Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(18):3417-3426. doi:10.7150/jca.24591 This issue Cite
Review
The Significance of Liquid Biopsy in Pancreatic Cancer
1. Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center, Shanghai 200032, P.R. China
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, P.R. China
3. Shanghai Pancreatic Cancer Institute, Shanghai 200032, P.R. China
4. Pancreatic Cancer Institute, Fudan University, Shanghai 200032, P.R. China
*Zi-Hao Qi and Hua-Xiang Xu contributed equally to this work.
Received 2017-12-27; Accepted 2018-6-16; Published 2018-9-8
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal types of cancer. The 5-year survival rate for PDAC remains low because it is always diagnosed at an advanced stage and it is resistant to therapy. A biomarker, which could detect asymptomatic premalignant or early malignant tumors and predict the response to treatment, will benefit patients with PDAC. However, traditional biopsy has its limitations. There is an urgent need for a tumor biomarker that could easily and repeatedly sample and monitor, in real time, the progress of tumor development. Liquid biopsy could be a tool to assess potential biomarkers. In this review, we focused on the latest discoveries and advancements of liquid biopsy technology in pancreatic cancer research and demonstrated how this technology is being used in clinical applications.
Keywords: Circulating tumor cells, Cell-free nucleic acid, Exosomes, Liquid biopsy, Pancreatic cancer
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal types of cancer. Over the past 30 years, although improvements have been developed in the detection and treatment of this disease, the 5-year survival rate remains 8%. This low survival rate is mainly because PDAC is usually diagnosed at an advanced stage and is resistant to therapy.[1-4] Most of the patients die from extensive metastatic disease.[5] A biomarker that could detect asymptomatic premalignant or early malignant tumors and predict the response to treatment will greatly benefit these patients. However, the widely used carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) in clinical practice have limited sensitivities and specificities.[6-8]
Because of tumor heterogeneity, a single biopsy sample is very limited, and only provides spatially and temporally limited information of a tumor.[9-15] S There is an urgent need for a better tumor biomarker that could be correlated with the tumor burden, correlated with changes in response to treatment or surgery, could reflect tumor progression and metastasis, and could indicate the response to treatment and tumor resistance or recurrence.[16-19] In addition, easy and repeated sampling should be an important characteristic of this biomarker.[17,19-23] Liquid biopsy is a method to diagnose cancer by detecting the circulating tumor cells (CTCs), cell-free circulating nucleic acids (cfNAs), and microvesicles such as exosomes containing nucleic acids and proteins, which are released into the body fluid, usually into the blood, from the primary or metastatic tumor. Unlike open surgical biopsy and needle biopsy, liquid biopsy is a noninvasive method for tumor diagnosis. It results in real time detection, which provides an effective prognosis (Figure 1).[20, 22-26]
The difference between liquid biopsy and traditional biopsy.
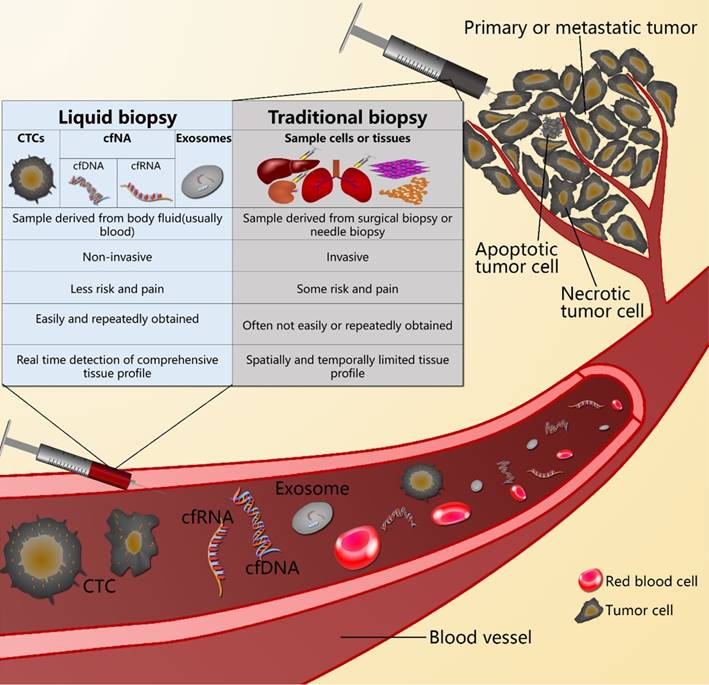
This review focused on the latest discoveries and advancements of liquid biopsy technology in pancreatic cancer research, and demonstrated the potential applications of this technology in clinical applications, including the early detection of pancreatic cancer, the assessment of prognosis, predicting response to treatments, the development of acquired resistance, and the early detection of disease recurrence.
2. Current types of liquid biopsy in pancreatic cancer
2.1. Circulating tumor cells
CTCs are cells that are released into peripheral blood from tumors of patients with all major solid carcinomas before metastasis. CTCs can be shed from the tumor in the early stages during tumor formation and development.[27] CTCs are rare, comprising as few as one cell per 109 hemocytes in the blood of cancer patients. But the number of CTCs in the blood of cancer patients is thought to be correlated with tumor development, treatment response, tumor recurrence, and long-term prognosis for many cancers.[27, 28] However, its role in pancreatic cancer has not yet been clearly determined, so it is an extensive field of research in pancreatic cancer.
