-2.44, P<0.001) (Figure 3A). And we also found that upregulation of CRNDE expression significantly related to short OS in the studies using qRT-PCR method (HR=1.45, 95% CI: 1.30-1.59, P<0.001) as well as microarray/RNA-seq method (HR=1.49, 95% CI: 1.26-1.72, P<0.001) (Figure 3B). Higher CRNDE expression predicted shorter OS in the studies with sample size >100 (HR=1.44, 95% CI: 1.32-1.57, P<0.001) as well as those with sample size <100 (HR=1.96, 95% CI: 1.23-2.69, P<0.001) (Figure 3C). In addition, the effect of CRNDE overexpression on predicting poor OS was found in the studies with NOS<7 (HR=1.45, 95% CI: 1.31-1.58, P<0.001) as well as those with NOS>7 (HR= 1.50, 95% CI: 1.23-1.78, P<0.001) (Figure 3D).
The independent prognostic value of CRNDE expression in different cancers were also evaluated based on the multivariate analysis in 10 studies with 1766 patients (Figure 2B). The pooled HRs showed that CRNDE expression was an independent prognostic factor for OS in cancer patients (HR=1.37, 95% CI: 1.22-1.52, P<0.001). In addition, the prognostic value of CRNDE expression for DFS or PFS were also evaluated in 4 studies including 1355 patients (Figure 2C). The pooled data suggested that elevated CRNDE expression predicted a poor DFS for in hepatocellular carcinoma and ovarian cancer (HR=1.46, 95% CI: 1.06-1.87, P<0.001). However, there was no significant association between CRNDE and PFS, maybe due to small sample size.
The main characteristics of the included studies in this meta-analysis
| Author | Year | Country | Tumor Type | TNM Stage | Sample Size | Cut-off Value | Follow-up (months) | Detection Method | Adjuvant therapy | Survival Analysis | Outcome Measure | NOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Szafron LM | 2015 | Poland | OC | IIb-IV | 135 | Mean | 38(median) | qRT-PCR | None | M | OS | DFS | 9 |
| Han Y | 2015 | China | CC | I-IV | 87 | 2-ΔΔCt=1 | 60(total) | qRT-PCR | None | U/M | OS | 9 | |
| Liu T | 2016 | China | CRC | I-IV | 142 | Median | 92 (total) | qRT-PCR | None | U/M | OS | 8 | |
| Jing SY | 2016 | China | Glioma | I-IV | 164 | Median | 70 (total) | qRT-PCR | None | U/M | OS | 8 | |
| Du DX | 2017 | China | GC | I-IV | 118 | Median | 60 (total) | qRT-PCR | None | U/M | OS | 9 | |
| Dai MY | 2017 | China | HCC | I-IV | 373 | Median | 120 (total) | Microarray | NA | U | OS | 8 | |
| Wang G | 2017 | China | PC | I-IV | 58 | NA | 50 (total) | qRT-PCR | NA | U | OS | 8 | |
| Huan JL | 2017 | China | BrC | I-IV | 103 | Mean | 60 (total) | qRT-PCR | None | U | OS | 8 | |
| Jiang HJ | 2017 | China | CRC | I-IV | 251 | NA | 57(median) | qRT-PCR | NA | U/M | OS | 9 | |
| Luo YH | 2017 | China | PTC | I-IV | 502 | NA | 200 (total) | RNA-seq | NA | M | OS | 6 | |
| Kiang KM1 | 2017 | China | Glioma | I-IV | 68 | Median | 200 (total) | Microarray | NA | U | OS | 6 | |
| Kiang KM2 | 2017 | China | Glioma | I-IV | 101 | Median | 200 (total) | Microarray | NA | U | OS | 6 | |
| Han P | 2017 | China | CRC | NA | 64 | Median | 50 (total) | qRT-PCR | None | U | OS | 8 | |
| Ding J | 2017 | China | CRC | I-IV | 80 | NA | NA | qRT-PCR | None | NA | NA | 6 | |
| Li XH | 2017 | China | NSCLC | I-IV | 137 | Mean | 60 (total) | qRT-PCR | None | U/M | OS | PFS | 9 |
| Zhang M | 2017 | China | NSCLC | I-IIIa | 66 | NA | 60 (total) | qRT-PCR | NA | U | OS | 8 | |
| Xia XL | 2017 | China | IHCC | I-IV | 118 | Mean | 60 (total) | qRT-PCR | None | U/M | OS | PFS | 8 |
| Tang QL | 2018 | USA | HCC | NA | 369 | Median | 120 (total) | RNA-req | NA | U | OS | DFS | 6 |
| Cheng JL | 2018 | China | BlC | I-IV | 54 | N/T=1 | NA | qRT-PCR | NA | NA | NA | 6 | |
| Li ZB | 2018 | China | BrC | I-IV | 76 | 2-ΔΔCt=1 | NA | qRT-PCR | None | NA | NA | 6 | |
| Ding CG1 | 2018 | China | RCC | I-IV | 112 | Median | 96 (total) | qRT-PCR | None | U/M | OS | 9 | |
| Ding CG2 | 2018 | USA | RCC | NA | 512 | Median | 150(total) | RNA-req | NA | U | OS | 6 | |
Abbreviations: OC, ovarian cancer; CC, cervical cancer; CRC, colorectal cancer; HCC, hepatocellular carcinoma; GC, gastric cancer; PC, pancreatic cancer; BrC, breast cancer; PTC, papillary thyroid cancer; NSCLC, non-small cell lung cancer; IHCC, intrahepatic cholangiocarcinoma; BlC, bladder cancer; RCC, renal cell carcinoma; NA, not available; T, tumor; N, normal; qRT-PCR quantitative real-time PCR; OS, overall survival; DFS, disease-free survival; PFS, progression‑free survival.
