Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(10):2161-2168. doi:10.7150/jca.26727 This issue Cite
Research Paper
HLA Polymorphisms And TKI-Induced Liver Injury in Patients with Cancer: A Meta-analysis
1. Department of Pharmacy, Peking University First Hospital, No. 6, Dahongluochang Street, Xicheng District, Beijing 100034, China
2. Department of General Surgery, Peking University First Hospital, No. 8, Xishikuda Street, Xicheng District, Beijing 100034, China
Received 2018-4-17; Accepted 2019-2-23; Published 2019-5-21
Abstract
Background: Tyrosine kinase inhibitors (TKIs) are widely used for patients with cancer, although liver injury has been observed in a small proportion of these patients. The aim of this study was to investigate the mechanisms underlying TKI-induced liver injury according to HLA polymorphisms.
Methods: We systematically searched three electronic databases (PubMed, PMC, EmBase, and the Cochrane library) to identify studies that evaluated the impact of HLA polymorphisms on the incidence of TKI-induced liver injury in cancer patients as of November 2018. The summary odds ratios (ORs) with 95% confidence intervals (CIs) were calculated using a random-effects model. Further, sensitivity analysis and publication bias assessments were also performed.
Results: In the final analysis, four studies that involved a total of 12,181 patients were included. Three of the studies included patients who received lapatinib, and the other study included patients who received pazopanib. Overall, carriers of HLA-DQA1*02:01 were associated with an increased risk of liver injury (OR: 6.85; 95%CI: 2.34-20.02; P<0.001). Furthermore, HLA-DQB1*02:02 was correlated with a greater risk of liver injury (OR: 5.61; 95%CI: 2.80-11.25; P<0.001). Finally, HLA-DRB1*07:01 was significantly correlated with a greater risk of liver injury (OR: 4.06; 95%CI: 2.33-7.09; P<0.001).
Conclusions: The findings of this meta-analysis confirmed that three polymorphisms of HLA were significantly associated with an increased risk of TKI-induced liver injury. Further work will be required to determine these relationships in patients with specific characteristics.
Keywords: Tyrosine kinase inhibitors, liver injury, HLA, polymorphisms
Introduction
Adverse drug reactions (ADRs) are widely found in all hospital admissions, which account for 6.5% of all patients and occur in 15% of inpatients [1]. The burden of ADRs is large in terms of the number of beds and economic costs [3]. To date, ADRs are known to show a large inter-individual variation and have even caused life-threatening episodes or fatalities in some patients. The overall prevalence of serious and fatal ADRs was 6.7% and 0.32% in inpatients, respectively [4]. The reason for the inter-individual differences in ADRs could be environmental (including co-administered drugs, foods, smoking, drinking, and underlying disease) and genetic factors. Currently, ADRs mostly result from unintended pharmacological or non-pharmacological interactions, which were mediated by T cells, and the clinical symptoms vary from a mild delayed rash to a life-threatening cutaneous pathology, including systemic or organ disease [5].
Tyrosine kinase inhibitors (TKIs) have already been approved for the treatment of various cancers. Although TKI has an acceptable safety profile, grade 3 alanine aminotransferase (ALT) elevation and serious liver injury with hyperbilirubinemia were observed in 1.6% and 0.2% of patients, respectively [6]. Previous studies have already demonstrated that HLA allele-DRB1*07:01 was a risk factor for lapatinib-induced liver injury. However, although the impact of the HLA allele-DRB1*07:01 was strong and accounted for greater than 80% of lapatinib-induced liver injuries, only 10% of carriers have reported liver injury after receiving lapatinib for one year [7-9]. Those authors stated that this could be accounted for by HLA antigen presentation to CD4+ T cells and activation of an inflammatory immune response, which could cause hepatocyte damage.