2.2. Cell-free nucleic acid
Cell-free nucleic acid (cfNA) includes cell-free DNA (cfDNA) and cell-free RNA (cfRNA). It contains information that can be used to examine total cfDNA levels, microsatellite instability, cancer-associated DNA methylation and integrity, loss of heterozygosity, somatic mutations, and expression levels of mRNAs, miRNAs, and other non-coding RNAs in the blood of cancer patients. The cfNAs can be released into the circulation by apoptosis, necrosis, and autophagy of the cells, or secreted by packaging into extracellular vesicles.[29-34] Circulating tumor DNA (ctDNA) is a special kind of DNA fragment released into the blood by apoptosis and necrosis of tumor cells. Unlike CTCs, ctDNAs are present at high concentrations in the blood of cancer patients. They are comprised of genomic DNA, mitochondrial DNA, and non-coding gDNA. The ctDNAs that originate from tumor cells can detect tumors, reflect tumor burden, and monitor response to therapies.[33, 35] It was found that a significant amount of cfNA in the plasma of pancreatic cancer patients is probably derived from apoptotic or necrotic tumor cells, and the cfNA differed between pancreatic cancer patients and healthy controls.[36-39]
2.3. Exosomes
Exosomes are membrane vesicles of endocytic origin ranging in size from 30-150 nm. They are secreted by both normal and cancerous cells into the extracellular space and into the blood circulation. These circulating extracellular vesicles are emerging as key players in intercellular communication between cancer cells and their microenvironment via horizontal transfer of information of their cargos, which include protein, DNA, mRNA, and miRNA. [40, 41] Compared with ctDNA, exosomes can be secreted by living cells. As a result, exosomes are present in the blood earlier than necrosis-caused release of cfNAs, and can be found in the early stages of tumors.[42]
3. Clinical application of liquid biopsy in pancreatic cancer
3.1. Early diagnoses and differential diagnoses
Pancreatic cancer cells were found be present in the circulation of patients before tumors could be detected. Circulating pancreas epithelial cells (CECs) were found only in patients with cystic lesions and pancreatic ductal adenocarcinoma. Moreover, the CEC count was significantly different across healthy donors, patients with PDAC, and patients with cystic lesions.[43] CTCs could be found in most patients with PDAC of any stage, whether localized, locally advanced, or metastatic; but no CTCs were detected in blood from healthy donors.[44] With different isolation and detection methods, the detection rates of CTCs in PDAC patients were in the range of 21%-100%, while it was nearly 0% in benign patients or healthy controls (Table 1).[43, 45-50] All of these studies suggested that CTCs could be used as markers in the early diagnosis and differential diagnosis of pancreatic cancer. Moreover, a combination of CA19-9 and CTC detection increased the detection pancreatic cancers.[49]
Circulating tumor cells in the diagnosis of pancreatic cancer.
| Isolation method | Detection method | positive criteria | Patient/control | Detection rate | Ref. |
|---|---|---|---|---|---|
| GEDI:Anti-EpCAM | DAPI+/ CD45-/CK+ | ≥3 cells/ml | 11 patients with PDAC of all stages | 8/11(73%) | [43] |
| 21 patients with cystic lesions of the pancreas | 7/21(33%) | ||||
| 19 patients without cysts or cancer | 0/19(0%) | ||||
| ScreenCell: size-based filtration | Cytological detection and detection of KRAS mutations | ≥2 cells/mL or KRAS mutations positive | 11 patients with pancreatic cancer | 8/11(73%) | [44] |
| 9 healthy controls | 0/9(0%) | ||||
| MACS:Anti-EpCAM | RT-PCR: h-TERT, CK20, CEA, C-MET. | Amplification of at least one tumor-associated marker mRNA | 25 pancreatic cancer patients | 20-25/25(80-100%) | [47] |
| 15 benign patients | 0-1/15(0-6.8%) | ||||
| Immunomagnetic enrichment: Anti-EpCAM/mucin1 | RT-PCR: KRT19, MUC1, EPCAM, CEACAM5, BIRC5. | Amplification of at least one tumor-associated marker mRNA | 34 pancreatic cancer patients before systemic therapy | 7/34(21%) | [48] |
| 40 healthy controls | 0/40(0%) | ||||
| CellSearch/Anti-EpCAM | DAPI+/ CD45-/CK+ /EpCAM+ | ≥1 cells/ml | 35 patients with PDAC of all stages | 7/35 (20%) | [45] |
| Immunol staining and FISH | DAPI+/ CD45-/CK+ /CEP8+ | ≥2 cells/ml | 33 patients with PDAC of all stages | 26/33(79%) | [49] |
| 3 solid pseudopapillary pancreatic tumor | 2/3(66%) | ||||
| 14 benign pancreatic tumor | 1/14(7%) | ||||
| 40 healthy individuals | 0/40(0%) | ||||
| Immunol staining and FISH | CD45-/DAPI+/CEP8>2 | ≥2 cells/3.2 mL | 95 patients with pancreatic cancer | 83/95(87%) | [50] |
| 45 healthy controls | 29/48(60%) |
GEDI: geometrically enhanced differential immunocapture; EpCAM: epithelial cell adhesion molecule;
MACS: Magnetic-activated cell separation
Early in 1998, it was found that cfDNAs in human blood plasma were different in patients with pancreatic cancer versus healthy controls. The cfDNAs had diagnostic values in PDAC patients, and could be useful in patients without detectable CTCs.[38] The total load of cfDNAs was higher in PDAC patients than that in patients with pancreatic neuroendocrine tumors or chronic pancreatitis [51], and different in patients with different stage tumors.[36] Previous studies also found mutated gene fragments present in the blood of cancer patients.[34] Zill et al. analyzed 54 genes in PDAC patients, and found that nearly 90% mutations of these genes in tumor biopsies were also detectable in the cfDNAs. They suggested that cfDNAs could be diagnostic markers with high sensitivity and specificity.[52] As one of the most important driven gene, K-RAS mutations have also been found frequently in pancreatic cancer patients, and were detected in nearly half of the PDAC patients in their plasma DNA.[53] Nearly no K-RAS mutations were detected in healthy control plasma. K-RAS Mutations in ctDNA can be an early diagnostic biomarkers.[54-56] In a study by Finkelstein analyzing biliary specimens, it was found that biliary specimens were enriched with DNA. Mutation analyses using these cfDNAs targeted 17 genomic sites (including K-RAS) associated with pancreaticobiliary cancer, showing mutations existed in most patients with malignancy, with no mutations found in benign control patients. In these 17 genomic sites, K-RAS mutations only occurred in patients with pancreatic cancer. Mutational profiling of cfDNA in biliary specimens could be also used as a biomarker with high sensitivity and specificity for PDAC patients.[55]