Forest plots of the HRs for the association between CRNDE expression and OS (A), independent predictive factor for OS (B), and DFS/PFS (C).
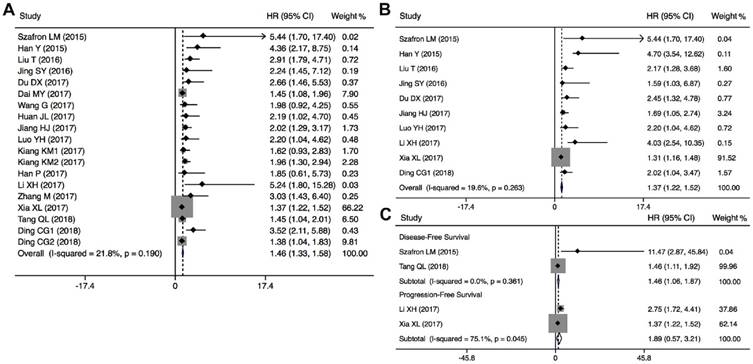
Forest plots of subgroup analysis for the HRs of OS by (A) cancer type, (B) detection method, sample size (C), and NOS (D).
Association between CRNDE expression and clinicopathological characteristics of solid cancers
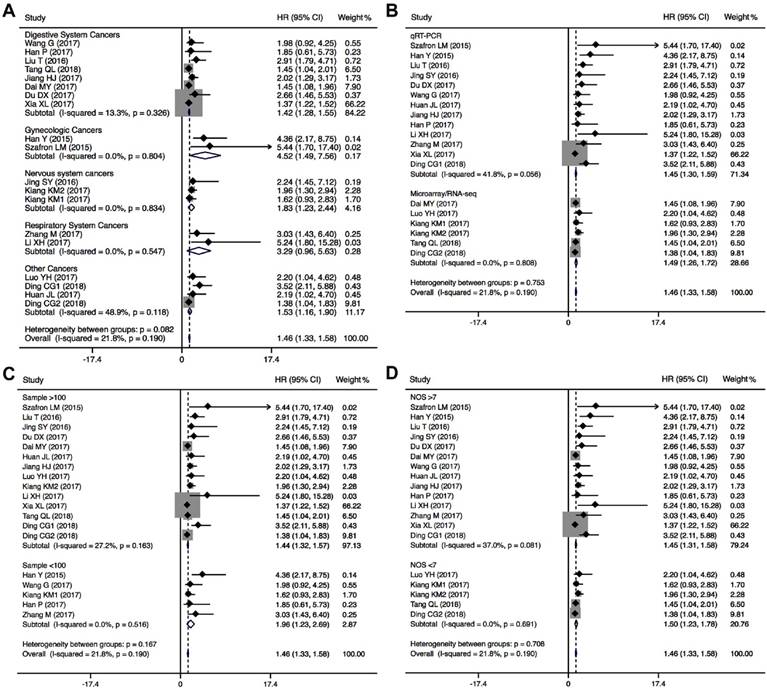
The association between CRNDE expression and clinicopathological characteristics are examined in 15 studies with 1861 cancer patients (Table 2). 12 studies with 1346 cancer patients were included to analysis the link between CRNDE and tumor size, and the pooled data found an obviously association between CRNDE and tumor size (OR=2.10, 95%CI: 1.68-2.63, P<0.001) (Figure 4A). According to the association between CRNDE and TNM stage, the analysis results of 13 studies with 1546 cancer patients showed that there was a significant relationship between CRNDE and TNM stage (OR=2.86, 95%CI: 2.29-3.56, P<0.001) (Figure 4B). As indicated in Figure 4C, 1141 cancer patients from 12 studies were included to assess the association between CRNDE and LNM, and the results demonstrated that the patients with high CRNDE expression were more susceptibility to develop LNM (OR=3.21, 95%CI: 2.01-5.13, P<0.001). In addition, 5 studies with 682 cancer patients were included to analysis the link between CRNDE and distant metastasis, and the results found an obviously association between CRNDE expression and distant metastasis (OR=4.36, 95%CI: 2.36-8.07, P<0.001). However, there was no significantly correlation between CRNDE and other clinicopathological features, including age (Z=1.63, P=0.103), gender (Z=0.75, P=0.451), and tumor differentiation (Z=1.27, P=0.203).
