Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(17):3208-3215. doi:10.7150/jca.24506 This issue Cite
Research Paper
Validation of Urine-based Gene Classifiers for Detecting Bladder Cancer in a Chinese Study
1. Department of Urology, Fudan University Shanghai Cancer Center, Shanghai, P.R. China
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, P.R. China
3. Department and Laboratory of Urology, Hospital Clínic, Institut d'Investigacions Biomèdiques August Pi iSunyer, Universitat de Barcelona, Barcelona, Spain
4. Fudan University Shanghai Cancer Center - Institut Merieux Laboratory, Cancer Institute,Fudan University Shanghai Cancer Center, Shanghai, P.R. China.
5. bioMerieux (Shanghai) Company Limited, Shanghai, P.R. China.
6. CIBERehd. Plataforma de Bioinformática. IDIBAPS
7. Department of Pathology, Rui Jin Hospital, School of Medicine, Shanghai Jiao-Tong University, Shanghai, P.R. China.
* Both authors have contributed equally to this work
Received 2017-12-22; Accepted 2018-4-15; Published 2018-8-6
Abstract
Background: Current standard methods used to detect and monitor bladder cancer (BC) are invasive or have low sensitivity. We have previously reported in an international European study four non-invasive tests for BC diagnosis based on the gene expression patterns of urine.
Objective: to validate the tests in an independent Asian cohort.
Design, setting and participants: Prospective blinded study in which consecutive voided urine samples from BC patients and controls (n=520) were collected in the Fudan University Shanghai Cancer Center from 2014-2016. Gene expression values were quantified using TaqMan Arrays. The same cut-off as previously reported for discrimination between tumours and controls was used in this validation study.
Results and limitations: Finally, a total of 257 tumour and 132 control urine samples were analysed. We found a high accuracy for the four gene classifiers in this independent Asian set, the classifiers composed of 5 and 10 genes achieved the best sensitivity (80.54% and 81.32%, respectively) maintaining a high specificity (91.67% and 85.61%, respectively). Sensitivity of 5-gene (GS_D5) and 10-gene (GS_D10) expression classifiers in recurrent BC cases (78 and 79%, respectively) is comparable to that of primary BC cases (82%). Cytology and NMP22 identified 67% and 40%, respectively, of tumours that have been diagnosed with our tests. In addition, influence of each studied gene was analyzed and showed similar gene rank between Chinese and Caucasian population.
Conclusions: Our study proves that our non-invasive diagnostic BC tests can be reproduced in independent cohorts and in an external laboratory. All the four gene classifiers have shown equal or superior performance to the current gold standard in the present and previously reported validation studies. Consequently, they may be taken for consideration as molecular tests applicable to clinical practice in the management of BC.
Patient summary: Our gene classifiers achieve sensitivities up to 90% in HR NMIBC and MIBC patients, while this achievement is comparatively lower in LR NMIBC ones.
Keywords: Bladder cancer, Biomarkers, Gene expression, Gene classifiers, Non-invasive, Chinese
Introduction
As in other countries, urinary bladder cancer (BC) remains the second most frequent cause of mortality among genitourinary cancers in China, including approximately 4.8/105 incidence and 2.2/105 mortality rate in male in 2012 [1, 2]. Although this two rates are not as high as those in western countries [3], the incidence and mortality rates of BC in China have increased gradually in the past few years [2].
The striking majority of malignant bladder tumours are urothelial cell carcinomas (UCC). Depending on the degree of tumour infiltration in the bladder wall, BC is classified as non-muscle invasive BC (NMIBC) accounting for 75% of tumours while the remainder are muscle invasive BC (MIBC) [4]. Although not typically life-threatening if detected early, NMIBC has up to 70% recurrence rate during the first two years after diagnosis, depending on the patient risk profile [5, 6]. This recurrence phenomenon means that NMIBC patients may undergo up to 15 invasive procedures during the first 5 years of follow up, depending on the patients risk profile [7].
Current approaches for detecting both primary and recurrent disease rely on invasive cystoscopy aided by voided urine cytology. Cytology is a noninvasive technique and has high specificity (90% to 96%) [8], but lacks sensitivity especially in low risk tumours (11% to 76%) [9]. Additionally, the inter-observer and intra-observer reproducibility of cytology is poor [10].