As a kind of important part of cfNAs, miRNA was proved to be as a sensitive and specific body fluid based biomarker for pancreatic cancer. (Table 2) Serum miRNAs were differentially found in pancreatic cancer patients compared with controls.[57] On the one hand, circulating miRNAs could therefore be used in the early diagnosis of pancreatic cancer, and the miR-155, miR-486-5p, miR-938, miR-744 and miR-223 levels were significantly higher in pancreatic cancer patients than healthy controls.[39, 58-60] On the other hand, circulating miRNAs could be used in differential diagnosis. Plasma miR-223 tended to discriminate the malignant potential between benign intraductal papillary mucinous neoplasm (IPMN) and malignant IPMN, and the progressive extent of invasiveness between malignant IPMN and PDAC.[60] miR-486-5p and miR-938 were able to discriminate PDAC patients from healthy controls and those with chronic pancreatitis.[58] Patients with pancreatic cancer and multifocal PanIN2/3 lesions had significantly higher serum levels of miR-196a and miR-196b than patients with PanIN1, pancreatic neuroendocrine tumors, chronic pancreatitis, or healthy controls.[61] In contrast to single miRNAs, a combined panel of single miRNAs may have more potential value.[62] In the study by Lee et al., they found that a combination of both miR-196a and miR-196b reached a sensitivity of 100% and specificity of 90% in diagnosing PC or high grade PanIN lesions.[61] And, a combination of four miRNAs (miR-21, miR-210, miR-155, and miR-196a) reached a sensitivity of 64% and a specificity of 89% to diagnose pancreatic cancer.[59] Not only in serum or plasma, but also in other body fluids, miRNAs have revealed their potential value. Elevated levels of the four miRNAs (miR-205, miR-210, miR-492, and miR-1247), together in pancreatic juice, predicted PDAC with a specificity of 88% and sensitivity of 87%.[63] In saliva samples, hsa-miR-21, hsa-miR-23a, hsa-miR-23b, and miR-29c were significantly up-regulated in the saliva of pancreatic cancer patients compared with control patients. [64] Even in stool samples, it was found that PDAC patients had significantly higher miR-21 and miR-155, and lower miR-216 levels compared to normal controls or chronic pancreatitis patients. A combination of miR-21 and miR-155 had a sensitivity of 93.33%, while a combination of miR-21, miR-155, and miR-216 would be best for detecting PDAC with a better balance of sensitivity and specificity.[65]
The candidate miRNA from body fluid in the diagnose of pancreatic cancer.
| Sample source | Patient/control characteristics and size | miRNA | Ref. |
|---|---|---|---|
| Peripheral blood | 94 PDAC patients 68 healthy controls | miR-744 | [39] |
| Peripheral blood | 197 PDAC patients 158 cancer-free controls | miR-20a, miR-21, miR-24, miR-25, miR-99a, miR-185, miR-191. | [57] |
| Peripheral blood | 192 PDAC patients 267 cancer-free controls | miR-486-5p, miR-938 | [58] |
| Peripheral blood | 39 PDAC patients 29 healthy controls | miR-155 | [59] |
| Peripheral blood | 71 PDAC patients 67 healthy controls | miR-223 | [60] |
| Peripheral blood | 19 PDAC patients 25 cancer-free controls | miR-196a, miR-196b | [61] |
| Pancreatic juice | 50 PDAC patients 38 cancer-free controls | miR-205, miR-210, miR-492, miR-1247 | [63] |
| Saliva | 7 PDAC patients 10 cancer-free controls | miR-21, miR-23a, miR-23b, miR-29c | [64] |
| Stool | 30 PDAC patients 25 cancer-free controls | miR-21, miR-155, miR-216 | [65] |
Cancer patients exhibit a significantly higher quantity of total exosomes than healthy individuals, because exosomes are secreted in large amounts during carcinogenesis. Compared with ctDNAs, exosomes can be secreted by living cells. This results in exosomes being present in blood earlier than necrosis-caused release of cell-free miRNAs, so they can be found in the early stages of tumors.[42] Serum-exosome protein or miRNA markers might be suitable candidates for the diagnosis of PDAC.[66, 67] A panel of pancreatic cancer-initiating cell protein markers (CD44v6, Tspan8, EPCAM, and CD104) were selected according to expression in pancreatic cancer-derived exosomes. The miR-1246, miR-4644, miR-3976, and miR-4306 were selected according to abundant recovery in microarrays of patients with pancreatic cancer. The selected panel of protein markers and miRNAs were significantly upregulated in most of pancreatic cancer serum-exosomes, but rarely found in healthy donors and patients with nonmalignant diseases. Thus, combined evaluation with a panel of protein and miRNA serum-exosome biomarkers could serve as a highly sensitive, minimally invasive pancreatic cancer diagnostic tool.[68] Exosomes contain RNA and protein, and a previous study investigated whether exosomes from pancreatic cancer cells and serum from patients with pancreatic ductal adenocarcinoma contained genomic DNA. Their results showed that exosomes contained >10 kb fragments of double-stranded genomic DNA. Mutations in K-RAS and p53 have been detected using genomic DNA from exosomes derived from pancreatic cancer cell lines and serum from patients with pancreatic cancer. These results indicated that serum-derived exosomes can be used to determine genomic DNA mutations for cancer diagnoses. It is a distinct source of tumor DNA that may be complementary to other liquid biopsy DNA sources.[69, 70] A recent study reported that a kind of protein named GPC1, located in the membranes of exosomes, could be used as a specific marker of cancer exosomes. Exosomes from 190 patients with PDAC exhibited higher levels of GPC1 than that in healthy donors. GPC1 positive exosomes were found before magnetic resonance imaging of detectable masses. The levels of GPC1 positive exosomes can distinguish patients with stage I-IV pancreatic cancer as compared with healthy donors, patients with pancreatic cancer precursor lesions, and patients with benign pancreatic disease. The receiver operating curves showed that GPC1 positive exosomes were nearly perfect markers with a sensitivity and specificity of 100%.[71] All these studies suggested that exosomes could be a biomarker in the early diagnosis and differential diagnosis of pancreatic cancer.
