Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(10):2342-2349. doi:10.7150/jca.30454 This issue Cite
Research Paper
Proteomic Analysis of the Molecular Mechanism of Lovastatin Inhibiting the Growth of Nasopharyngeal Carcinoma Cells
1. NHC Key Laboratory of Carcinogenesis, Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, Hunan 410013, China
2. The Key Laboratory of Carcinogenesis and Cancer Invasion of the Chinese Ministry of Education, Cancer Research Institute and School of Basic Medical Science, Central South University, Changsha 410078, China
3. Hunan Key Laboratory of Nonresolving Inflammation and Cancer, Disease Genome Research Center, The Third Xiangya Hospital, Central South University, Changsha, Hunan, China.
4. Department of Cancer Biology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio 44195, USA
*Yongzhen Mo and Yumin Wang contributed equally to this work.
Received 2018-10-5; Accepted 2019-4-15; Published 2019-5-26
Abstract
Metabolic abnormalities are one of the essential features of tumors. Increasingly more studies have shown that lovastatin, a lipid-reducing drug, has visible inhibitory effects on tumors, but it has not been reported in nasopharyngeal carcinoma. In this paper, we explored the effects of lovastatin on the growth of nasopharyngeal carcinoma cells and its possible molecular mechanisms. After treating nasopharyngeal carcinoma cells with different concentrations of lovastatin, we found that lovastatin can inhibit the growth of nasopharyngeal carcinoma in a time- and dose-dependent manner. To explore the molecular mechanism of how lovastatin inhibits the growth of nasopharyngeal carcinoma, we examined the proteome of nasopharyngeal carcinoma cells treated at different time points using an LC/MS whole-proteomic strategy. The molecular network of differentially expressed proteins was constructed using IPA software. It was found that lovastatin inhibited the growth of nasopharyngeal carcinoma cells mainly by affecting the EIF2 and the mTOR pathways, which regulate cell metabolism and apoptosis. The results of this study provide a robust basis for further research on the molecular mechanism of lovastatin's inhibition of nasopharyngeal carcinoma cells and provide a reference for the clinical use of lovastatin in the treatment of nasopharyngeal carcinoma.
Keywords: lovastatin, nasopharyngeal carcinoma, LC/MS, EIF2 pathway, mTOR pathway
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor originating from the nasopharyngeal epithelium. The disease occurs in the southern part of China and has apparent regional clustering, especially in Guangdong, Hong Kong and Hunan[1-5]. At present, the treatment of nasopharyngeal carcinoma is mainly based on concurrent chemo-radiotherapy[6-10]. The NCCN Guidelines recommend a standard procedure for locally advanced nasopharyngeal carcinoma to emphasize concurrent chemotherapy with single-agent cisplatin chemotherapy. However, due to the high incidence of digestive tract reaction and the ototoxicity and nephrotoxicity of cisplatin, the type and severity of adverse reactions make the patient's quality of life extremely poor[11-15]. Therefore, finding alternative chemotherapy drugs to improve the prognosis and quality of life of patients with locally advanced nasopharyngeal carcinoma has become an urgent problem to be solved. However, the traditional technological route to develop new drugs has a long cycle and is costly; therefore, the advantages of in-depth studies of approved drugs, in order to explore the full spectrum of uses or repositioning of existing drugs, have been widely recognized by the biomedical industry[16-18].
More importantly, many studies have shown that the combination of statins with different drugs can inhibit the proliferation of various tumor cells, such as prostate cancer, and promote the occurrence of apoptosis[19]. These epidemiological and clinical trial studies have shown that statins may play an essential role in the treatment of tumors. Recently, an extensive sample of epidemiological studies showed a negative correlation between cancer mortality and statin use rates, and the authors speculated that it could be used as a potential antitumor metastatic drug[20-22]. Lovastatin, which is the most widely used statin, is currently the first-line treatment for hypercholesterolemia and mixed hyperlipidemia. However, the potential of lovastatin in nasopharyngeal carcinoma treatment has not been reported. Therefore, we mainly studied the effect of lovastatin on the proliferation of nasopharyngeal carcinoma cells and explore the molecular mechanism.
Materials and Methods
Lovastatin was purchased from Sigma, USA; RPMI1640 medium, fetal bovine serum, 0.25% trypsin and penicillin, streptomycin were purchased from Gibco, USA; 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and dimethyl sulfoxide (DMSO) were obtained from Sigma, USA.