Publication bias and sensitivity analysis
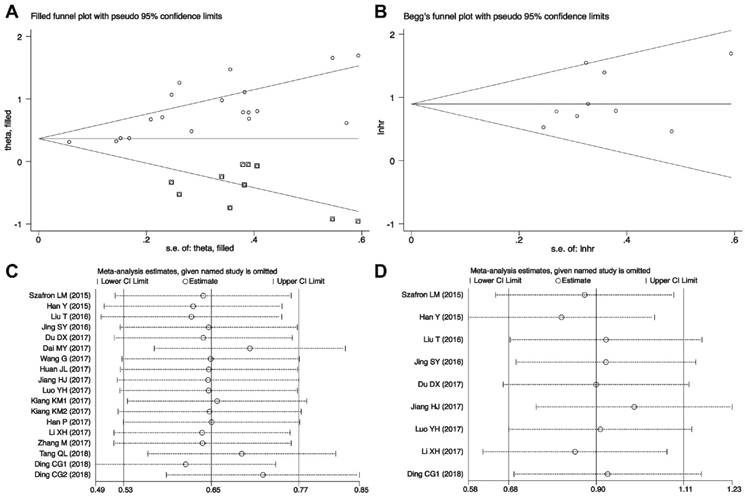
To evaluate the publication bias, the Begg's funnel plots and Egger's linear regression tests were applied in this meta-analysis. In the analysis of evaluating the association between CRNDE expression on OS, visual inspection of the Begg's funnel plot revealed asymmetry (Figure 5A), and Egger's test suggested the probable evidence of publication bias (t=2.61, P=0.018). Furthermore, to assess the impact of potential publication bias, the trim and fill analysis was performed with the fixed-effects model. Ten which conservatively imputes hypothetical negative unpublished studies to mirror the positive studies that cause funnel plot asymmetry. The imputed studies produce a symmetrical funnel plot (Figure 5A). The pooled analysis incorporation the hypothetical studies continued to show a statistically significant association between CRNDE expression on OS in solid cancers (corrected HR=1.44, 95% CI: 1.33-1.55, P<0.001). According to evaluating the independent prognostic value of CRNDE expression for OS in cancers, visual inspection of the Begg's funnel plot revealed symmetry (Figure 5B), and Egger's linear regression test revealed no publication bias for the analysis of OS (t=1.24, P=0.253). Due to the small number of studies, the publication bias was not analyzed in the DFS and PFS groups.
Forest plots of the included studies evaluating the correlation between CRNDE expression and clinicopathological characteristics. (A) tumor size; (B) TNM stages; (C) LNM; (D) distant metastases.
Publication bias and sensitivity analysis for OS in this meta-analysis. (A) Begg's funnel plots of the included studies for OS; (B) Begg's funnel plots of the included studies for independent predictive factor for OS; (C) Sensitivity analysis of the included studies for OS; (D) Sensitivity analysis of the included studies for independent predictive factor for OS. HR, hazard ratios; CI, confidence interval; SE, standard error.
Correlation between CRNDE expression and clinicopathological characteristics of cancers
| Clinical parameters | No. of studies | No. of patients | OR (95% CI) | P-value | Heterogeneity | |
|---|---|---|---|---|---|---|
| I2 | P-value | |||||
| Age (elder vs. younger) | 15 | 1861 | 1.17(0.97-1.40) | 0.103 | 9.6% | 0.346 |
| Gender (male vs. female) | 12 | 1595 | 1.02(0.83-1.25) | 0.876 | 0.0% | 0.594 |
| Tumor size (larger vs. smaller) | 12 | 1346 | 2.10(1.68-2.63) | <0.001 | 35.8% | 0.104 |
| Tumor differentiation (poor vs. well) | 11 | 1168 | 1.41(0.83-2.39) | 0.203 | 74.0% | <0.001 |
| TNM stage (III+IV vs. I+II) | 13 | 1546 | 2.86(2.29-3.56) | <0.001 | 38.8% | 0.075 |
| Lymph node metastasis (present vs. absent) | 12 | 1141 | 3.21(2.01-5.13) | <0.001 | 64.0% | 0.002 |
| Distant metastasis (present vs. absent) | 5 | 682 | 4.36(2.36-8.07) | <0.001 | 1.4% | 0.398 |
Sensitivity analyses demonstrated that the study by Xia XL et al.[30] was the top one with heterogeneity in the OS group, and their removal changed the results into more significant ones with no heterogeneity (HR=1.64, 95% CI: 1.43-1.85; I2=9.5%, P=0.341) (Figure 5C). And as indicated in Figure 5D, the pooled HR for the independent prognostic value of CRNDE in cancers was not significantly affected by the exclusion of any of the studies.