3.2. Predicting response to treatments and assessment of prognosis
Because CTCs counts may be a surrogate of cancer metastatic, prognostic values of CTC were studied by many studies. These studies found the presence or absence of CTCs can be an independent prognostic factor. CTC positivity either before treatment or after treatment was associated with poor overall survival (OS).[72-74] CTCs can also be used as a biomarker of response to treatment. In patients with fluorouracil chemotherapy, the detection rate of CTCs dramatic declined after the first cycle of therapy. And, apoptotic CTCs could be detected in these patients. These apoptotic CTCs could be used to monitor the efficacy of chemotherapy in pancreatic cancer patients.[75, 76] Serial monitoring of the CTC burden may therefore be used as a minimally invasive approach to predict and monitor treatment response for guided therapeutic regimens. Recently study found that, not only CTC numbers but also the CTCs phenotype represents a useful tool for predicting prognoses and therapeutic responses to therapy among patients with pancreatic cancer. CTCs expressing markers of tumor-initiating cells, such as CD133 and CD44, were significantly associated with worse survival.[77]
The candidate miRNA from body fluid as prognostic and predictive markers for pancreatic cancer.
| Sample source | miRNA | Prognostic | Predictive of treatment efficacy | Ref. |
|---|---|---|---|---|
| Peripheral blood | miR-21 (↑) | (-) | (-) | [57] |
| Pancreatic juice | miR-205 (↑), miR-210 (↑), miR-492 (↑), miR-1247 (↑) | (-) | N.A | [63] |
| Peripheral blood | miR-196a (↑), miR-196b (↑) | (-) | (-) | [61] |
| Peripheral blood | miR-744 | (-) | (-) | [39] |
| Peripheral blood | miR-21 (↑), miR-210 (↑), miR-221 (↑), miR-7 (↑) | (-) | (-) | [84] |
| Plasma exosome | miR-451a (↑) | (-) | N.A | [85] |
(↑): Upregulated; (-): Bad prognosis/response to treatment; N.A: Not available.
Numerous studies have investigated the prognostic potential of cfNAs. In recent studies about ctDNA, it was found that ctDNA could be used as an independent prognostic marker for monitoring treatment efficacy and disease progression in pancreatic cancer patients. Furthermore, it arises as an indicator of shorter survival in resected or metastatic patients when detected after surgery or chemotherapy.[78-80]Among the patients who were ctDNA positive before chemotherapy, 90% experienced disease progression during follow-up, compared to 25% ctDNA negative patients.[81] Higher levels of plasma ctDNA were significantly associated with lower overall survival times.[82] The survival of patients with K-RAS mutations in ctDNAs was significantly shorter than that of patients without mutations.[45, 79] The difference was especially evident in patients with a G12V mutation.[83]
Some plasma miRNAs have recently been used to predict response to treatments and assess the prognosis for pancreatic cancer (Table 3).[84,85] The miR-21 levels in serum were significantly associated with overall survival of pancreatic cancer patients.[57] Elevated levels of circulating miR-205, miR-210, miR-492, and miR-1247 in pancreatic fluids were associated with decreased overall survival.[63] The presence of miR-196a and miR-196b in patients with pancreatic cancer predicted a curative resection if their levels dropped to normal levels after surgery[61], and the levels of miR-744 were significantly reduced in postoperative samples. A high level of plasma miR-744 in postoperative pancreatic cancer patients indicated a poor prognosis, which was correlated with lymph node metastasis, recurrences, and chemotherapy-resistance.[39]
Tumor exosomes have a supportive role in the survival and growth of tumor cells and are involved in promoting host tissue invasion and subsequent metastasis and evasion from the immune response. Zech et al. reported that exosomes from the rat pancreatic adenocarcinoma lines BSp73ASML (ASML) bind to and are taken up by all leukocyte subpopulations in vivo and in vitro. ASML exosomes can affect leukocyte proliferation via reduced CD44v6, ZAP70, and ERK1/2 phosphorylation. [86] The role of exosomes in the immune system has been the objective of additional studies. Another study reported that pancreatic cancer-derived exosomes downregulated TLR4 and downstream cytokines in dendritic cells via miR-203.[87] However, some studies have reported a possible apoptotic function of tumor-derived nanoparticles on tumor cells. Human pancreatic tumor nanoparticles counteracted the constitutively activated PI3K/AKT survival pathway to induce apoptosis of tumor cells.[88] However, in pancreatic cancer, there has been no definitive clinical study about the role of exosomes in the prognosis of pancreatic cancer. Another study of pancreatic cancer suggested that the protein and mRNA of four inhibitors of apoptosis (IAPs)—survivin, XIAP, cIAP1, and cIAP2—were found in exosomes, and that both cellular and exosomal IAPs should be investigated for their possible roles in drug resistance in pancreatic cancer. In contrast, other studies have suggested that exosomes play an antitumor role through immune cells. They suggested that human pancreatic tumor exosomes counteracted the constitutively activated PI3K/AKT survival pathway to induce apoptosis of tumor cells. Their study provided evidence of an apoptotic function of tumor-derived nanoparticles on tumor cells [88], although there has been no clinical study to support this possibility, so additional studies are necessary.
3.3. Monitoring metastasis and tumor recurrence
The detection and increase in the number of CTCs have been shown be predictors of forthcoming metastasis in many cancers. Although the role of CTCs in pancreatic cancer patients has been extensively studied, there is still no definitive evidence of their role. Some studies correlated PDAC metastases with CTCs. They found that, the presence of CTCs in portal vein of the patients undergoing pancreatic resection was associated with a higher rate of liver metastases [89, 90]. In consideration of that CTCs disseminate from the primary tumor and travel through the bloodstream. They may mediate the hematogenous spread of cancer to distant organs. It seems to that reduced dissemination of circulating tumor cells will benefit the patients with pancreatic cancer. However, study showed that reducing dissemination of circulating tumor cells with no-touch isolation surgical technique (approaches permit tumor resection without any grasping or squeezing) [91] has no benefits on the patients with pancreatic cancer compared with standard pancreaticoduodenectomy.[92] This may be caused by the early releasing of CTCs into peripheral blood from primary tumors.[27] It was found that patients with occult metastatic disease had significantly more CTCs than patients with local disease only[93]. And, dynamically monitoring revealed that CTCs count decreased in 3 days after surgery, but increased in 10 days after surgery in most patients. CTCs count increased patients after surgery showed high metastasis rate.[49] So, high CTCs count in peripheral blood before surgery or CTCs count increased after surgery may represent undetectable metastatic disease has been present before surgery.