Invasive cystoscopy is associated with significant discomfort, possible infection and trauma. Moreover, cystoscopy misses up to 15% of the papillary and up to 30% of the flat lesions [11, 12]. In an effort to reduce the frequency of cystoscopies conducted, several noninvasive biomarkers such as nuclear matrix protein 22 (NMP22) test, have been approved by the Food and Drug Administration (FDA), albeit with performance rates remaining insufficient to replace or to guide current diagnostic methods [13]. Since the genetic nature of bladder tumours is heterogeneous, one possible reason of the lack performance of the assays is that they focus on a single or a limited number of biomarkers [14, 15]. In the last decade, several cancer-associated gene classifiers, obtained from voided urine, have been described with high diagnostic performances by different groups [14, 16-19]. Although the promising results presented, these multiple gene classifiers require large-scale prospective validations to prove its repeated efficiency and widespread application.
In this study, we have tested four gene expression classifiers previously developed and validated in Caucasian population [20], in an independent Asiatic cohort, to confirm its widespread clinical application. In addition, we have analyzed whether there is a difference in the influence of the genes included in the study to diagnostic performances between Chinese and Caucasian population.
Materials and Methods
Clinical sampling and processing
A total of 520 consecutive urine samples from patients with BC (336) and controls (184) were consecutively collected between September 2014 and March 2016 in the Fudan University Shanghai Cancer Center (FUSCC) and Shanghai 8th people hospital after obtaining Institutional Review Board approval and patients' informed consents. Of the 520 urine samples collected, 96 samples (49 from the cancer and 47 from the control group) were excluded from study because they did not fulfill RNA quality criteria (a Cq value of GUSB>23; see materials and methods). Twenty-seven samples were excluded for incomplete and incorrect clinical information and eight samples were excluded for repeated testing (Figure 1). Thus, 389 urine specimens were finally analyzed, including 257 samples from patients treated with transurethral resection of the bladder (TURB) for primary or recurrent BC who had histologically confirmed tumours and 132 from controls with non-neoplastic urological disease (Table 1). Grade and stage of the tumours were determined according to WHO criteria [21] and the TNM classification [22], respectively. Tumours were classified according to their risk in 3 categories, including low risk NMIBC: Ta and T1 LG without associated CIS, high risk NMIBC: Ta or T1 LG with associated CIS, Ta or T1 HG, or Tis and MIBC: T2, T3 or T4 LG and HG with or without associated CIS. Voided urine samples (20 to 100 ml) were collected in sterile containers containing 4 ml 0.5 M EDTA (pH 8.0). Urine samples were immediately stored at 4ºC and processed within the next 24 hours. Samples were centrifuged at 1,000×g for 10 minutes at 4ºC. Cell pellets were suspended in 1 ml TRIzol reagent and frozen at -80C until RNA extraction.
Gene Expression Quantification
RNA extraction, complementary DNA synthesis and gene expression quantification were performed in the FUSCC - Institut Merieux Laboratory as previously described [16, 17]. All the 16 target genes and two endogenous controls (GUSB and PPIA) analyzed in our previously reported studies were also analyzed in the present study [16]. Before gene expression quantification of the 16 target genes, an aliquot of 1 μl of preamplified cDNA was applied to verify the actual amount of endogenous control GUSB by quantitative PCR (qPCR) and standard reaction and amplification conditions. Those samples that provide GUSB cycle quantification (Cq) values lower than 18, were diluted with water to ensure a homogeneous amount of cDNA in all the samples and the correct quantification of mRNAs. Whereas those samples with a Cq value higher that 23 were excluded from the study. Real-time quantitative PCR data was processed with SDS 2.4. Previous defined gene thresholds [16] were used for all genes to record Cq values.
Data Analysis
Relative expression values (DCq) for the genes contained in the four evaluated predictive models (GS_D2, GS_D5, GS_D10 and GS_D12; Supplementary Table 1S) [13] were used to calculate the risk for the sample of presenting BC. Raw data obtained from the qPCR platform was sent to Hospital Clinic (Barcelona, Spain) to be analized using a previously defined algorithm for each model which classify samples as tumours or controls All the researchers from the Hospital Clinic involved in this analysis of samples were blinded to the patients' clinical data, ensuring the reliability of the results. R-software was used for all calculations. Receiver Operating Characteristic (ROC) curves were generated using the Diagnosis Med (http://CRAN.R-project.org/packageZDiagnosisMed) and pROC package [23]. Gene influence analysis was performed using R package globaltest [17].