Cell culture
The NPC cell lines 5-8F and HNE2 used in this study were preserved in our laboratory (Cancer Research Institute, Central South University). Culture conditions: Cells in RPMI1640 liquid medium containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin were cultured in a constant temperature incubator at 37 °C with 5% CO2. The cells were spread over the bottom of the culture dish, trypsinized and passaged to reach the logarithmic growth phase for experiments.
MTT assay for assessing the inhibition of cell proliferation by different drug concentrations
The NPC cell lines 5-8F and HNE2 were seeded in 96-well plates at a density of 1000 cells per well. After the cells were attached to the wall, lovastatin was added: 1) the negative control group was the lovastatin-free cell group; 2) in the lovastatin drug treatment group, the final lovastatin concentrations in the wells were 20, 40, 60, 80, 100, and 120 μM, with five replicate wells in each group. Then, 20 μL of MTT (5 mg/mL) was added to the cells for 24 h, and incubation was continued for 4 h. Next, the supernatant was aspirated, and 200 μL of DMSO was added to each well. The mixture was incubated for 10 min on a shaker at room temperature, and a microplate reader was used to measure the absorbance at 490 nm. Based on the absorbance, the cell activity curves of the nasopharyngeal carcinoma cells were plotted, and the IC50 value of lovastatin was calculated.
Time-effect during the detection of the lovastatin IC50 on cell inhibition in MTT assays
Logarithmic-stage 5-8F and HNE2 nasopharyngeal carcinoma cells were seeded in 96-well plates at a density of 1000 cells per well. After the cells were attached to the walls, the drug was added separately: 1) the negative control group received no drug solution; 2) the lovastatin drug treatment group received a lovastatin dose at the of IC50 concentration and were treated for the following times: 0, 24, 48, 72, 96 h. Five replicate wells were set up for each group, 20 μL of MTT (5 mg/mL) was added at the end of the experiment and incubation was continued for 4 h. Next, the supernatant was aspirated, 200 μL of DMSO was added to each well, and the plates were shaken at room temperature for 10 min. A microplate reader was then used to measure the absorbance at 490 nm. The time-dependent effect of lovastatin at the IC50 concentration on cell growth inhibition was determined.
Mass spectrometry
To determine the mechanism by which lovastatin works, we used the Ultimate 3000 RSLC nano system of the LTQ Orbitrap Velos Pro mass spectrometer (Thermo Scientific, Bremen, Germany) to recover proteins after lovastatin treatment for 0, 6, 12, 18, and 24 h. Total protein was subjected to proteomic analysis. The peptides were diluted with 0.1% trifluoroacetic acid (TFA) and then transferred to an analytical column; 5 μL of the sample was injected per analysis. The mass spectrometer was operated in a data-dependent mode. Full-scan MS spectra were obtained in an Orbitrap analyzer at a mass resolution of 60,000 (mass range 350-2000 m/z). For label-free analysis, the ten most abundant tandem mass spectrum peaks were obtained in a linear ion trap by peptide cleavage using collision-induced dissociation (CID). The normalized collision energy (NCE) was set to 35%, and the isolation width was 2 m/z. Proteins were identified using Proteome Discoverer software (Thermo Scientific, Waltham, MA, USA). The original Thermo file was imported and used to search in the UniProt KB / Swiss-Prot database (version 2014_02) [23, 24].
Proteomics data analysis
For database searches, the mass tolerances of the precursor and fragment ions were set to 10 ppm and 0.8 Da, respectively. Peptides with a false positive rate <1% (q value <0.01) were discarded. Proteins that met the following criteria were considered to be differentially expressed proteins: i) proteins identified based on ≥ 2% peptides with ≥ 95% confidence, and ii) mean change in protein ≥ 1.50 or ≤ 0.67 (Student's t-test, p < 0.05). IPA bioinformatics software was used to obtain differentially expressed proteins to determine altered signaling pathways after lovastatin treatment.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 (GraphPad, La Jolla, CA, USA). Student's t tests were performed to evaluate significant differences between any two groups of data. All data were represented as the mean ± standard deviation (s.d.) unless otherwise stated. P<0.05 was considered to indicate statistical significance.