CTCs demonstrate potential as a preoperative biomarker for identifying patients at high risk of occult metastatic disease. The potential mechanism may be that CTCs exhibit a very high expression of stromal-derived extracellular matrix proteins, whose knockdown in cancer cells suppresses cell migration and invasiveness. The aberrant expression by CTCs of stromal extracellular matrix genes points to their contribution of micro-environmental signals for the spread of cancer to distant organs.[94] And, some CTCs with the specific phenotype could predictive recurrence, CTCs are an exciting potential strategy for understanding the biology of metastases.[74]
A promising clinical application for cfNAs is the early detection of relapse after potentially curative treatments. In recent studies, higher levels of plasma cfNAs in pancreatic patients were found to significantly correlate with metastasis and recurrences. [82] Sausen et al. reported that detection of ctDNA after resection predicted clinical relapse and poor outcomes. Recurrences were detected approximately 6.5 months earlier by ctDNA than with computed tomography imaging.[95] Higher levels of miR-205, miR-210, and miR-744 were also correlated with lymph node metastasis and recurrence.[39, 63]
Exosomes could function as a language that mediates neighboring or long-distance cell-cell communication. The potential role of exosomes in cancer metastasis was studied by many investigators. Gesierich el al. reported an abundance of tetraspanin D6.1A in exosomes derived from pancreatic tumor cells that initiated an angiogenic loop in the organs distant from the tumor.[96] Marhaba et al. studied rat pancreatic adenocarcinoma, and found that cancer cells could abundantly deliver exosomes, which functioned as long-distance intercellular communicators. These exosomes were the main components in forming the pre-metastatic niche.[97] This result was consistent with another study, which also suggested that exosomes played an important role in mutant CD44-mediated pre-metastatic niche formation.[98] A recent study also showed that PDAC-derived exosomes induced liver pre-metastatic niche formation in naive mice and increased liver metastatic burden.[99] It also reported that macrophage migration inhibitory factor (MIF) was highly expressed in PDAC-derived exosomes. Compared with patients whose pancreatic tumors did not progress, there were significantly higher exosome levels from stage I PDAC patients who later developed liver metastasis. These findings suggested that exosomal MIF levels could serve as an early biomarker for liver pre-metastatic niche formation and a prognostic factor for metastatic risk in patients with pre-tumoral lesions, such as pancreatitis, PanINs, IPMNs, and pre-metastatic PDACs.[99] Together, these studies showed that tumor derived exosomes may participate in primary tumor cells organ-specific metastasis. Further studies are required to determine the effects of approaches that inhibit exosomes' function on pre-metastatic niche formation and its therapeutic potential in pancreatic cancer.
4. Current limitations and future perspective
Liquid biopsy is a well-known method to monitor circulating subcellular structures, including CTCs, exosomes, and other biomolecules, such as cell-free nucleic acids. All these biomarkers are obtained from body fluids and are characteristic of the tissue from which they originate. Extensive basic and clinical research has shown that liquid biopsy can be used as a diagnostic, prognostic, and to measure treatment markers for pancreatic cancer because of its less invasive nature.
Liquid biopsy research has experienced exponential development. Some platforms for liquid biopsy have already received government approval for clinical use. The Cell-Search System for CTC enumeration[100] and liquid biopsy tests for determining patients suitable for EGFR-targeted therapy by analysis of their cfDNA[101] have already been granted FDA approval. However, several problems remain to be solved before their applications to clinical practice can occur. One of the important limitations is the lower sensitivity and specificity when compared with traditional biopsies. This is because liquid biopsy detects alterations in body fluids rather than the tumor itself. Both CTCs and ctDNAs are rare in the blood of cancer patients. CTCs may be intermixed with billions of hemocytes. Although ctDNA and tumor-derived exosomes are present at high concentrations in the blood of cancer patients, abundant cfDNA and exosomes can be derived from cellular sources other than the tumor. Liquid biopsy depends on ctDNA that can detect physiologically occurring but clinically inconsequential mutational events. Another limitation is the lack of a consensus about the methodology for the detection and assessment of CTCs and circulating cfNAs. Different research groups have used different methods, making the widespread use of this technology challenging.
In spite of these limitations, liquid biopsy still seems to be the ideal tool to identify biomarkers for pancreatic cancer. The repeatable and less invasive nature of liquid biopsies facilitate the evolutional analyses of cancers in real time, which may be difficult with tissue biopsies because of anatomical and clinical difficulties. The most urgent improvement in liquid biopsy involves the improvement of sensitivity and specificity. With technological development and analytical advances, a wide range of assays have been developed. Large prospective studies need to be carried out to assess the feasibility of these new methods. Another issue that needs to be solved is the establishment of standardized techniques and consensus among all processes for the detection, extraction, and assessment of liquid biopsies. This will facilitate a standardization to compare data obtained from different investigators, in order to translate the technology from research to clinical practice.[102]
5. Conclusion
PDAC is one of the most lethal types of cancer, with a low 5-year survival rate. This is mainly caused by a late diagnosis. Considering the limitations of traditional biopsies, there is an urgent need for new tumor biomarkers. As reviewed here, analyses of liquid biopsy samples provide noninvasive clinical tools for early detection, for prognosis, and for the monitoring of responses of primary tumor and metastatic diseases to treatments. By taking multiple or serial biopsies, it allows us to overcome the limitations of traditional biopsies. Real time monitoring of the tumor facilitates characterization of treatment effects and allows selection of the optimal therapy according to changes in the therapeutic responses. Many questions still remain, and liquid biopsy still needs additional studies to help develop the optimal clinical applications for the diagnosis and treatment of PDAC.