Several studies have investigated the role of HLA polymorphisms (HLA-DQA1*02:01, HLA-DQB1*02:02, and HLA-DRB1*07:01) and the risk of TKI-induced liver injury [7,9-11]. Therefore, we conducted this meta-analysis to summarize available studies to determine associations between HLA polymorphisms and the risk of TKI-induced liver injury in patients with various cancers.
Methods
Data Sources, Search Strategy, and Selection Criteria
This review was conducted and reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis Statement issued in 2009 [12]. Any study that investigated the relationship between HLA polymorphisms and TKI-induced liver injury in patients with various cancers was eligible for inclusion, and no restrictions were placed on language or publication status (i.e., published, in press, or in progress). We systematically searched PubMed, PMC, EmBase, and the Cochrane library databases for studies published prior to November 2018 and used (“human leukocyte antigen” or “human leukocyte antigens” or “HLA”) AND (“afatinib” OR “alectinib” OR “axitinib” OR “bosutinib” OR “brigatinib” OR “cabozantinib” OR “ceritinib” OR “cobimetinib” OR “crizotinib” OR “dabrafenib” OR “dasatinib” OR “erlotinib” OR “everolimus” OR “gefitinib” OR “ibrutinib” OR “imatinib” OR “lapatinib” OR “lenvatinib” OR “neratinib” OR “nilotinib” OR “nintendanib” OR “osimertinib” OR “palbociclib” OR “pazopanib” OR “ponatinib” OR “regorafenib” OR “ribociclib” OR “ruxolitinib” OR “sirolimus” OR “sorafenib” OR “sunitinib” OR “temsirolimus” OR “tofacitinib” OR “trametinib” OR “vandetanib” OR “vemurafenib” OR “tyrosine kinase inhibitor” OR “TKI”) AND (“adverse drug reaction” OR “adverse event” OR “ADR”) as the search terms. Details of the search strategy used for PubMed are presented in Table S1. Reference lists of published studies and reviews were screened for additional references. The study topic, study design, exposure, disease status, intervention, and outcome variables of these studies were used to identify any potential included studies.
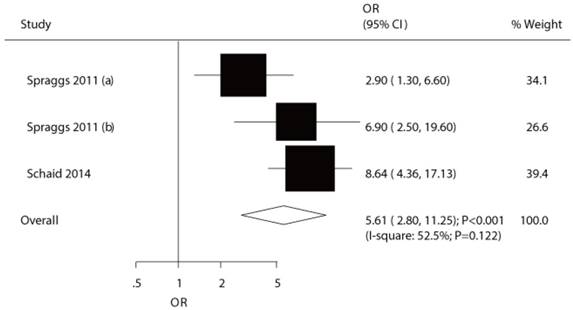
The literature search and study selection were carried out by two authors who independently used a standardized approach; any inconsistencies between these two authors were settled by group discussion until a consensus was reached. A study was eligible for inclusion if the following criteria were met: (1) the study had an observational design (cross-sectional, cohort, and case controlled study); (2) all patients received TKI therapies; (3) the study included patients with various cancers; (4) all studies reported HLA polymorphisms as exposure; and (5) all studies had to have reported the effect estimate of TKI-induced liver injury. Exclusion criteria are listed below: (1) duplicated studies; (2) reviews, case report, communication, and comments; (3) studies reported no desirable data; and (4) studies included duplicate populations.
Data Collection and Quality Assessment
Data extraction was conducted using a standardized data extraction sheet by two authors; any inconsistencies were examined and adjudicated independently by an additional author who referred to the original studies. Data collected included the first author's name, publication year, sample size, mean age, number of men and women, number of Caucasians and Asians, cancer sites, intervention, and reported outcomes. The Newcastle-Ottawa Scale was employed to assess methodological quality, which is comprehensive and has been partially validated for evaluating the quality of observational studies [13]. This method consisted of selection (4 items), comparability (1 item), and outcomes (3 items).