Results
Validation of Four Gene Expression classifiers
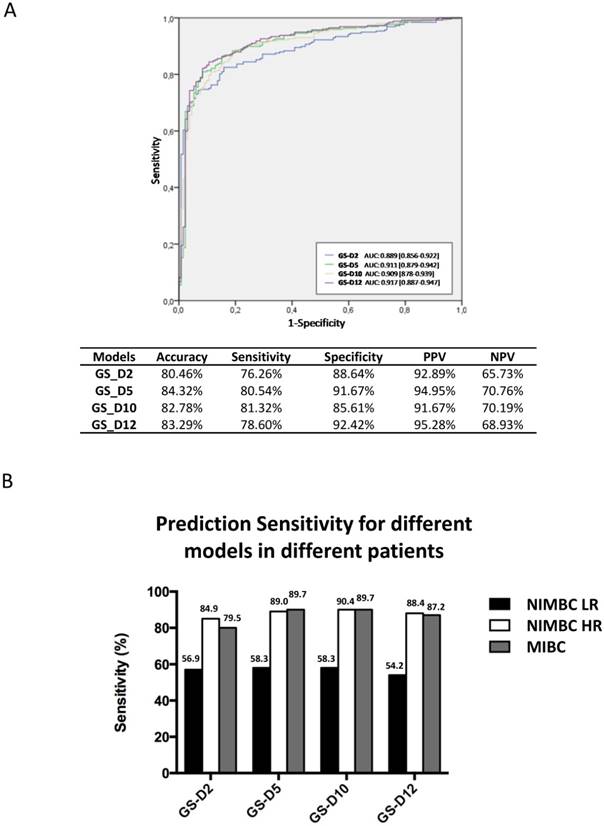
A total of 389 urine samples were finally analyzed (Figure 1; Table 1). The performances of four diagnostic classifiers in the Chinese set are listed in Figure 2A. All the four gene classifiers achieved high diagnostic accuracy (80.46%-84.32%; AUC=0.889-0.917). GS_D5 achieved the best diagnostic accuracy (84.32%; AUC=0.911), with 80.54% SN and 91.67% SP. GS_D10 has the best SN (81.32%), while GS_D12 has the best SP (92.42%). Similarly to that of European studies, SN increased through the BC risk groups. It was lower in low risk NMIBC (54%-58%), while SN are up to ~90% for high risk bladder cancer patients except for GS_D2 (85%) (Figure 2B). Therefore, the study in the Chinese cohort has confirmed the performance results previously obtained in European Studies.
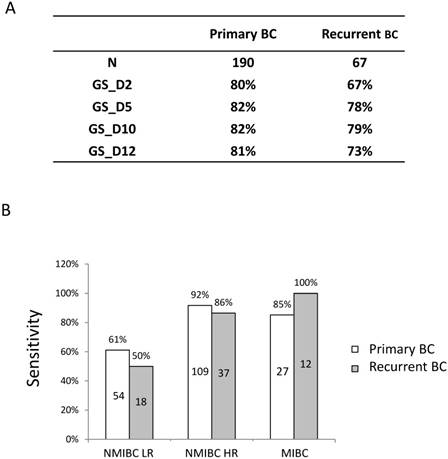
Sensitivity in primary and recurrent BC cases
Among the 257 cancer patients, 67 of them are recurrent and 190 are primary tumours. GS_D10 showed the best sensitivity, both in primary and recurrent cases (Figure 3A). Interestingly, the sensitivity of GS_D10 in recurrent cases (79%) is comparable to that of primary tumours (82%). Furthermore, sensitivities of GS_D10, which showed best sensitivity among the classifiers, in LR NMIBC, HR NMIBC and MIBC patients for primary and recurrent cases are shown in Figure 3B. There are no significant differences between primary and recurrent cases in all the comparisons (LR NMIBC: P=0.4233; HR NMIBC: P=0.3459; MIBC: P=0.2916).
Clinical and histopathological variables for the patients and controls included in the study.