Acknowledgements
Grant Support: This work was supported by grants from the National Science Foundation for Distinguished Young Scholars of China (81625016) and the Natural Science Foundation of China (81472670, 81402397, and 81402398). The funding agencies had no role in the study design, data collection and analyses, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ryan DP, Hong TS, Bardeesy N. Pancreatic Adenocarcinoma. NEW ENGL J MED. 2014;371(11):1039-1049
2. Kamisawa T, Wood LD, Itoi T. et al. Pancreatic cancer. The Lancet. 2016;388(10039):73-85
3. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7-30
4. Long J, Luo GP, Xiao ZW. et al. Cancer statistics: current diagnosis and treatment of pancreatic cancer in Shanghai, China. CANCER LETT. 2014;346(2):273-277
5. Iacobuzio-Donahue CA, Fu B, Yachida S. et al. DPC4 Gene Status of the Primary Carcinoma Correlates With Patterns of Failure in Patients With Pancreatic Cancer. J CLIN ONCOL. 2009;27(11):1806-1813
6. Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. European Journal of Surgical Oncology (EJSO). 2007;33(3):266-270
7. Ni XG, Bai XF, Mao YL. et al. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. European Journal of Surgical Oncology (EJSO). 2005;31(2):164-169
8. Gattani AM, Mandeli J, Bruckner HW. Tumor markers in patients with pancreatic carcinoma. CANCER-AM CANCER SOC. 1996;78(1):57-62
9. Welch DR. Tumor Heterogeneity-A 'Contemporary Concept Founded on Historical Insights and Predictions. CANCER RES. 2016;76(1):4-6
10. Khoo BL, Chaudhuri PK, Ramalingam N. et al. Single-cell profiling approaches to probing tumor heterogeneity. INT J CANCER. 2016;139(2):243-255
11. Cortez E, Gladh H, Braun S. et al. Functional malignant cell heterogeneity in pancreatic neuroendocrine tumors revealed by targeting of PDGF-DD. Proceedings of the National Academy of Sciences. 2016;113(7):E864-E873
12. Roberts NJ, Norris AL, Petersen GM. et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. CANCER DISCOV. 2016;6(2):166-175
13. Parker JS, Perou CM. Tumor Heterogeneity: Focus on the Leaves, the Trees, or the Forest? CANCER CELL. 2015;28(2):149-150
14. Gerlinger M, Rowan AJ, Horswell S. et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. NEW ENGL J MED. 2012;366(10):883-892
15. Koay EJ, Amer AM, Baio FE. et al. Toward stratification of patients with pancreatic cancer: Past lessons from traditional approaches and future applications with physical biomarkers. CANCER LETT. 2016;381(1):237-243
16. Kaczkowski B, Tanaka Y, Kawaji H. et al. Transcriptome Analysis of Recurrently Deregulated Genes across Multiple Cancers Identifies New Pan-Cancer Biomarkers. CANCER RES. 2016;76(2):216-226
17. Crowley E, Di Nicolantonio F, Loupakis F. et al. Liquid biopsy: monitoring cancer-genetics in the blood. NAT REV CLIN ONCOL. 2013;10(8):472-484
18. Diaz Jr LA, Williams RT, Wu J. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. NATURE. 2012;486(7404):537-540
19. Brooks JD. Translational genomics: The challenge of developing cancer biomarkers. GENOME RES. 2012;22(2):183-187
20. Joosse SA, Pantel K. Tumor-Educated Platelets as Liquid Biopsy in Cancer Patients. CANCER CELL. 2015;28(5):552-554
21. Robertson EG, Baxter G. Tumour seeding following percutaneous needle biopsy: The real story!. CLIN RADIOL. 2011;66(11):1007-1014
22. Bagcchi S. Urine test can detect early stage pancreatic cancer. The Lancet Oncology. 2015;16(9):e431
23. Adams DL, Martin SS, Alpaugh RK. et al. Circulating giant macrophages as a potential biomarker of solid tumors. Proc Natl Acad Sci U S A. 2014;111(9):3514-3519
24. Schwarzenbach H, Nishida N, Calin GA. et al. Clinical relevance of circulating cell-free microRNAs in cancer. NAT REV CLIN ONCOL. 2014;11(3):145-156
25. Best MG, Sol N, Kooi I. et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. CANCER CELL. 2015;28(5):666-676
26. Melo SA, Luecke LB, Kahlert C. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. NATURE. 2015;523(7559):177-182
27. Ntouroupi TG, Ashraf SQ, McGregor SB. et al. Detection of circulating tumour cells in peripheral blood with an automated scanning fluorescence microscope. BRIT J CANCER. 2008;99(5):789-795
28. Cristofanilli M, Budd GT, Ellis MJ. et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. NEW ENGL J MED. 2004;351(8):781-791
29. Zhang A, Zhang J, Kaipainen A. et al. Long non-coding RNA: A newly deciphered “code” in prostate cancer. CANCER LETT. 2016;375(2):323-330
30. Casey M, Sweeney KJ, Brown JAL. et al. Exploring circulating micro-RNA in the neoadjuvant treatment of breast cancer. INT J CANCER. 2016;139(1):12-22
31. Ali S, Suresh R, Banerjee S. et al. Contribution of microRNAs in understanding the pancreatic tumor microenvironment involving cancer associated stellate and fibroblast cells. AM J CANCER RES. 2015;5(3):1251-1264
32. Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. NAT REV CANCER. 2011;11(6):426-437
33. Mehra N, Penning M, Maas J. et al. Circulating Mitochondrial Nucleic Acids Have Prognostic Value for Survival in Patients with Advanced Prostate Cancer. CLIN CANCER RES. 2007;13(2):421-426
34. Nawroz H, Koch W, Anker P. et al. Microsatellite alterations in serum DNA of head and neck cancer patients. NAT MED. 1996;2(9):1035-1037
35. Umetani N, Giuliano AE, Hiramatsu SH. et al. Prediction of Breast Tumor Progression by Integrity of Free Circulating DNA in Serum. J CLIN ONCOL. 2006;24(26):4270-4276
36. Bettegowda C, Sausen M, Leary RJ. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. SCI TRANSL MED. 2014;6(224):224r
37. Gornik I, Wagner J, Gasparovic V. et al. Free serum DNA is an early predictor of severity in acute pancreatitis. CLIN BIOCHEM. 2009;42(1-2):38-43