Results
Lovastatin inhibits 5-8F and HNE2 nasopharyngeal carcinoma cells in a dose-dependent manner
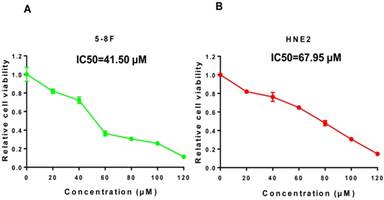
We treated 5-8F and HNE2 nasopharyngeal carcinoma cells for 48 h with lovastatin at concentrations of 20, 40, 60, 80, 100, 120 μM and measured cell viability using MTT. As shown in Figure 1A and 1B, the numbers of viable 5-8F and HNE2 cells were significantly decreased in a dose-dependent manner after lovastatin treatment, indicating that different concentrations of lovastatin have specific inhibitory effects on cell growth. It was also found that the IC50 values of lovastatin were 41.50 μM for 5-8F cells and 67.95 μM for HNE2 cells.
The time-dependence of lovastatin on nasopharyngeal carcinoma cell inhibition
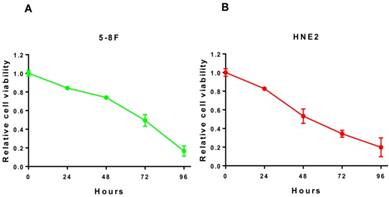
To further investigate whether the inhibitory effect of lovastatin on the proliferation of nasopharyngeal carcinoma cells is time-dependent, we used the IC50 concentration obtained in Figure 1 to treat HNE2 and 5-8F cells for 24, 48, 72, and 96 h and examined the cells to obtain a survival curve. As shown in Figure 2A and 2B, as the treatment time increased, the proliferation rate of the cells decreased significantly, and there were significant differences between the different time points. It can be seen that with longer exposure times to lovastatin, the inhibitory effect on the proliferation of 5-8F cells and HNE2 cells gradually increased. These results indicated that lovastatin had a significant time-dependent effect on the inhibition of nasopharyngeal carcinoma cell proliferation.
Identification of differentially expressed proteins after treatment of nasopharyngeal carcinoma cells with lovastatin using liquid chromatography-tandem mass spectrometry
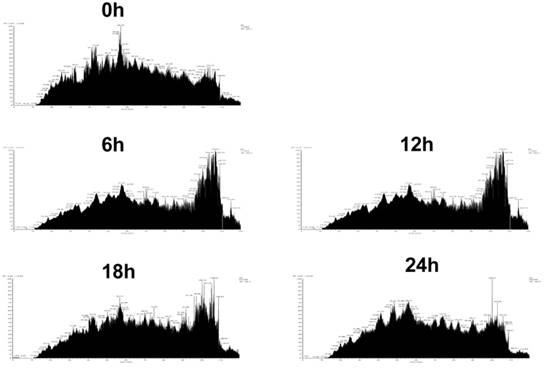
To further explore the mechanism of how lovastatin inhibits the proliferation of nasopharyngeal carcinoma cells and to find proteins and signaling pathways related to this inhibitory effect, we used whole-proteome mass spectrometry performed at 0, 6, 12, 18, and 24 h before and after lovastatin treatment of HNE2 nasopharyngeal carcinoma cells. The ion current maps at five time points are shown in Figure 3. The results showed a significant change in the ion chromatogram of the mass spectrometry after drug treatment, suggesting a change in the relevant proteins and signaling pathways in the cells after drug treatment (Figure 3).
Lovastatin inhibits 5-8F and HNE2 nasopharyngeal carcinoma cells in a dose-dependent manner. We treated 5-8F and HNE2 nasopharyngeal carcinoma cells with lovastatin for 48 h at concentrations of 20, 40, 60, 80, 100, and 120 μM and measured cell viability with MTT. As shown in Figure 1A and 1B, the numbers of viable 5-8F and HNE2 cells were significantly decreased in a dose-dependent manner after lovastatin treatment, indicating that different concentrations of lovastatin have a specific inhibitory effects on cell growth. The IC50 values of lovastatin were 41.50 μM for the 5-8F cell line and 67.95 μM for the HNE2 cell line.
The time-dependent component on the inhibition of nasopharyngeal carcinoma cells by lovastatin. We used the IC50 concentration obtained in Figure 1 to treat HNE2 and 5-8F cells for 24, 48, 72, and 96 h and then examined the cells to obtain cell viability curves. As shown in Figure 2A and 2B, as the treatment time increased, the proliferation rate of the cells decreased significantly, and there were significant differences between the different time points. It can be seen that with prolonged lovastatin treatment, the inhibitory effect on the proliferation of 5-8F cells and HNE2 cells is gradually increased.