Statistical Analysis
We examined the relationship between HLA polymorphisms and risk of TKI-induced liver injury based on the effect estimate and 95% CI published in each study. The summary odds ratios were calculated to investigate the impact of HLA polymorphisms on the risk of TKI-induced liver injury in various cancers using random-effects models [14,15]. Heterogeneity among included studies was evaluated using the Q statistic; P-values < 0.10 were considered to indicate significant heterogeneity [16,17]. Sensitivity analysis was conducted by removing each individual study from the overall analysis [18]. Subgroup analysis was not conducted because a small number of studies were included. Funnel plots, Egger tests, and Begg tests were calculated to evaluate publication biases [19,20]. If significant publication bias was observed, the trim and fill method was used to calculate the adjusted outcomes [21]. All reported P-values were two-sided, and those P-values that were less than 0.05 were considered to be statistically significant. Statistical analyses were performed using STATA software (version 12.0; Stata Corporation, College Station, TX, USA).
Results
Literature Search
The results of the study selection process are shown in Figure 1. Our initial search identified 1489 publications that reported an association of HLA alleles and adverse reactions to TKI agents. After screening the titles, 224 duplicate studies were excluded. A further 1244 publications were excluded after scanning the abstracts and a further 17 publications were excluded after reviewing the full study reports. Ultimately, 4 studies were included in our final meta-analysis [7,9-11]. The eligible studies included discovery and confirmatory analysis using a dataset from previous trials. A manual search of the reference lists of these studies did not yield any new eligible studies. General characteristics of the included studies are presented in Table 1.
Study Characteristics
The 4 studies included a total of 12,181 patients with various cancers. All of the included studies were published between 2011 and 2017, and 502-8381 individuals were included in each study. Three studies included patients with breast cancer [7,9,11], and the other study included patients with various cancers [10]. Furthermore, three studies included patients who received lapatinib [7,9,11], and one study included patients who received pazopanib [10]. Two of the included studies had a score of 8 [9,11], while the remaining two studies had a score of 7 [7,10].
Baseline characteristic of studies included in the systematic review and meta-analysis
| First author | Publication year | Sample size | Mean age (years) | Men/Women | Caucasian/Asian | Cancer sites | Intervention | NOS score |
|---|---|---|---|---|---|---|---|---|
| Xu [10] | 2015 | 2190 | 58 | 916/2174 | 1655/439 | Various | Pazopanib | 7 |
| Spraggs [11] | 2017 | 8381 | NA | 0/8381 | NA | Breast cancer | Lapatinib | 8 |
| Spraggs [7] | 2011 | 502 | 56.8 | 0/512 | 366/118 | Breast cancer | Lapatinib | 7 |
| Schaid [9] | 2014 | 1108 | 51.7 | 0/1108 | 782/288 | Breast cancer | Lapatinib | 8 |
Flow diagram of the literature search and trial selection process.
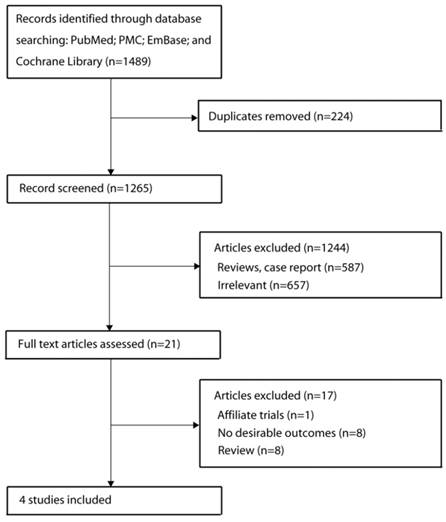
The relationship between HLA-DQA1*02:01 and TKI-induced liver injury.