38. Giacona MB, Ruben GC, Iczkowski KA. et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. PANCREAS. 1998;17(1):89-97
39. Miyamae M, Komatsu S, Ichikawa D. et al. Plasma microRNA profiles: identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. BRIT J CANCER. 2015;113(10):1467-1476
40. Choi D, Kim D, Kim Y. et al. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. PROTEOMICS. 2013;13(10-11):1554-1571
41. Thakur BK, Zhang H, Becker A. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. CELL RES. 2014;24(6):766-769
42. Smyth TJ, Redzic JS, Graner MW. et al. Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2014;1838(11):2954-2965
43. Rhim AD, Thege FI, Santana SM. et al. Detection of Circulating Pancreas Epithelial Cells in Patients With Pancreatic Cystic Lesions. GASTROENTEROLOGY. 2014;146(3):647-651
44. Kulemann B, Pitman MB, Liss AS. et al. Circulating Tumor Cells Found in Patients With Localized and Advanced Pancreatic Cancer. PANCREAS. 2015;44(4):547-550
45. Earl J, Garcia-Nieto S, Martinez-Avila JC. et al. Circulating tumor cells (CTC) and KRAS mutant circulating free DNA (cfDNA) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC CANCER. 2015;15(1):797
46. Kulemann B, Liss AS, Warshaw AL. et al. KRAS mutations in pancreatic circulating tumor cells: a pilot study. TUMOR BIOL. 2016;37(6):7547-7554
47. Zhou J, Hu L, Yu Z. et al. Marker expression in circulating cancer cells of pancreatic cancer patients. J SURG RES. 2011;171(2):631-636
48. de Albuquerque A, Kubisch I, Breier G. et al. Multimarker gene analysis of circulating tumor cells in pancreatic cancer patients: a feasibility study. ONCOLOGY-BASEL. 2012;82(1):3-10
49. Zhang Y, Wang F, Ning N. et al. Patterns of circulating tumor cells identified by CEP8, CK and CD45 in pancreatic cancer. INT J CANCER. 2015;136(5):1228-1233
50. Liu H, Sun B, Wang S. et al. Circulating Tumor Cells as a Biomarker in Pancreatic Ductal Adenocarcinoma. CELL PHYSIOL BIOCHEM. 2017;42(1):373-382
51. Sikora K, Bedin C, Vicentini C. et al. Evaluation of cell-free DNA as a biomarker for pancreatic malignancies. Int J Biol Markers. 2015;30(1):e136-e141
52. Zill OA, Greene C, Sebisanovic D. et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. CANCER DISCOV. 2015;5(10):1040-1048
53. Uemura T, Hibi K, Kaneko T. et al. Detection of K-ras mutations in the plasma DNA of pancreatic cancer patients. J GASTROENTEROL. 2004;39(1):56-60
54. Botezatu I, Serdyuk O, Potapova G. et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. CLIN CHEM. 2000;46(8 Pt 1):1078-1084
55. Finkelstein SD, Bibbo M, Loren DE. et al. Molecular analysis of centrifugation supernatant fluid from pancreaticobiliary duct samples can improve cancer detection. Acta Cytol. 2012;56(4):439-447
56. Cohen JD, Javed AA, Thoburn C. et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proceedings of the National Academy of Sciences. 2017;114(38):10202-10207
57. Liu R, Chen X, Du Y. et al. Serum MicroRNA Expression Profile as a Biomarker in the Diagnosis and Prognosis of Pancreatic Cancer. CLIN CHEM. 2012;58(3):610-618
58. Le Large TYS, Meijer LL, Mato Prado M. et al. Circulating microRNAs as diagnostic biomarkers for pancreatic cancer. EXPERT REV MOL DIAGN. 2015;15(12):1525-1529
59. Wang J, Chen J, Chang P. et al. MicroRNAs in Plasma of Pancreatic Ductal Adenocarcinoma Patients as Novel Blood-Based Biomarkers of Disease. CANCER PREV RES. 2009;2(9):807-813
60. Komatsu S, Ichikawa D, Miyamae M. et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. EXPERT OPIN BIOL TH. 2015;15(6):773-785
61. Slater EP, Strauch K, Rospleszcz S. et al. MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer. TRANSL ONCOL. 2014;7(4):464-471
62. Greystoke A, Ayub M, Rothwell DG. et al. Development of a circulating miRNA assay to monitor tumor burden: From mouse to man. MOL ONCOL. 2016;10(2):282-291
63. Wang J, Raimondo M, Guha S. et al. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. J CANCER. 2014;5(8):696-705
64. Humeau M, Vignolle-Vidoni A, Sicard F. et al. Salivary MicroRNA in Pancreatic Cancer Patients. PLOS ONE. 2015;10(6):e130996
65. Yang JY, Sun YW, Liu DJ. et al. MicroRNAs in stool samples as potential screening biomarkers for pancreatic ductal adenocarcinoma cancer. AM J CANCER RES. 2014;4(6):663-673
66. San Lucas FA, Allenson K, Bernard V. et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. ANN ONCOL. 2016;27(4):635-641
67. Lau C, Kim Y, Chia D. et al. Role of Pancreatic Cancer-derived Exosomes in Salivary Biomarker Development. J BIOL CHEM. 2013;288(37):26888-26897
68. Madhavan B, Yue S, Galli U. et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. INT J CANCER. 2015;136(11):2616-2627
69. Kahlert C, Melo SA, Protopopov A. et al. Identification of Double-stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and p53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J BIOL CHEM. 2014;289(7):3869-3875
70. Allenson K, Castillo J, San Lucas FA. et al. High Prevalence of Mutant KRAS in Circulating Exosome-derived DNA from Early Stage Pancreatic Cancer Patients. ANN ONCOL. 2017;28(4):x4
71. Melo SA, Luecke LB, Kahlert C. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. NATURE. 2015;523(7559):177-182
72. Bidard FC, Huguet F, Louvet C. et al. Circulating tumor cells in locally advanced pancreatic adenocarcinoma: the ancillary CirCe 07 study to the LAP 07 trial. ANN ONCOL. 2013;24(8):2057-2061
73. Okubo K, Uenosono Y, Arigami T. et al. Clinical impact of circulating tumor cells and therapy response in pancreatic cancer. European Journal of Surgical Oncology (EJSO). 2017;43(6):1050-1055
74. Poruk KE, Valero V, Saunders T. et al. Circulating Tumor Cell Phenotype Predicts Recurrence and Survival in Pancreatic Adenocarcinoma. ANN SURG. 2016;264(6):1073-1081