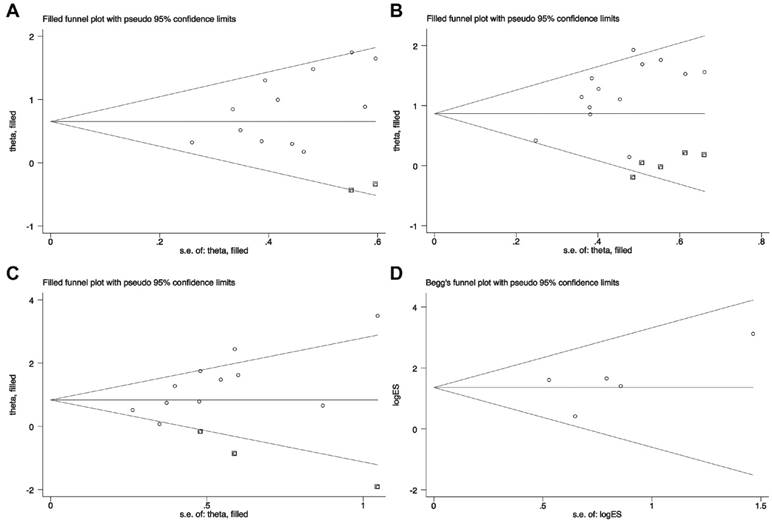
In addition, we also evaluated the association between CRNDE and tumor size, TNM stages, LNM or distant metastases. Visual inspection of the Begg's funnel plots revealed obvious asymmetry in the studies investigating CRNDE on tumor size (t=2.30, P=0.045, Figure 6A), TNM stages (t=2.99, P=0.012, Figure 6B), and LNM (t=3.04, P=0.014, Figure 6C). The corrected OR regarding the association between CRNDE and tumor size (corrected OR=1.92, 95% CI: 1.54-2.39, P<0.001), TNM stages (corrected OR=2.38, 95% CI: 1.94-2.91, P<0.001), and LNM (corrected OR=2.31, 95% CI: 1.40-3.80, P=0.001), which showed that the correction for potential publication bias had an influence on the stability of the result. And Begg's funnel plots suggested that there was no evidence of publication bias among the studies investigating CRNDE on distant metastases (t=0.99, P=0.397) (Figure 6D).
Discussion
Long noncoding RNAs (lncRNAs), which are encoded by a vast less explored region of the human genome, may hold missing drivers of cancer and have gained attention recently as a potentially crucial layer of carcinogenesis and cancer development. Recently, more evidence has emerged that lncRNAs are dysregulated in various types of human cancers, and has led to pioneering efforts to develop lncRNA-based clinical applications such as diagnostic, therapeutic, or prognostic biomarkers, drug sensitizers, and therapeutic targets.
As a newly characterized oncogene, CRNDE is an important lncRNA in human malignancies. Recent studies have revealed the multilayered biological function of CRNDE involve in cell proliferation, chemo-resistance, invasion and metastasis, which result in the cancer initialization and development [43-45]. Collective evidence has indicated that dysregulation of CRNDE is associated with adverse clinical characteristics such as tumor size, tumor differentiation, TNM stage, lymph node metastasis and distant metastasis, and poor prognosis in many solid cancers [46]. However, the perplexity and inconsistence arise from a wide range of studies due to heterogeneity. In this present study, we found that elevated CRNDE expression predicted a shorter OS in thirteen types of solid cancers, and severed as an independent predictive factor for patients' prognosis. Furthermore, CRNDE was positively associated with tumor size, TNM stage, lymph node metastasis, and distant metastasis. To sum up, our findings provided convincing evidences to support CRNDE as a negative, unfavorable prognostic biomarker for human solid cancers.