HLA-DQA1*02:01
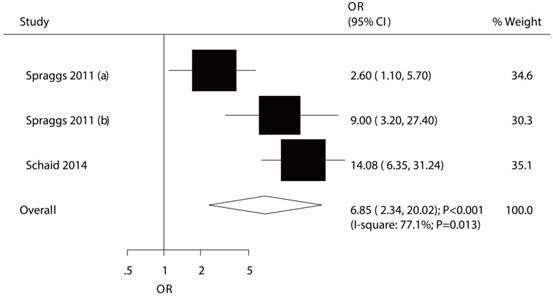
The association between HLA-DQA1*02:01 and TKI-induced liver injury was studied in a total of 3 cohorts in 2 studies [7,9]. The study conducted by Spraggs et al indicated significant increased risk of TKI-induced liver injury in patient's carrier HLA-DQA1*02:01 in exploratory study (OR: 2.60) and confirmatory study (OR: 9.00) [7]. Further, Schaid et al indicated HLA-DQA1*02:01 carriage as a predictor of lapatinib-induced liver injury as compared with HLA-DQA1*02:01 noncarrier (OR: 14.08) [9]. The summary OR showed that HLA-DQA1*02:01 was associated with an increased risk of TKI-induced liver injury (OR: 6.85; 95%CI: 2.34-20.02; P<0.001; Figure 2), but potential evidence of significant heterogeneity was observed (P = 0.013). As a result, a sensitivity analysis was conducted and after each study was sequentially excluded, the conclusion was not affected (Figure S1). Finally, no significant publication bias was detected across the included studies (P-value for Egger: 0.937; P-value for Begg: 1.000; Figure 3).
HLA-DQB1*02:02
The potential association between HLA-DQB1*02:02 and TKI-induced liver injury was investigated in a total of 3 cohorts in 2 studies [7,9]. Both significant differences were observed for exploratory study (OR: 2.90) and confirmatory study (OR: 6.90) in the study conducted by Spraggs et al [7]. Moreover, Schaid et al indicated the risk of lapatinib-induced liver injury was significantly increased in patient's carrier HLA-DQB1*02:02 (OR: 8.64) [9]. We noted that HLA-DQB1*02:02 was associated with an increased risk of TKI-induced liver injury (OR: 5.61; 95%CI: 2.80-11.25; P<0.001; Figure 4), and moderate heterogeneity was observed (P=0.122). After the sequential exclusion of each study from all of the pooled analyses, the conclusion was unaffected (Figure S2). No significant publication bias was detected across the included studies (P-value for Egger: 0.838; P-value for Begg: 1.000; Figure 3).
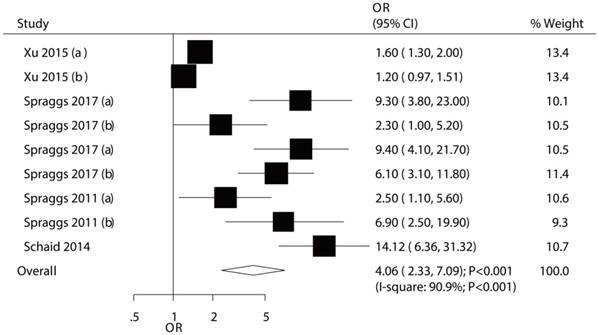
HLA-DRB1*07:01
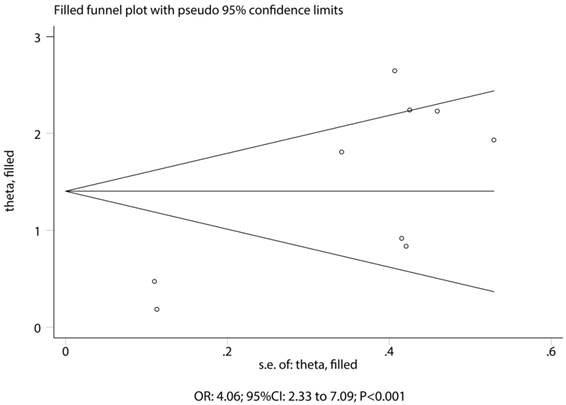
The association between HLA-DRB1*07:01 and TKI-induced liver injury was investigated in a total of 9 cohorts in 4 studies [7,9-11]. All of included cohorts reported significantly associations between HLA-DRB1*07:01 and TKI-induced liver injury except the confirmatory study conducted by Xu et al [10]. HLA-DRB1*07:01 was associated with an increased risk of TKI-induced liver injury (OR: 4.06; 95%CI: 2.33-7.09; P<0.001; Figure 5). Heterogeneity was observed in the magnitude of the effect across trials (P<0.001); however, the conclusion was not affected after the sequential exclusion of each study from the pooled analyses (Figure S3). Although the Begg test (P=0.466; Figure 3) showed no significant publication bias, significant publication bias was detected using the Egger test (P=0.002; Figure 3). These conclusions were not changed after adjusting for publication bias using the trim and fill method (Figure 6) [21].