| Variable | Tumor (N=257) | Control (N=132) |
|---|---|---|
| Sex (%) | ||
| Male | 211 (82.1) | 105 (79.5) |
| Female | 46 (17.9) | 27 (20.5) |
| Age (yr) | ||
| Mean | 62.1 | 63.6 |
| Range | 24-89 | 29-90 |
| Grade | ||
| NMIBC LR | 72 | |
| NMIBC HR | 146 | |
| MIBC | 39 | |
| Urological condition | ||
| Normal | 10 | |
| BPH | 73 | |
| Urinary tract infection | 11 | |
| Calculus | 27 | |
| Others | 11 |
Sensitivity comparison of 4 gene classifiers and cytology
| Grade | Overall | LR | HR | MIBC |
|---|---|---|---|---|
| N | 154/257 | 42 | 89 | 23 |
| cytology | 55% | 19% | 67% | 74% |
| GS_D2 | 78% | 57% | 87% | 83% |
| GS_D5 | 81% | 60% | 90% | 87% |
| GS_D10 | 82% | 60% | 91% | 87% |
| GS_D12 | 79% | 57% | 88% | 83% |
Cytology and NMP22 results BC samples
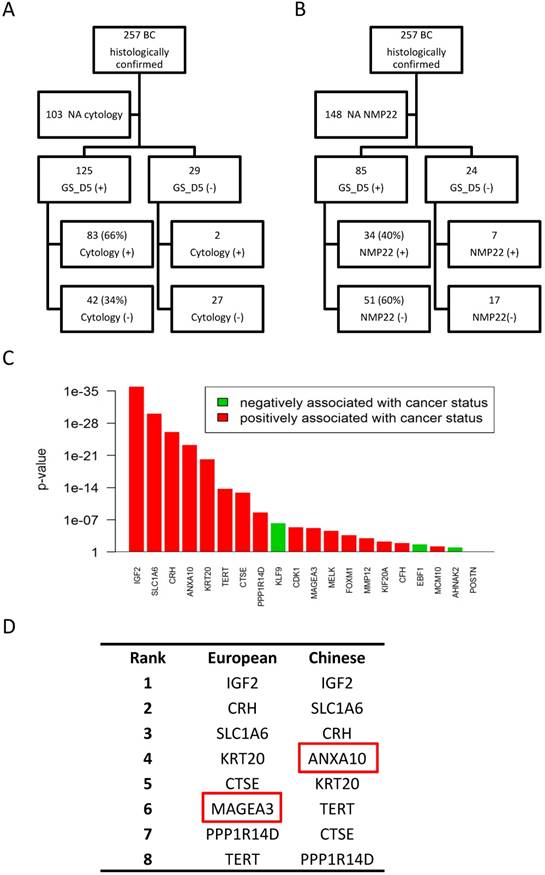
Cytology results were available for 154 (60%) of the 257 BC patients included in the study. We compared sensitivity of four gene classifiers to those of cytology (Table 2). In this subset of patients, overall sensitivity of cytology was 55%, much lower than that of the four gene classifiers (78%-82%). SN of the cytology in LR NMIBC, HR NMIBC and MIBC patients was 19%, 67% and 74% respectively, lower of those sensitivities of four gene models (Table 2). Further comparison analysis of 5-gene classifier (GS_D5) and cytology results showed that all positive cytologies were confirmed by the GS_D5 except in two cases (2%). On the contrary, of all the patients diagnosed GS_D5, cytology only detected BC in 66% of them (Figure 4A).
NMP22 test have been done for 109 (42%) of the 257 BC patients included in our study. Overall, sensitivity of NMP22 test in this subset of patients was 38% while SN of the gene classifiers ranged from 74% to 88%. SN of the NMP22 in LR NMIBC, HR NMIBC and MIBC patients was 21%, 45% and 42% respectively, more than half lower of those sensitivities of four gene models (Table 3). Furthermore, GS_D5 detects BC in 34 (83%) of all 41 positive NMP22 cases. On the contrary, of all the patients diagnosed by GS_D5, NMP22 tests only detect BC in 40% of them (Figure 4B).
Sensitivity comparison of 4 gene classifiers and NMP22 tests
| Overall | LR | HR | MIBC | |
|---|---|---|---|---|
| N | 109/257 | 29 | 60 | 19 |
| NMP22 | 38% | 21% | 45% | 40% |
| GS_D2 | 76% | 48% | 88% | 80% |
| GS_D5 | 78% | 55% | 86% | 87% |
| GS_D10 | 74% | 45% | 86% | 80% |
| GS_D12 | 76% | 52% | 85% | 87% |
Influence of each studied gene in Chinese population
IGF2, SLC1A6 and CRH are the most influential genes in both Caucasian and Chinese populations. All the high influent genes included in the top 8 genes are almost the same; except for MAGEA3, that has more influence in the European cohort, while ANXA10 ranks higher in the Chinese validation set (Figure 4D). Those data were consistent with the good performances presented by the four gene classifiers on the Chinese validation cohort.