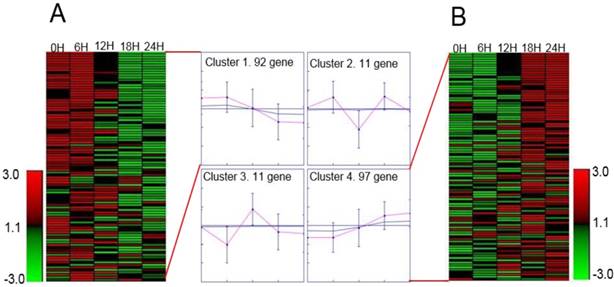
We selected proteins with specific peptides >2 and area values at five-time points for protein identification. There were 211 proteins conform our standard for lovastatin treatment after. We used GENE protein software for SOM analysis of the 211 proteins. We found that the levels of 189 proteins changed significantly after lovastatin treatment: 97 of them were upregulated and 92 were downregulated (Figure 4).
Lovastatin inhibits the growth of nasopharyngeal carcinoma cells by regulating cell metabolism and apoptosis pathways
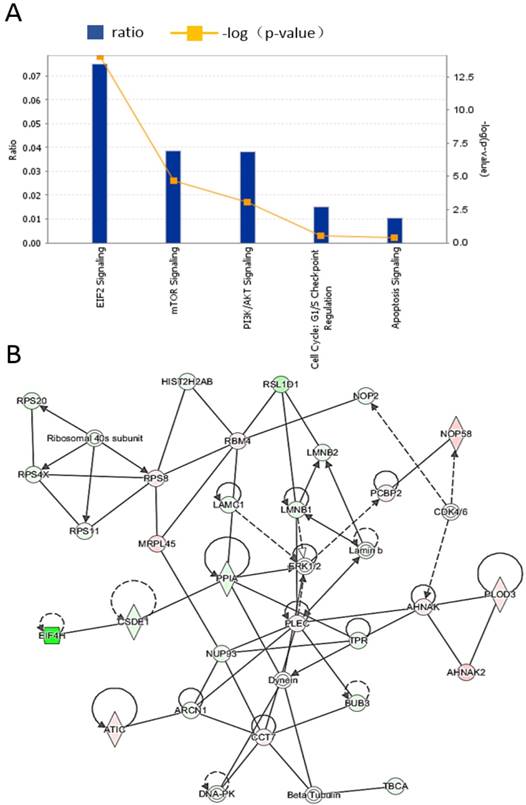
To further elucidate the functions of these differentially expressed proteins and their signaling pathways, we used IPA software to analyze the differentially expressed proteins identified in the above mass spectrometry experiments. The results showed that the differentially regulated proteins were mainly enriched in the EIF2, mTOR, and PI3K/AKT pathways. The pathways are depicted as shown in Figure 5A. Among the proteins, the most abundant proteins are from the EIF2 pathway, and Figure 5B shows the interactions between different proteins in the EIF2 pathway.
Identification of differentially expressed proteins by mass spectrometry after treatment of nasopharyngeal carcinoma cells with lovastatin. Mass spectrometry was performed on the whole proteome of HNE2 nasopharyngeal carcinoma cells at 0, 6, 12, 18, and 24h after lovastatin treatment. The total ion chromatography at five time points are shown.
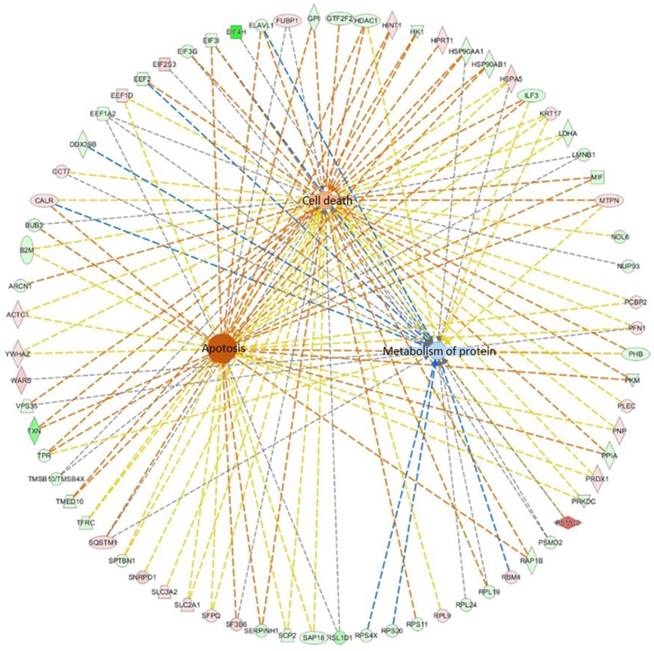
Further, we used IPA software to analyze the cellular functions regulated by these differentially regulated proteins. We found that these proteins mainly regulate cell metabolism, cell death, and apoptosis, as shown in Figure 6. The interactions between differentially regulated proteins involved in apoptosis are shown. Our results indicate that lovastatin inhibits the growth of nasopharyngeal carcinoma cells primarily by altering cellular metabolism and apoptotic pathways.