Although CRNDE has been suggested to serve as oncogenic lncRNA, the underlying molecular mechanisms involved in CRNDE interactions are complex and diverse (Table 3). Recent discoveries have revealed that CRNDE could provide specific functional scaffolds for regulatory complexes, such as the polycomb repressive complex 2 (PRC2) and CoREST chromatin‑modifying complexes. CRNDE functions as an oncogene to interact with PRC2 to repress p21 transcription, contributing to the radio-resistant phenotype formation of lung cancer cells [47]. Furthermore, CRNDE was also discovered to act as a scaffold of deleted in malignant brain tumors 1 (DMBT1) and cellular inhibitor of apoptosis protein 1 (C-IAP1) complexes, which could accelerate cancer development by promoting the PI3K/AKT pathway [39, 48]. CRNDE can epigenetically silence dual-specificity phosphatase 5 (DUSP5) and CDKN1A expression through binding to EZH2 [19]. CRNDE also modulate the levels of CDK4/6 and CCNE1 expression, contributing to the cell cycle transition from G0/G1 stage to S stage[48]. In addition, CRNDE could promote tumorigenesis through the EGFR, mTOR, Notch1, Ras/MAPK and toll-like receptor pathway [17, 25, 38, 49-52].
Begg's funnel plots of the included studies for clinicopathological characteristics. (A) tumor size; (B) TNM stages; (C) LNM; (D) distant metastases.
Roles of CRNDE in cancer initiation and progression
| Cancer type | Molecular Mechanisms | Functions | Reference |
|---|---|---|---|
| Ovarian Cancer | decrease accumulation of TP53 protein | prognostic biomarker | [34] |
| Cervical Cancer | NA | prognostic biomarker and correlate with FIGO stage, invasion depth and LNM, promote cell growth and metastasis | [35, 43] |
| Colorectal Cancer | epigenetically silence DUSP5/CDKN1A, downregulate E2F1 by targeting miR-136, upregulated by insulin/IGF pathway, activate Ras/MAPK pathway by binding to hnRNPUL2, regulate Wnt/β-catenin pathway by suppressing miR-181a-5p or binding to miR-217 with TCF7L2 | prognostic biomarker and correlate with tumor size, TNM stage, LNM and distant metastasis, promote cell proliferation, cell cycling, metastasis and chemo-resistance, and inhibit cell apoptosis | [16-19, 40, 54, 55] |
| Glioma | upregulated by EGFR and mTOR pathway, downregulate miR-136-5p to increase Bcl-2 and Wnt2, inhibit miR-384 /PIWIL4/STAT3 axis, downregulate XIAP and PAK7 by binding to miR-186 | prognostic biomarker and correlate with tumor size, WHO grade and recurrence, promote cell growth, proliferation, migration and invasion, and inhibit cell apoptosis | [24, 25, 49, 51, 57, 58] |
| Gastric Cancer | activate PI3K/AKT pathway, downregulate E2F3 by targeting miR-145 | prognostic biomarker and correlate with invasion depth, TNM stage and LNM, promote cell proliferation, migration and invasion | [29, 59] |
| HCC | activate PI3K/Akt-Wnt/β-catenin pathway, regulate miR-203/ BCAT1 axis, regulate NF-kB and p-AKT by inhibition of miR-384 | diagnostic and prognostic biomarker, promotes HCC cell proliferation, growth and metastasis, and inhibit cell apoptosis | [20, 21, 45, 56] |
| Pancreatic Cancer | upregulate IRS1 by sponging miR-384 | prognostic biomarker and correlate with tumor size, TNM stage, differentiation and LNM, promote cell proliferation, migration and invasion | [31] |
| Breast Cancer | activate Wnt/β-catenin pathway by directly repressing miR-136 | diagnostic and prognostic biomarker and correlate with tumor size and TNM stage, promote cell proliferation, migration and invasion | [27, 28] |
| PTC | increase PTN expression by competitively binding miR-384 | prognostic biomarker , promotes cell proliferation, invasion and migration | [32, 60] |
| NSCLC | downregulate p21 by interacting with PRC2/ EZH2, regulate CDK4/ 6 and CCNE1 expression, activate PI3K/AKT pathway | prognostic biomarker and correlate with cancer type, TNM stage and distant metastasis, promote cell proliferation and growth | [22, 23, 48] |
| IHCC | NA | prognostic biomarker and correlate with tumor size, TNM stage, differentiation and LNM, promote cell proliferation, migration, invasion and EMT | [30] |
| Bladder Cancer | NA | correlate with TNM stage, promote cell proliferation and migration, inhibit apoptosis | [33] |
| RCC | upregulate Vimentin, ZEB1/2 expression, downregulate E-cadherin expression, activate Wnt/β-catenin pathway | prognostic biomarker and correlate with tumor size, TNM stage, LNM, pathological grade and metastasis , promote cell migration and invasion | [26, 53] |