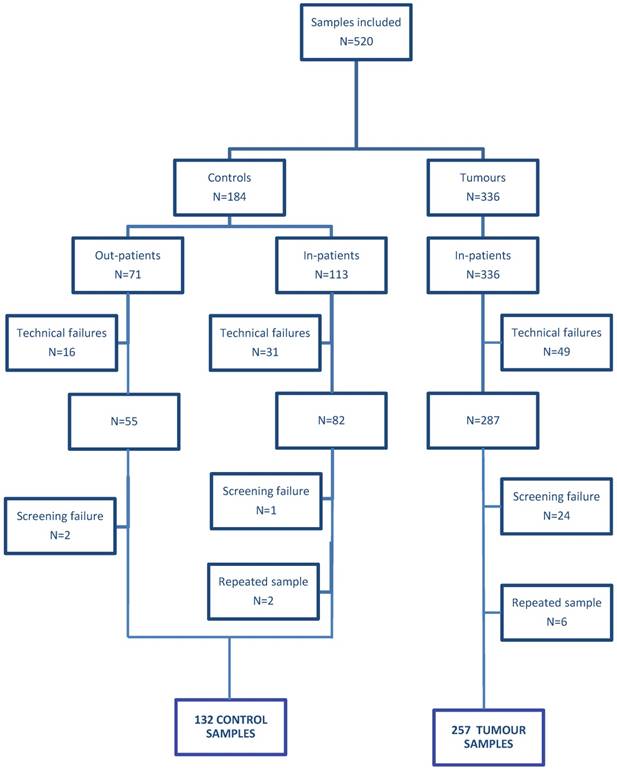
Flow diagram of participants satisfying the criteria for inclusion. Technical failure: samples that yielded insufficient RNA and samples that did not meet the GUSB RNA quality control; see material and methods. In-patients: patients samples collected in FUSCC; Out-patients: samples collected in Shanghai 8th people hospital.
Diagnostic performance of 4 gene expression classifiers in the Chinese validation cohort. A) ROC curves and overall diagnostic performances of the 4 diagnostic gene expression classifiers in the Chinese cohort. B) SN of 4 gene expression classifiers in BC risk groups. Abbreviation: AUC, area under curve. PPV, positive predictive value. NPV, negative predictive value
Discussion
Currently, cystoscopy is considered the gold standard method to diagnose and monitor BC, but misses up to 15% of the papillary and up to 30% of the flat recurrences [11, 12]. Furthermore, cystoscopy is expensive, invasive and bothersome to patients. Urine cytology, on the other hand, has a high specificity (SP=96%), but lacks sensitivity (SN=44%) especially in low risk tumours [24]. Additionally, the interobserver and intraobserver reproducibility of cytology is poor [10]. The combination of both techniques achieves a high SN and SP in the diagnosis and monitoring of the disease (SN: 71%, SP: 96%) [25]. Nevertheless, the invasiveness of cystoscopy has led to the search for biomarkers in urine. This is especially important in the surveillance of BC patients. The high recurrence rate of BC leads to a life-time surveillance with frequent invasive procedures which are associated to significant pain, anxiety and financial cost to the BC patients. Our current and previous studies demonstrated that the non-invasive urine biomarkers tests achieved accuracy values (SN up to 80% and SP up to 90%) for BC diagnosis in the range of that achieved for the gold standard. Furthermore, the good diagnostic performances were observed not only in BC patients with primary tumours but also on recurrent cases. Therefore, urine biomarkers tests could be a potential tool that aid to reduce the frequency of cystoscopies in selected patients.
Diagnostic performance of 4 the gene expression classifiers in primary and recurrent BC. A) SN of 4 gene expression classifiers in primary and recurrent BC. B) SN of the 10-gene expression classifier (GS_D10) between primary and recurrent cases in BC risk groups. Numbers in the boxes indicate the patient numbers for each group.
A number of molecular tests have been developed to achieve this goal. Some of these tests include NMP-22 [25-28] bladder tumour antigen (BTA) [29], Survivin [30], RNA [19, 31] or MicroRNA profiling [32] and fluorescence in situ hybridization analysis for chromosomal abnormalities [9]. However, most of the biomarkers reported above have limited sensitivity, thus by themselves, have not proven to be accurate enough to replace cystoscopy or even cytology. In a meta-analysis of 57 studies none of them achieved a SN >69%, except 78% SN for ImmunoCyt (Scimedx, SP: 78%), with an overall moderate SP ranged of 74%~88% [9]. Consistently, in our study, the diagnostic performance of NMP22 (BladderChek, Alere) test is not sufficient to skip one cystoscopy resulting in a safe approach. In contrast to the moderate performance of one singe marker [33, 34], multiplex biomarkers panel usually show better performances [12-16, 18, 20, 31, 35, 36]. Urquidi et al reported a 14-gene panel in 2012 with 90% SE and 100% SP for BC diagnosis, although more high grade patients included in the cohort [19]. Recently, a DNA methylation signature (A 150 CpG loci biomarker panel) identified by high-throughput DNA sequencing showed a perfect performance (98% SE and 97% SP) on primary BC detection [37]. However, for routine clinical diagnostic use of those promising panels, they need to be tested further and prospective validated in heterogeneous patient populations. So far, our method is the first test validated in both Caucasian and Chinese populations.