Discussion
Lovastatin is a 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitor that affects many physiological processes in the body by inhibiting the synthesis of mevalonate (MVA) and its downstream products[25, 26]. The effects of lovastatin in lowering cholesterol and improving the endocardial environment are widely applied in the treatment of hyperlipidemia and cardiovascular diseases. MVA is a precursor or substrate for many essential products such as cholesterol, polyester, coenzyme Q, farnesyl pyrophosphate (FPP), and geranyl pyrophosphate (GGPP). MVA is involved in cell membrane synthesis, glycoprotein synthesis, and intracellular signal transduction, cell cycle regulation and other critical biological functions[27, 28]. Therefore, HMG-CoA reductase, as the rate-limiting enzyme in intracellular MVA synthesis, is widely involved in critical cellular functions, such as cell signal transduction, glycoprotein synthesis, cell cycle progression, and cell membrane integrity[29-33]. In recent years, studies have found that in addition to their lipid-lowering effects, statins have multiple antitumor effects, including antiproliferative, proapoptotic, and anti-invasive activities as well as the ability to sensitize cells to radiochemotherapy[34-36]. Lovastatin early in the market and is a competitive inhibitor of hydroxymethylglutaryl coenzyme A (HMG-CoA) reductase. The drug is mainly used for the clinical treatment of hyperlipidemia and has been subjected to lengthy clinical study, which has generated a large body of data, and its therapeutic effects are well known[37]. In recent years, increasingly more studies have reported that lovastatin can inhibit cell proliferation, promote apoptosis and induce differentiation in tumor cells, but it has fewer toxic side effects for normal cells[38]. Marcelli et al. found that lovastatin treatment of prostate cancer LNCa cells for 48 h can induce apoptosis[39]. There are also reports that lovastatin can be combined with 5-fluorouracil or cisplatin as a chemotherapeutic drug to increase the chemosensitivity of colon cancer cells and to induce apoptosis[40].
Heatmap representation of differentially expressed proteins. We selected proteins with specific peptides >2 and area values at five-time points for protein identification. There were 211 proteins that met these conditions after lovastatin treatment. We used GENE protein software for the SOM analysis of the 211 proteins. We found that the levels of 189 proteins changed significantly after lovastatin treatment, and shown is the heatmap representation of the differentially expressed proteins after treatment of nasopharyngeal carcinoma cells with lovastatin. Proteins are listed by their respective gene names, and the fold changes in protein expression are shown. Overall, 92 proteins were downregulated (A), and 97 proteins were upregulated (B).
Nasopharyngeal carcinoma is a tumor known as "Chinese cancer". It has a low incidence in most parts of the world, but it is highly prevalent in southern China and Southeast Asia[41-45]. Currently, the treatment for nasopharyngeal carcinoma is mainly radiotherapy, supplemented with chemotherapy. However, differences in genetic backgrounds between patients with nasopharyngeal carcinoma lead to large individual differences in sensitivity to radiotherapy and chemotherapy[46, 47], which results in poor prognosis and quality of life. Many studies have shown that lovastatin has a growth-inhibiting effect on a variety of tumors, but no information has been reported on its role or mechanism of inhibition in nasopharyngeal carcinoma. Therefore, this study used nasopharyngeal carcinoma as a research model, and lovastatin was used to treat 5-8F and HNE2 nasopharyngeal carcinoma cells to assess its effect on the growth of nasopharyngeal carcinoma; furthermore, its functional mechanism was explored using proteomics.