| TSCC | upregulate Kras, CDC42 and IRS1 by inhibiting miR-384 | promote cell proliferation and invasion | [61] |
| Osteosarcoma | activate Notch1 pathway | correlate with advanced stage and metastasis, promote cell growth, proliferation, invasion and migration | [52] |
| Multiple Myeloma | suppressing miR-451 expression | correlate with tumor progression and poor survival, promote cell proliferation, inhibit cell cycle arrest in the G0/G1 phase and apoptosis | [44] |
| Gallbladder Cancer | activate PI3K/AKT pathway by recruiting DMBT1 and c-IAP1 | promote gallbladder cancer carcinogenesis | [39] |
Recent studies have indicated that CRNDE can act as competitive endogenous RNA (ceRNA) to regulate miRNAs, and play key roles in the occurrence and development of human cancers. CRNDE functions as a ceRNA to promote malignant progression of serval cancers through competitive sponging of microRNA-136, miR-181a-5p and miR-217 to activate Wnt/β-catenin pathway [18, 28, 53-55]. CRNDE also has ability to upregulate IRS1, piwi-like RNA- mediated gene silencing 4 (PIWIL4), NF-kappaB, p-AKT, Bcl-2, Wnt2 and E2F3 expression by sponging miR-384, miR-145, miR-136-5p [31, 56-61]. To fully appreciate the functions of CRNDE, further studies are required to construct complete functional cell type-specific lncRNA expression maps in different steps of cancer progression.
Some limitations of this meta-analysis should be acknowledged due to the discrete data across these clinical studies. The criteria for calculating the cut-off value were not the same in different studies. The inclusion of only studies that reported HR or survival curves might have resulted in the potential for selection bias. Furthermore, some of the HRs were calculated by reconstructing survival curves rather than directly obtained from the primary studies, thus a calculation bias might be present. In addition, the inclusion of a relatively larger number of studies in China might have decreased the applicability of our results across different ethnicities. The data collection may be incomplete because data from non-English and non-Chinese language papers were not included. The publication bias of meta-analysis about the relationship between CRNDE and clinicopathological features was significant, which might be caused by some unpublished studies with negative results. In this meta-analysis, only summarized data rather than individual patient data were used. Therefore, it is possible that our results might overestimate the prognostic effects of abnormal CRNDE expression on survival in different types of cancers. We believed that more clinical studies should be conducted to evaluate potential prognostic role of CRNDE expression in other types of cancer that have not been included.
In conclusion, this meta-analysis showed that elevated CRNDE expression was significantly associated with shorter overall survival in patients with solid cancers, and might act as a novel effective prognostic biomarker and therapeutic target for solid cancers. Furthermore, CRNDE could promote cancer cell proliferation, invasion and metastasis, and suppresses apoptosis through various complicated mechanisms. Thus, CRNDE may potentially be used as a novel biomarker for predicting poor prognosis of patients with solid cancers. More clinical studies on other different types of human cancers that have not yet been investigated needed to be conducted. In the near further, the lncRNA profile of each human cancer type should be systematically investigated to improve clinical outcomes for cancer patients by engendering a personalized approach to medicine.
Acknowledgements
This work was supported by grant from National Natural Science Foundation of China (No. 81402029) and Scientific research project of Health and Family Planning Commission of Sichuan Province (No. 16PJ429).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-32
3. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155-9
4. Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1:93-109
5. Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613-20
6. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391-407
7. Isin M, Dalay N. LncRNAs and neoplasia. Clin Chim Acta. 2015;444:280-8
8. Bhartiya D, Kapoor S, Jalali S, Sati S, Kaushik K, Sachidanandan C. et al. Conceptual approaches for lncRNA drug discovery and future strategies. Expert Opin Drug Discov. 2012;7:503-13
9. Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS. LncRNAs and cancer. Oncol Lett. 2016;12:1233-9
10. Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219
11. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703-19
12. Zeng S, Xiao YF, Tang B, Hu CJ, Xie R, Yang SM. et al. Long Noncoding RNA in Digestive Tract Cancers: Function, Mechanism, and Potential Biomarker. Oncologist. 2015;20:898-906
13. Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y. et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881-90
14. Lin CY, Xu HM. Novel perspectives of long non-coding RNAs in esophageal carcinoma. Carcinogenesis. 2015;36:1255-62
15. Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT. et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer. 2011;2:829-40
16. Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437-48
17. Jiang H, Wang Y, Ai M, Wang H, Duan Z, Wang H. et al. Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017;8:e2862
18. Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY. et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16:9
19. Ding J, Li J, Wang H, Tian Y, Xie M, He X. et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997
20. Tang Q, Zheng X, Zhang J. Long non-coding RNA CRNDE promotes heptaocellular carcinoma cell proliferation by regulating PI3K/Akt /beta-catenin signaling. Biomed Pharmacother. 2018;103:1187-93
21. Dai M, Chen S, Wei X, Zhu X, Lan F, Dai S. et al. Diagnosis, prognosis and bioinformatics analysis of lncRNAs in hepatocellular carcinoma. Oncotarget. 2017;8:95799-809
22. Zhang M, Gao C, Yang Y, Li G, Dong J, Ai Y. et al. Long Noncoding RNA CRNDE/PRC2 Participated in the Radiotherapy Resistance of Human Lung Adenocarcinoma Through Targeting p21 Expression. Oncol Res. 2018;26:1245-55
23. Xiaohua Li DT, Wei Tang, Xiaoping Dan, Sheng Hu, Zhenliang Xiao. Expression and clinical significance of lncRNA CRNDE in patients with non-small cell lung cancer. Chin J Cancer Biother. 2017;24:889-93
24. Jing SY, Lu YY, Yang JK, Deng WY, Zhou Q, Jiao BH. Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20:3992-6
25. Kiang KM, Zhang XQ, Zhang GP, Li N, Cheng SY, Poon MW. et al. CRNDE Expression Positively Correlates with EGFR Activation and Modulates Glioma Cell Growth. Target Oncol. 2017;12:353-63
26. Ding C, Han F, Xiang H, Xia X, Wang Y, Dou M. et al. LncRNA CRNDE is a biomarker for clinical progression and poor prognosis in clear cell renal cell carcinoma. J Cell Biochem. 2018
27. Zibo Li JZ, Lin Zhou, Fanghui Yang. Value of detection of long no-coding RNA CRNDE in the diagnosis of breast cancer. Pract Prev Med. 2018;25:276-8
28. Huan J, Xing L, Lin Q, Xui H, Qin X. Long noncoding RNA CRNDE activates Wnt/beta-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am J Transl Res. 2017;9:1977-89
29. Du DX, Lian DB, Amin BH, Yan W. Long non-coding RNA CRNDE is a novel tumor promoter by modulating PI3K/AKT signal pathways in human gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21:5392-8
30. Xia XL, Xue D, Xiang TH, Xu HY, Song DK, Cheng PG. et al. Overexpression of long non-coding RNA CRNDE facilitates epithelial-mesenchymal transition and correlates with poor prognosis in intrahepatic cholangiocarcinoma. Oncol Lett. 2018;15:4105-12
31. Wang G, Pan J, Zhang L, Wei Y, Wang C. Long non-coding RNA CRNDE sponges miR-384 to promote proliferation and metastasis of pancreatic cancer cells through upregulating IRS1. Cell Prolif. 2017:50
32. Luo YH, Liang L, He RQ, Wen DY, Deng GF, Yang H. et al. RNA-sequencing investigation identifies an effective risk score generated by three novel lncRNAs for the survival of papillary thyroid cancer patients. Oncotarget. 2017;8:74139-58
33. Cheng J, Chen J, Zhang X, Mei H, Wang F, Cai Z. Overexpression of CRNDE promotes the progression of bladder cancer. Biomed Pharmacother. 2018;99:638-44
34. Szafron LM, Balcerak A, Grzybowska EA, Pienkowska-Grela B, Podgorska A, Zub R. et al. The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget. 2015;6:43897-910
35. Yi Han ML, Qiaoru Let, Yanna Li. Expression and clinical significance of long non-coding RNA CRNDE in cervical cancer. Chin J Clin Oncol. 2015;42:705-8
36. Song H, Han LM, Gao Q, Sun Y. Long non-coding RNA CRNDE promotes tumor growth in medulloblastoma. Eur Rev Med Pharmacol Sci. 2016;20:2588-97
37. Dong R, Liu XQ, Zhang BB, Liu BH, Zheng S, Dong KR. Long non-coding RNA-CRNDE: a novel regulator of tumor growth and angiogenesis in hepatoblastoma. Oncotarget. 2017;8:42087-97