Similar to previous results in European studies, the four gene classifiers showed comparatively lower SN (54 to 58%) in LR NMIBC patients than those of HR ones (80 to 90%). Actually, 30 of the 46 misclassified tumour samples by GS_D5, one of the best performer in all series, are LR NMIBC. However, the SN of four gene classifiers in LR NMIBC is still 40% higher than that of cytology and 30% higher than that of NMP22 tests. On the other hand, considering high recurrence rate and life threaten of HR NMIBC, the performance of the tests in this subgroup of tumours is of great importance. Our gene classifiers achieve sensitivities up to 90% in HR NMIBC and MIBC patients, that is 10-20% higher than those of cytology and 40-50% higher than those of NMP22 test in the present cohort. One limitation of our study is the lack of cytology and NMP22 test in the control group that does not allow for a direct calculation of the SP of cytology and NMP22 test in our own series. But total SP of our gene classifiers are in the range as that of reported for cytology [7].
Another limitation of the current study is that the prevalence of tumours in this cohort is higher than the disease prevalence in urologic practice [7], so evaluation of the validation study cohort is likely to provide an overly optimistic assessment of the positive predictive value (PPV) and an overly pessimistic assessment of the negative predictive value (NPV). For an estimated real prevalence of 10% in urologic practice, NPV would be above 90% for all gene classifiers.
Although these gene classifiers have been already validated in patients from multiple centers, this was the first time they were tested outside Europe. Thus, we wondered if there is any heterogeneity of BC gene expression between the two populations that could affect the performance of the gene classifiers. Therefore, we analyzed the influence of each gene included in the four classifiers (Figure 4C). Surprisingly, ranks of the gene influences on the BC diagnosis were very similar between Caucasian and Chinese population, although there are minor variations on the priority of those genes to predict BC between the two populations. We found that MAGEA3, one of the two genes included in GS_D2, seems to be more important in Caucasian population, while ANXA10 shows higher influence in Chinese population. This could possibly explain why when comparing to the GS_D5, GS_D10 and GS_D12 models, GS_D2 showed comparatively lower SN (75.49%) in the Chinese cohort. This minor discrepancy could be affected by the different enrolment of the two cohorts, such as BC grades, primary or recurrent cases distribution, or it could be explained by the heterogeneity of BC gene expression between the two populations. Further studies of the classifiers on Asiatic cohorts may be provide more evidence about it.
Performance comparison of the 4 gene expression classifiers with cytology and NMP22 results. Comparison of the 5-gene classifier (GS_D5) results with cytology (A) and NMP22 results (B). C) Influence on BC diagnosis of each studied gene in the Chinese validation set. D) Comparison of influence top-ranked genes between Caucasian and Chinese populations.
Taken together, this blinded and independent study proves that our non-invasive diagnostic BC tests can be reproduced in the Chinese cohort and in an external laboratory. All the four gene expression classifiers have shown equal or superior performance to the current gold standard in the present and previously reported validation studies. Consequently, they may be taken for consideration as a molecular test applicable to clinical practice in the management of BC.
Supplementary Material
Supplementary table.
Acknowledgements
This work was supported by bioMerieux, Fina Biotech and Biofina Diagnostics. We thank all the patients who participated in this study and all the staff and nurses from the Urology Department of the Fudan University Shanghai Cancer Center for collaborating in collecting urine samples.
Competing Interests
Lourdes Mengual, Juan José Lozano, and Antonio Alcaraz are inventors of patents WO 2008/113870 and WO 2014/118334 applied by Fina Biotech S.L. All other authors declared no conflicts of interest.