Discussion
This is first meta-analysis to evaluate the potential role of HLA polymorphisms and risk of TKI-induced liver injury. This meta-analysis was based on observational studies and investigated any potential relationship between HLA polymorphisms and the risk of TKI-induced liver injury. This comprehensive quantitative study included 12,181 patients with various cancers from 4 individual studies with broad range patient characteristics. The findings from this meta-analysis suggest that each of the three genotypes in HLA polymorphisms were correlated with an increased risk of TKI-induced liver injury. None of the results changed upon excluding each individual study. This finding could help determine the specific safety profile of each drug based on its predicted treatment effects derived from host genomic data.
Spraggs et al found that breast cancer patients who were homozygous carriers of HLA-DRB1*07:01 were associated with an increased risk and severity of lapatinib-induced aminotransferase elevation compared with subjects who were heterozygous carriers of HLA-DRB1*07:01 [11]. The reasons for this association were that perturbation of HLA-mediated presentation of antigens by lapatinib could have caused immune-mediated hepatocyte injury [22,23]. Furthermore, HLA-DRB1*07:01 protein could link CD4+ T-cell activity to the initiation of hepatic cell damage. In addition, Spraggs et al indicated that lapatinib could induce adaptive immune responses, and metabolites of lapatinib could bind covalently to proteins to form haptens, which could result in T-cell driven immune activation and inflammatory tissue damage [7,24,25]. Finally, in breast cancer patients, Schaid et al concluded that HLA-DRB1*07:01 could regarded as a predictor of the risk of lapatinib-induced liver injury, suggesting an immune pathology [9]. They point out that HLA-DRB1*07:01 or DQA1*02:01 protein isoforms could present antigens to CD4+ T-cell that were associated with an immune-mediated attack on hepatocytes [6].
Publication biases for the outcomes that were investigated.
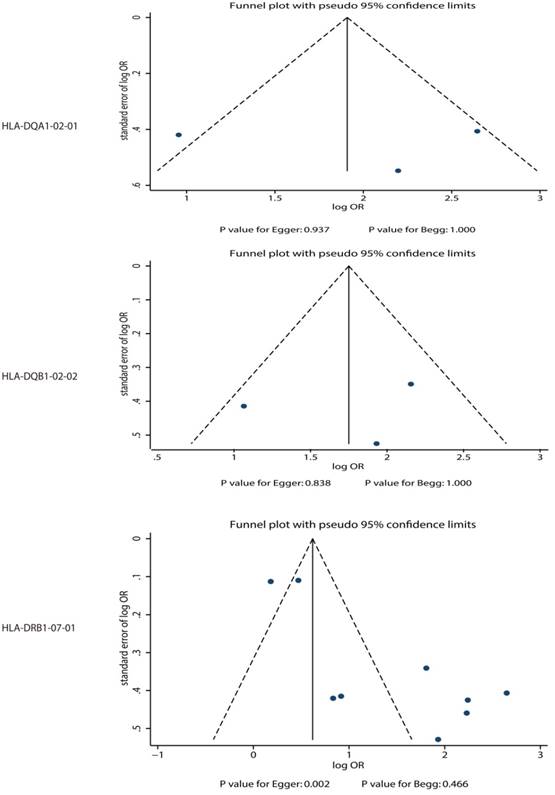
The relationship between HLA-DQB1*02:02 and TKI-induced liver injury.