75. Ren C, Han C, Zhang J. et al. Detection of apoptotic circulating tumor cells in advanced pancreatic cancer following 5-fluorouracil chemotherapy. CANCER BIOL THER. 2014;12(8):700-706
76. Torphy RJ, Tignanelli CJ, Kamande JW. et al. Circulating Tumor Cells as a Biomarker of Response to Treatment in Patient-Derived Xenograft Mouse Models of Pancreatic Adenocarcinoma. PLOS ONE. 2014;9(2):e89474
77. Poruk KE, Blackford AL, Weiss MJ. et al. Circulating Tumor Cells Expressing Markers of Tumor-Initiating Cells Predict Poor Survival and Cancer Recurrence in Patients with Pancreatic Ductal Adenocarcinoma. CLIN CANCER RES. 2017;23(11):2681-2690
78. Berger AW, Schwerdel D, Ettrich TJ. et al. Targeted deep sequencing of circulating tumor DNA in metastatic pancreatic cancer. ONCOTARGET. 2018;9(2):2076-2085
79. Perets R, Greenberg O, Shentzer T. et al. Mutant KRAS Circulating Tumor DNA Is an Accurate Tool for Pancreatic Cancer Monitoring. The Oncologist. 2018:2017-2467
80. Pietrasz D, Pécuchet N, Garlan F. et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. CLIN CANCER RES. 2017;23(1):116-123
81. Tjensvoll K, Lapin M, Buhl T. et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. MOL ONCOL. 2016;10(4):635-643
82. Singh N, Gupta S, Pandey RM. et al. High Levels of Cell-Free Circulating Nucleic Acids in Pancreatic Cancer are Associated With Vascular Encasement, Metastasis and Poor Survival. CANCER INVEST. 2015;33(3):78-85
83. Kinugasa H, Nouso K, Miyahara K. et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. CANCER-AM CANCER SOC. 2015;121(13):2271-2280
84. TIAN X, SHIVAPURKAR N, WU Z. et al. Circulating microRNA profile predicts disease progression in patients receiving second-line treatment of lapatinib and capecitabine for metastatic pancreatic cancer. ONCOL LETT. 2016;11(3):1645-1650
85. Takahasi K, Iinuma H, Wada K. et al. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J HEPATO-BIL-PAN SCI. 2018;25(2):155-161
86. Zech D, Rana S, Büchler MW. et al. Tumor-exosomes and leukocyte activation: an ambivalent crosstalk. CELL COMMUN SIGNAL. 2012;10(1):37
87. Shin SJ, Smith JA, Rezniczek GA. et al. Unexpected gain of function for the scaffolding protein plectin due to mislocalization in pancreatic cancer. Proceedings of the National Academy of Sciences. 2013;110(48):19414-19419
88. Ristorcelli E, Beraud E, Verrando P. et al. Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. The FASEB Journal. 2008;22(9):3358-3369
89. Catenacci DVT, Chapman CG, Xu P. et al. Acquisition of Portal Venous Circulating Tumor Cells From Patients With Pancreaticobiliary Cancers by Endoscopic Ultrasound. GASTROENTEROLOGY. 2015;149(7):1794-1803
90. Bissolati M, Sandri MT, Burtulo G. et al. Portal vein-circulating tumor cells predict liver metastases in patients with resectable pancreatic cancer. TUMOR BIOL. 2015;36(2):991-996
91. Hirota M, Kanemitsu K, Takamori H. et al. Pancreatoduodenectomy using a no-touch isolation technique. The American Journal of Surgery. 2010;199(5):e65-e68
92. Gall TMH, Jacob J, Frampton AE. et al. Reduced Dissemination of Circulating Tumor Cells With No-Touch Isolation Surgical Technique in Patients With Pancreatic Cancer. JAMA SURG. 2014;149(5):482
93. Court CM, Ankeny JS, Sho S. et al. Circulating Tumor Cells Predict Occult Metastatic Disease and Prognosis in Pancreatic Cancer. ANN SURG ONCOL. 2018;25(4):1000-1008
94. Ting DT, Wittner BS, Ligorio M. et al. Single-Cell RNA Sequencing Identifies Extracellular Matrix Gene Expression by Pancreatic Circulating Tumor Cells. CELL REP. 2014;8(6):1905-1918
95. Sausen M, Phallen J, Adleff V. et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. NAT COMMUN. 2015;6(1):7686
96. Gesierich S, Berezovskiy I, Ryschich E. et al. Systemic Induction of the Angiogenesis Switch by the Tetraspanin D6.1A/CO-029. CANCER RES. 2006;66(14):7083-7094
97. Marhaba R, Klingbeil P, Nuebel T. et al. CD44 and EpCAM: cancer-initiating cell markers. CURR MOL MED. 2008;8(8):784-804
98. Jung T, Castellana D, Klingbeil P. et al. CD44v6 dependence of premetastatic niche preparation by exosomes. NEOPLASIA. 2009;11(10):1093-1105
99. Costa-Silva B, Aiello NM, Ocean AJ. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. NAT CELL BIOL. 2015;17(6):816-826
100. Onstenk W, Gratama JW, Foekens JA. et al. Towards a personalized breast cancer treatment approach guided by circulating tumor cell (CTC) characteristics. CANCER TREAT REV. 2013;39(7):691-700
101. Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Annals of Translational Medicine. 2017;5(3):46
102. Perez-Gracia JL, Sanmamed MF, Bosch A. et al. Strategies to design clinical studies to identify predictive biomarkers in cancer research. CANCER TREAT REV. 2017;53:79-97
Author contact
![]() Corresponding authors: Professor Xian-Jun Yu, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center; 270 Dong An Road, Shanghai 200032, P.R. China. E-mail: yuxianjunorg. Tel: +86-21-64175590, Fax: +86-21-64031446 and Professor Liang Liu, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center; 270 Dong An Road, Shanghai 200032, P.R. China. E-mail: liuliangorg. Tel: +86-18017317395, Fax: +86-21-64031446
Corresponding authors: Professor Xian-Jun Yu, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center; 270 Dong An Road, Shanghai 200032, P.R. China. E-mail: yuxianjunorg. Tel: +86-21-64175590, Fax: +86-21-64031446 and Professor Liang Liu, Department of Pancreatic Surgery, Fudan University Shanghai Cancer Center; 270 Dong An Road, Shanghai 200032, P.R. China. E-mail: liuliangorg. Tel: +86-18017317395, Fax: +86-21-64031446

 Global reach, higher impact
Global reach, higher impact