References
1. Chen W, Zheng R, Zuo T, Zeng H, Zhang S, He J. National cancer incidence and mortality in China, 2012. Chinese journal of cancer research = Chung-kuo yen cheng yen chiu. 2016;28:1-11
2. Pang C, Guan Y, Li H, Chen W, Zhu G. Urologic cancer in China. Japanese journal of clinical oncology. 2016;46:497-501
3. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359-86
4. Nielsen ME, Smith AB, Meyer AM, Kuo TM, Tyree S, Kim WY. et al. Trends in stage-specific incidence rates for urothelial carcinoma of the bladder in the United States: 1988 to 2006. Cancer. 2014;120:86-95
5. Babjuk M, Burger M, Zigeuner R, Shariat SF, van Rhijn BW, Comperat E. et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2013. European urology. 2013;64:639-53
6. Donat SM. Evaluation and follow-up strategies for superficial bladder cancer. The Urologic clinics of North America. 2003;30:765-76
7. Babjuk M, Bohle A, Burger M, Capoun O, Cohen D, Comperat EM, Hernandez V, Kaasinen E, Palou J, Roupret M, van Rhijn BW, Shariat SF, Soukup V. et al. EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur Urol. 2016
8. Glas AS, Roos D, Deutekom M, Zwinderman AH, Bossuyt PM, Kurth KH. Tumor markers in the diagnosis of primary bladder cancer. A systematic review. The Journal of urology. 2003;169:1975-82
9. Lokeshwar VB, Habuchi T, Grossman HB, Murphy WM, Hautmann SH, Hemstreet GP 3rd. et al. Bladder tumor markers beyond cytology: International Consensus Panel on bladder tumor markers. Urology. 2005;66:35-63
10. Sherman AB, Koss LG, Adams SE. Interobserver and intraobserver differences in the diagnosis of urothelial cells. Comparison with classification by computer. Anal Quant Cytol. 1984;6:112-120
11. Grossman HB, Gomella L, Fradet Y, Morales A, Presti J, Ritenour C, Nseyo U, Droller MJ, PC 302/01 Study Group. A phase III, multicenter comparison of hexaminolevulinate fluorescence cystoscopy and white light cystoscopy for the detection of superficial papillary lesions in patients with bladder cancer. J Urol. 2007;178:62-67
12. Fradet Y, Grossman HB, Gomella L, Lerner S, Cookson M, Albala D, Droller MJ. A comparison of hexaminolevulinate fluorescence cystoscopy and white light cystoscopy for the detection of carcinoma in situ in patients with bladder cancer: a phase III, multicenter study. J Urol. 2007;178:68-73
13. Chou R, Gore JL, Buckley D, Fu R, Gustafson K, Griffin JC. et al. Urinary Biomarkers for Diagnosis of Bladder Cancer: A Systematic Review and Meta-analysis. Annals of internal medicine. 2015;163:922-31
14. Holyoake A, O'Sullivan P, Pollock R, Best T, Watanabe J, Kajita Y. et al. Development of a multiplex RNA urine test for the detection and stratification of transitional cell carcinoma of the bladder. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:742-9
15. Xie XY, Yang X, Zhang JH, Liu ZJ. Analysis of hTERT expression in exfoliated cells from patients with bladder transitional cell carcinomas using SYBR green real-time fluorescence quantitative PCR. Annals of clinical biochemistry. 2007;44:523-8
16. Mengual L, Burset M, Ribal MJ, Ars E, Marin-Aguilera M, Fernandez M. et al. Gene expression signature in urine for diagnosing and assessing aggressiveness of bladder urothelial carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2624-33
17. Mengual L, Ribal MJ, Lozano JJ, Ingelmo-Torres M, Burset M, Fernandez PL. et al. Validation study of a noninvasive urine test for diagnosis and prognosis assessment of bladder cancer: evidence for improved models. The Journal of urology. 2014;191:261-9
18. O'Sullivan P, Sharples K, Dalphin M, Davidson P, Gilling P, Cambridge L. et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. The Journal of urology. 2012;188:741-7
19. Urquidi V, Goodison S, Cai Y, Sun Y, Rosser CJ. A candidate molecular biomarker panel for the detection of bladder cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:2149-58
20. Ribal MJ, Mengual L, Lozano JJ, Ingelmo-Torres M, Palou J, Rodriguez-Faba O. et al. Gene expression test for the non-invasive diagnosis of bladder cancer: A prospective, blinded, international and multicenter validation study. European journal of cancer. 2016;54:131-8