The relationship between HLA-DRB1*07:01 and TKI-induced liver injury.
Trim and fill plot for the relationship between HLA-DRB1*07:01 and TKI-induced liver injury.
Other HLA polymorphisms associated with TKI-induced liver injury have been reported. Those HLA polymorphisms were not included in our meta-analysis because only one study reported such relationships. For example, Xu et al included discovery and confirmatory analysis to show the impact of four-digit HLA alleles on alanine aminotransferase elevation in pazopanib-treated patients with various cancers. They found that HLA-B*57:01 conferred a greater risk of alanine aminotransferase elevation in patients who received pazopanib than in non-carriers [10]. They suggest that HLA-B*57:01 carriers should be excluded as recipients of pazopanib.
This meta-analysis has some limitations. First, a relatively small number of studies were included, and stratified analysis was not conducted. Second, the study included patients who received various drugs and had cancer at different sites. Third, publication bias was observed as this meta-analysis was based on published studies. Fourth, although three of the included studies reported the number of Caucasians and Asians, the impact of HLA polymorphisms on TKI-induced liver injury according to ethnicity was not assessed in all of the included studies. Finally, details of the stratified analyses were not evaluated because pooled data were used.
In conclusion, the results of this meta-analysis validated that the HLA alleles HLA-DQA1*02:01, HLA-DQB1*02:02, and HLA-DRB1*07:01 significantly contribute to the risk of TKI-induced liver injury. This result could support the clinical treatment of patients with TKI-induced liver injury. Future studies should be conducted to validate the predicted value of HLA polymorphisms on liver injury.
Abbreviations
TKIs: tyrosine kinase inhibitors; ORs: odds ratios; CIs: confidence intervals; ADRs: adverse drug reactions; ALT: alanine aminotransferase.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by grants from the National Key Technologies R&D Program (No. 2016YFC0904900), National Natural Science Foundation of China (No. 81573504 and No. 81673509), Beijing Natural Science Foundation (No. 7171012) and National Science and Technology Major Projects for "Major New Drugs Innovation and Development" (No. 2017ZX09304028 and No. 2017ZX09101001).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Pirmohamed M, James S, Meakin S, Green C, Scott AK, Walley TJ, Farrar K, Park BK, Breckenridge AM. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18,820 patients. BMJ. 2004;329:15-19
2. Davies EC, Green CF, Taylor S, Williamson PR, Mottram DR, Pirmohamed M. Adverse drug reactions in hospital in- patients: a prospective analysis of 3,695 patient- episodes. PLoS One. 2009;4:e4439
3. Davies EC, Green CF, Mottram DR, Pirmohamed M. Adverse drug reactions in hospitals: a narrative review. Curr Drug Saf. 2007;2:79-87
4. Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279:1200-1205
5. Fan WL, Shiao MS, Hui RC, Su SC, Wang CW, Chang YC, Chung WH. HLA Association with Drug-Induced Adverse Reactions. J Immunol Res. 2017;2017:3186328
6. Moy B, Rappold E, Williams L, Kelly T, Nicolodi L. Hepatobiliary abnormalities in patients with metastatic cancer treated with lapatinib. J Clin Oncol. 2009;27:1043
7. Spraggs CF, Budde LR, Briley LP, Bing N, Cox CJ, King KS, Whittaker JC, Mooser VE, Preston AJ, Stein SH. et al. HLA-DQA1*02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J Clin Oncol. 2011;29:667-673