In our results, we found that lovastatin has an inhibitory effect on the growth of nasopharyngeal carcinoma and that there are concentration- and time-dependent effects. Furthermore, we used liquid chromatography-tandem mass spectrometry to analyze the molecular mechanism of lovastatin's inhibition of nasopharyngeal carcinoma. By examining the differentially regulated proteins before and after lovastatin treatment, we found that the differentially regulated proteins were mainly enriched for proteins from the EIF2 pathway, which is the endoplasmic reticulum stress activation signaling pathway[48]. The endoplasmic reticulum is an organelle that is involved in protein folding, homeostasis, and lipid synthesis[49]. A variety of factors, including oxidative stress, ischemia, and balance disorders, can affect endoplasmic reticulum function, leading to unfolded proteins and errors. The folding proteins accumulate in the endoplasmic reticulum. When the endoplasmic reticulum is stressed, three endoplasmic reticulum transmembrane sensors are activated to initiate an adaptive response. These three transmembrane sensors include protein kinase-like endoplasmic reticulum kinase (PERK), inositol kinase (IRE1), and transcriptional activator 6 (ATF6). These three sensors are maintained in their inactive state by binding to the ends of glycodelin. When unfolded proteins accumulate in the endoplasmic reticulum, these sensors are released, allowing their oligomerization to initiate the reaction. After the initiation of endoplasmic reticulum stress, PERK phosphorylates eukaryotic translation initiation factor 2 (eIF2), reducing the protein load in the endoplasmic reticulum. When the response is insufficient to control the levels of unfolded and misfolded proteins in the endoplasmic reticulum, the endoplasmic reticulum-triggered apoptotic signaling pathway is activated. Direct degradation of structural and functional proteins in cells causes apoptosis [50-53]. In addition to the EIF2 pathway, other pathways that were significantly changed were the mTOR and PI3K/AKT pathways. The mammalian target of rapamycin (mTOR) is an atypical serine/ threonine protein kinase, which, similar to phosphatidylinositol 3 kinase (PI3K), is a member of the protein kinase family. PI3K kinase dysfunction causes diseases such as tumorigenesis and immune disorders. mTOR is relatively conservatively involved in biological processes, such as gene transcription, protein translation, ribosome synthesis, and apoptosis, and thus plays an essential role in many physiological processes such as cell growth, senescence, and metabolism. Numerous studies have found that a variety of tumors are accompanied by abnormal regulation of the mTOR signaling pathway [54-59]. Studies have shown that the PI3K / Akt / mTOR signaling pathways are inactivated in breast cancer, leukemia, gastrointestinal stromal tumors, hepatocellular carcinoma, lung cancer, esophageal cancer, etc. Activation of the PI3K/Akt/mTOR signaling pathway promotes a variety of stimulation- induced apoptotic processes[60-62]. In our results, we identified molecular mechanisms by which lovastatin inhibits the growth of nasopharyngeal carcinoma cells, but whether other molecular mechanisms are also involved requires further study. On the other hand, in our study, we only performed in vitro cell experiments, and whether the results of in vivo experiments will be consistent with our in vitro experiments requires further investigation. In summary, we found that lovastatin can inhibit the growth of nasopharyngeal carcinoma cells and that its molecular mechanism is mainly through regulation of the EIF2 and mTOR pathways, which affect metabolism and apoptosis in nasopharyngeal carcinoma cells, to regulate the occurrence and development of nasopharyngeal carcinoma.
Lovastatin inhibits the growth of nasopharyngeal carcinoma cells by regulating cell metabolism and apoptosis pathways. IPA software was used to analyze the differentially expressed proteins identified by the mass spectrometry. The differentially regulated proteins were mainly enriched in proteins from the EIF2, mTOR, and PI3K/AKT pathways (A). Among them, proteins involved in the EIF2 signaling pathway are the most abundant (B).
The interactions between differential proteins that regulate cell metabolism, cell death, and apoptosis. The differentially expressed proteins mainly regulate cell metabolism, cell death, and apoptosis.