21. Lopez-Beltran A, Sauter G, Gasser T. et al. Tumours of the urinary system. In: (ed.) Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Pathology and genetics of tumours of the urinary system and male genital organs. World Health Organization classification of tumours. Lyon: IARC Press. 2004
22. Sobin LH, Wittekind CH. TNM classification of malignant tumours. International Union Against Cancer. 6th ed. New York: Jonh Wiley & Sons. 2002
23. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics. 2011;12:77
24. Brown FM. Urine cytology. It is still the gold standard for screening? Urol Clin North Am. 2000;27:25-37
25. Mowatt G, Zhu S, Kilonzo M, Boachie C, Fraser C, Griffiths TR. et al. Systematic review of the clinical effectiveness and cost-effectiveness of photodynamic diagnosis and urine biomarkers (FISH, ImmunoCyt, NMP22) and cytology for the detection and follow-up of bladder cancer. Health technology assessment. 2010;14:1-331 iii-iv
26. Shariat SF, Marberger MJ, Lotan Y, Sanchez-Carbayo M, Zippe C, Ludecke G. et al. Variability in the performance of nuclear matrix protein 22 for the detection of bladder cancer. The Journal of urology. 2006;176:919-26 discussion 26
27. Chang YH, Wu CH, Lee YL, Huang PH, Kao YL, Shiau MY. Evaluation of nuclear matrix protein-22 as a clinical diagnostic marker for bladder cancer. Urology. 2004;64:687-92
28. Pichler R, Tulchiner G, Fritz J, Schaefer G, Horninger W, Heidegger I. Urinary UBC Rapid and NMP22 Test for Bladder Cancer Surveillance in Comparison to Urinary Cytology: Results from a Prospective Single-Center Study. I.Int J Med Sci. 2017;14(9):811-819
29. D'Costa JJ, Goldsmith JC, Wilson JS, Bryan RT, Ward DG. A Systematic Review of the Diagnostic and Prognostic Value of Urinary Protein Biomarkers in Urothelial Bladder Cancer. Bladder Cancer. 2016;2(3):301-317
30. Chang Y, Xu J, Zhang Q. Microplate magnetic chemiluminescence immunoassay for detecting urinary survivin in bladder cancer. Oncol Lett. 2017;14(4):4043-4052
31. Urquidi V, Netherton M, Gomes-Giacoia E, Serie D, Eckel-Passow J, Rosser CJ. et al. Urinary mRNA biomarker panel for the detection of urothelial carcinoma. Oncotarget. 2016;7(25):38731-38740
32. Sapre N, Macintyre G, Clarkson M, Naeem H, Cmero M, Kowalczyk A. et al. A urinary microRNA signature can predict the presence of bladder urothelial carcinoma in patients undergoing surveillance. Br J Cancer. 2016Feb16;114(4):454-462 Published online 2016 Jan 26. doi: 10.1038/bjc.2015.472
33. Tilki D, Burger M, Dalbagni G, Grossman HB, Hakenberg OW, Palou J. et al. Urine markers for detection and surveillance of non-muscle-invasive bladder cancer. European urology. 2011;60:484-92
34. Xylinas E, Kluth LA, Rieken M, Karakiewicz PI, Lotan Y, Shariat SF. Urine markers for detection and surveillance of bladder cancer. Urologic oncology. 2014;32:222-9
35. Frantzi M, van Kessel KE, Zwarthoff EC, Marquez M, Rava M, Malats N. et al. Development and Validation of Urine-based Peptide Biomarker Panels for Detecting Bladder Cancer in a Multi-center Study. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:4077-86
36. Chen LM, Chang M, Dai YF, Chai KX, Dyrskjøt L, Sanchez-Carbayo M. et al. External Validation of a Multiplex Urinary Protein Panel for the Detection of Bladder Cancer in a Multicenter Cohort. Cancer Epidemiol Biomarkers Prev. 2014;23(9):1804-1812
37. Feber A, Dhami P, Dong LQ, Winter P, Tan WS, Martínez-Fernández M. et al. UroMark—a urinary biomarker assay for the detection of bladder cancer. Clin Epigenetics. 2017;9:8
Author contact
![]() Corresponding and senior authors: Antonio Alcaraz, MD, Phone: +34-93-2275545; E-mail: AALCARAZcat, Ji Liang, PhD, Phone: +86-21-64438380; E-mail: jill.liangcom and Dingwei Ye, MD, Phone: +86-21-64175590; E-mail: dwyelicom
Corresponding and senior authors: Antonio Alcaraz, MD, Phone: +34-93-2275545; E-mail: AALCARAZcat, Ji Liang, PhD, Phone: +86-21-64438380; E-mail: jill.liangcom and Dingwei Ye, MD, Phone: +86-21-64175590; E-mail: dwyelicom

 Global reach, higher impact
Global reach, higher impact