38. Li Z, Tang Y, Xing W, Dong W, Wang Z. LncRNA, CRNDE promotes osteosarcoma cell proliferation, invasion and migration by regulating Notch1 signaling and epithelial-mesenchymal transition. Exp Mol Pathol. 2017
39. Shen S, Liu H, Wang Y, Wang J, Ni X, Ai Z. et al. Long non-coding RNA CRNDE promotes gallbladder carcinoma carcinogenesis and as a scaffold of DMBT1 and C-IAP1 complexes to activating PI3K-AKT pathway. Oncotarget. 2016;7:72833-44
40. Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta. 2014;1843:372-86
41. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16
42. Oremus M, Oremus C, Hall GB, McKinnon MC, Ect, Cognition Systematic Review T. Inter-rater and test-retest reliability of quality assessments by novice student raters using the Jadad and Newcastle-Ottawa Scales. BMJ open. 2012:2
43. Meng Y, Li Q, Li L, Ma R. The long non-coding RNA CRNDE promotes cervical cancer cell growth and metastasis. Biol Chem. 2017;399:93-100
44. Meng YB, He X, Huang YF, Wu QN, Zhou YC, Hao DJ. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol Res. 2017;25:1207-14
45. Ji D, Jiang C, Zhang L, Liang N, Jiang T, Yang B. et al. LncRNA CRNDE promotes hepatocellular carcinoma cell proliferation, invasion, and migration through regulating miR-203/ BCAT1 axis. J Cell Physiol. 2018
46. Bach DH, Lee SK. Long noncoding RNAs in cancer cells. Cancer Lett. 2018;419:152-66
47. Zhang M, Gao C, Yang Y, Li G, Dong J, Ai Y. et al. Long Noncoding RNA CRNDE/PRC2 Participated in the Radiotherapy Resistance of Human Lung Adenocarcinoma Through Targeting p21 Expression. Oncol Res. 2017
48. Liu XX, Xiong HP, Huang JS, Qi K, Xu JJ. Highly expressed long non-coding RNA CRNDE promotes cell proliferation through PI3K/AKT signalling in non-small cell lung carcinoma. Clin Exp Pharmacol Physiol. 2017;44:895-902
49. Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122-8
50. Li H, Li Q, Guo T, He W, Dong C, Wang Y. LncRNA CRNDE triggers inflammation through the TLR3-NF-kappaB-Cytokine signaling pathway. Tumour Biol. 2017;39:1010428317703821
51. Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu Y. et al. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6:25339-55
52. Li Z, Tang Y, Xing W, Dong W, Wang Z. LncRNA, CRNDE promotes osteosarcoma cell proliferation, invasion and migration by regulating Notch1 signaling and epithelial-mesenchymal transition. Exp Mol Pathol. 2018;104:19-25
53. Shao K, Shi T, Yang Y, Wang X, Xu D, Zhou P. Highly expressed lncRNA CRNDE promotes cell proliferation through Wnt/beta-catenin signaling in renal cell carcinoma. Tumour Biol. 2016
54. Yu B, Ye X, Du Q, Zhu B, Zhai Q, Li XX. The Long Non-Coding RNA CRNDE Promotes Colorectal Carcinoma Progression by Competitively Binding miR-217 with TCF7L2 and Enhancing the Wnt/beta-Catenin Signaling Pathway. Cell Physiol Biochem. 2017;41:2489-502
55. Gao H, Song X, Kang T, Yan B, Feng L, Gao L. et al. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205-16
56. Chen Z, Yu C, Zhan L, Pan Y, Chen L, Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299-309
57. Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C. et al. CRNDE Promotes Malignant Progression of Glioma by Attenuating miR-384/PIWIL4/STAT3 Axis. Mol Ther. 2016;24:1199-215
58. Li DX, Fei XR, Dong YF, Cheng CD, Yang Y, Deng XF. et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget. 2017;8:88163-78
59. Hu CE, Du PZ, Zhang HD, Huang GJ. Long Noncoding RNA CRNDE Promotes Proliferation of Gastric Cancer Cells by Targeting miR-145. Cell Physiol Biochem. 2017;42:13-21
60. Sun H, He L, Ma L, Lu T, Wei J, Xie K. et al. LncRNA CRNDE promotes cell proliferation, invasion and migration by competitively binding miR-384 in papillary thyroid cancer. Oncotarget. 2017;8:110552-65
61. Ren Y, He W, Chen W, Ma C, Li Y, Zhao Z. et al. CRNDE promotes cell tongue squamous cell carcinoma cell growth and invasion through suppressing miR-384. J Cell Biochem. 2018
Author contact
![]() Corresponding author: Shikai Zhu, MD, PhD. Organ Transplant Center, Hospital of University of Electronic Science and Technology of China and Sichuan Provincial People's Hospital, Chengdu 610072, China. E-mail: zhushikaiedu.cn.
Corresponding author: Shikai Zhu, MD, PhD. Organ Transplant Center, Hospital of University of Electronic Science and Technology of China and Sichuan Provincial People's Hospital, Chengdu 610072, China. E-mail: zhushikaiedu.cn.

 Global reach, higher impact
Global reach, higher impact