8. Spraggs CF, Parham LR, Hunt CM, Dollery CT. Lapatinib-Induced Liver Injury Characterized by Class II HLA and Gilbert's Syndrome Genotypes. Clin Pharmacol Ther. 2012;91:647-652
9. Schaid DJ, Spraggs CF, McDonnell SK, Parham LR, Cox CJ, Ejlertsen B, Finkelstein DM, Rappold E, Curran J, Cardon LR. et al. Prospective validation of HLADRB1*07:01 allele carriage as a predictive risk factor for lapatinib-induced liver injury. J Clin Oncol. 2014;32:2296-303
10. Xu CF, Johnson T, Wang X, Carpenter C, Graves AP, Warren L, Xue Z, King KS, Fraser DJ, Stinnett S. et al. HLA-B*57:01 Confers Susceptibility to Pazopanib Associated Liver Injury in Patients with Cancer. Clin Cancer Res. 2015;22:1371-7
11. Spraggs CF, Parham LR, Briley LP, Warren L, Williams LS, Fraser DJ, Jiang Z, Aziz Z, Ahmed S, Demetriou G. et al. Characterisation of the HLA-DRB1*07:01 biomarker for lapatinib-induced liver toxicity during treatment of early-stage breast cancer patients with lapatinib in combination with trastuzumab and/or taxanes. Pharmacogenomics J. 2018;18:480-486
12. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Plos Med. 2009;6:e1000097
13. Wells G, Shea B, O'Connell D. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Ottawa (ON): Ottawa Hospital Research Institute. 2009 Available:http://www.ohri.ca/programs/clinical_ epidemiology/oxford.htm
14. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-88
15. Ades AE, Lu G, Higgins JP. The interpretation of random-effects metaanalysis in decision models. Med Decis Making. 2005;25:646-54
16. Deeks JJ, Higgins JPT, Altman DG. Analyzing data and undertaking meta-analyses. In: Higgins J, Green S, eds. Cochrane Handbook for Systematic Reviews of Interventions 5.0.1. Oxford, UK: The Cochrane Collaboration. 2008 chap: 9
17. Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-60
18. Tobias A. Assessing the influence of a single study in meta-analysis. Stata Tech Bull. 1999;47:15-17
19. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-34
20. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088-1101
21. Duvall S, Tweedie R. A nonparametric “trim and fill” method for assessing publication bias in meta-analysis. J Am Stat Assoc. 2000;95:89-98
22. Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, Miles JJ, Kjer-Nielsen L, Gras S, Williamson NA. et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554-558
23. Monshi MM, Faulkner L, Gibson A, Jenkins RE, Farrell J, Earnshaw CJ, Alfirevic A, Cederbrant K, Daly AK, French N. et al. Human leukocyte antigen (HLA)-B*57:01- restricted activation of drug-specific T cells provides the immunological basis for flucloxacillin-induced liver injury. Hepatology. 2013;57:727-739
24. Andrade RJ, Lucena MI, Alonso A, García-Cortes M, García-Ruiz E, Benitez R, Fernández MC, Pelaez G, Romero M, Corpas R. et al. HLA class II genotype influences the type of liver injury in drug-induced idiosyncratic liver disease. Hepatology. 2004;39:1603-1612
25. Kaplowitz N. Idiosyncratic drug hepatotoxicity. Nat Rev Drug Discov. 2005;4:489-499
Author contact
![]() Corresponding author: Yimin Cui, Department of Pharmacy, Peking University First Hospital, No. 6, Dahongluochang Street, Xicheng District, Beijing 100034, China. Telphone: + 86-10-66110802; Fax: +86-10-66110802; Email address: cuiymcom
Corresponding author: Yimin Cui, Department of Pharmacy, Peking University First Hospital, No. 6, Dahongluochang Street, Xicheng District, Beijing 100034, China. Telphone: + 86-10-66110802; Fax: +86-10-66110802; Email address: cuiymcom

 Global reach, higher impact
Global reach, higher impact