Acknowledgements
This study was supported by grants from The National Natural Science Foundation of China (81572787, 81672683, 81672993, 81672688, 81702907, 81772901, 81772928, 81803025 and 81872278), the Overseas Expertise Introduction Project for Discipline Innovation (111 Project, No. 111-2-12), and the Natural Science Foundation of Hunan Province (2016JC2035, 2017SK2105, 2018JJ3704, 2018JJ3815, 2018SK21210 and 2018SK21211).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wei F, Wu Y, Tang L. et al. Trend analysis of cancer incidence and mortality in China. Sci China Life Sci. 2017;60:1271-5
2. Fan C, Tang Y, Wang J. et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J Cancer. 2018;9:2852-64
3. Yang L, Tang Y, He Y. et al. High Expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97-103
4. Tu C, Zeng Z, Qi P. et al. Identification of genomic alterations in nasopharyngeal carcinoma and nasopharyngeal carcinoma-derived Epstein-Barr virus by whole-genome sequencing. Carcinogenesis. 2018;39:1517-28
5. He B, Li W, Wu Y. et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353
6. Wei F, Tang L, He Y. et al. BPIFB1 (LPLUNC1) inhibits radioresistance in nasopharyngeal carcinoma by inhibiting VTN expression. Cell Death Dis. 2018;9:432
7. Tu C, Zeng Z, Qi P. et al. Genome-Wide Analysis of 18 Epstein-Barr Viruses Isolated from Primary Nasopharyngeal Carcinoma Biopsy Specimens. J Virol. 2017;91:pii e00301-17
8. Tang L, Wei F, Wu Y. et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J Exp Clin Cancer Res. 2018;37:87
9. Fan C, Wang J, Tang Y. et al. Long non-coding RNA LOC284454 promotes migration and invasion of nasopharyngeal carcinoma via modulating the Rho/Rac signaling pathway. Carcinogenesis. 2018;40:380-91
10. Lian Y, Xiong F, Yang L. et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J Exp Clin Cancer Res. 2018;37:253
11. Tang Y, He Y, Shi L. et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001-11
12. Wang W, Yi M, Zhang R. et al. Vimentin is a crucial target for anti-metastasis therapy of nasopharyngeal carcinoma. Mol Cell Biochem. 2018;438:47-57
13. Zhou Y, Liao Q, Li X. et al. HYOU1, Regulated by LPLUNC1, Is Up-Regulated in Nasopharyngeal Carcinoma and Associated with Poor Prognosis. J Cancer. 2016;7:367-76
14. Yan Q, Zeng Z, Gong Z. et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766-82
15. Wang Y, Xue D, Li Y. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer. 2016;7:991-1001
16. Boguski MS, Mandl KD, Sukhatme VP Drug discovery. Repurposing with a difference. Science. 2009;324:1394-5
17. Xiong F, Deng S, Huang HB. et al. Effects and mechanisms of innate immune molecules on inhibiting nasopharyngeal carcinoma. Chin Med J (Engl). 2019;132:749-52
18. Duan S, Guo W, Xu Z. et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol Cancer. 2019;18:29
19. Ahern TP, Lash TL, Damkier P. et al. Statins and breast cancer prognosis: evidence and opportunities. Lancet Oncol. 2014;15:e461-8
20. Wu C, Li M, Meng H. et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62:640-7
21. Wei F, Jing YZ, He Y. et al. Cloning and characterization of the putative AFAP1-AS1 promoter region. J Cancer. 2019;10:1145-53
22. Zhang Y, Xia M, Jin K. et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer. 2018;17:45
23. Wei F, Wu Y, Tang L. et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Br J Cancer. 2018;118:233-47
24. Bo H, Gong Z, Zhang W. et al. Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget. 2015;6:20404-18
25. Chen CC, Liu TY, Huang SP. et al. Differentiation and apoptosis induction by lovastatin and gamma-tocotrienol in HL-60 cells via Ras/ERK/NF-kappaB and Ras/Akt/NF-kappaB signaling dependent down-regulation of glyoxalase 1 and HMG-CoA reductase. Cell Signal. 2015;27:2182-90
26. Yi M, Li J, Chen S. et al. Emerging role of lipid metabolism alterations in Cancer stem cells. J Exp Clin Cancer Res. 2018;37:118
27. Bo H, Fan L, Gong Z. et al. Upregulation and hypomethylation of lncRNA AFAP1-AS1 predicts a poor prognosis and promotes the migration and invasion of cervical cancer. Oncol Rep. 2019;41:2431-39
28. Tang Y, He Y, Zhang P. et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17:77
29. Zhong Y, Du Y, Yang X. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79
30. Zhou R, Wu Y, Wang W. et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134-42
31. Deng X, Xiong F, Li X. et al. Application of atomic force microscopy in cancer research. J Nanobiotechnology. 2018;16:102
32. He R, Liu P, Xie X. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145
33. Wang M, Zhao J, Zhang L. et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;8:761-73
34. Mamane Y, Petroulakis E, Rong L. et al. eIF4E-from translation to transformation. Oncogene. 2004;23:3172-9
35. Jiang X, Wang J, Deng X. et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10
36. Fan C, Tang Y, Wang J. et al. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16:130
37. Mulder KC, Mulinari F, Franco OL. et al. Lovastatin production: From molecular basis to industrial process optimization. Biotechnol Adv. 2015;33:648-65
38. Chae YK, Yousaf M, Malecek MK. et al. Statins as anti-cancer therapy; Can we translate preclinical and epidemiologic data into clinical benefit? Discov Med. 2015;20:413-27
39. Tang Y, He Y, Shi L. et al. Co-expression of AFAP1-AS1 and PD-1 predicts poor prognosis in nasopharyngeal carcinoma. Oncotarget. 2017;8:39001-11
40. Feleszko W, Zagozdzon R, Golab J. et al. Potentiated antitumour effects of cisplatin and lovastatin against MmB16 melanoma in mice. Eur J Cancer. 1998;34:406-11
41. He Y, Jing Y, Wei F. et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018;9:235
42. Zou G, Ren B, Liu Y. et al. Inhibin B suppresses anoikis resistance and migration through the transforming growth factor-beta signaling pathway in nasopharyngeal carcinoma. Cancer Sci. 2018;109:3416-27
43. Song Y, Li X, Zeng Z. et al. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783-99
44. Xiao K, Yu Z, Li X. et al. Genome-wide Analysis of Epstein-Barr Virus (EBV) Integration and Strain in C666-1 and Raji Cells. J Cancer. 2016;7:214-24
45. Wang Y, Mo Y, Yang X. et al. Long non-coding RNA AFAP1-AS1 is a novel biomarker in various cancers: a systematic review and meta-analysis based on the literature and GEO datasets. Oncotarget. 2017;8:102346-60
46. Xiong W, Zeng ZY, Xia JH. et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64:1972-4
47. Yu J, Liu Y, Gong Z. et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. 2017;8:16621-32
48. Tang Y, Wang J, Lian Y. et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol Cancer. 2017;16:42
49. Wang JP, Tang YY, Fan CM. et al. The role of exosomal non-coding RNAs in cancer metastasis. Oncotarget. 2018;9:12487-502
50. Moon HS, Kim B, Gwak H. et al. Autophagy and protein kinase RNA-like endoplasmic reticulum kinase (PERK)/eukaryotic initiation factor 2 alpha kinase (eIF2alpha) pathway protect ovarian cancer cells from metformin-induced apoptosis. Mol Carcinog. 2016;55:346-56
51. Xia M, Zhang Y, Jin K. et al. Communication between mitochondria and other organelles: a brand-new perspective on mitochondria in cancer. Cell Biosci. 2019;9:27
52. Yang L, Tang Y, Xiong F. et al. LncRNAs regulate cancer metastasis via binding to functional proteins. Oncotarget. 2018;9:1426-43
53. Bo H, Fan L, Li J. et al. High Expression of lncRNA AFAP1-AS1 Promotes the Progression of Colon Cancer and Predicts Poor Prognosis. J Cancer. 2018;9:4677-83
54. Foster KG and Fingar DC Mammalian target of rapamycin (mTOR). conducting the cellular signaling symphony. J Biol Chem. 2010;285:14071-7
55. Liang F, Li Q, Li X. et al. TSC22D2 interacts with PKM2 and inhibits cell growth in colorectal cancer. Int J Oncol. 2016;49:1046-56
56. Wang YA, Li XL, Mo YZ. et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer. 2018;17:168
57. Yu J, Liu Y, Guo C. et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8:523-30
58. Li Q, Chen P, Zeng Z. et al. Yeast two-hybrid screening identified WDR77 as a novel interacting partner of TSC22D2. Tumour Biol. 2016;37:12503-12
59. Wang Y, Mo Y, Gong Z. et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25
60. Chow S, Minden MD, Hedley DW. Constitutive phosphorylation of the S6 ribosomal protein via mTOR and ERK signaling in the peripheral blasts of acute leukemia patients. Exp Hematol. 2006;34:1183-91
61. Chan SM, Weng AP, Tibshirani R. et al. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278-86
62. Hou G, Xue L, Lu Z. et al. An activated mTOR/p70S6K signaling pathway in esophageal squamous cell carcinoma cell lines and inhibition of the pathway by rapamycin and siRNA against mTOR. Cancer Lett. 2007;253:236-48
Author contact
![]() Corresponding authors: zengzhaoyangedu.cn or guocdeedu.cn
Corresponding authors: zengzhaoyangedu.cn or guocdeedu.cn

 Global reach, higher impact
Global reach, higher impact