Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(16):2930-2937. doi:10.7150/jca.24425 This issue Cite
Research Paper
Clinicopathologic characteristics and survival outcome in patients with advanced lung adenocarcinoma and KRAS mutation
1. Department of Pathology, Zhejiang Cancer Hospital
2. Department of Medical Oncology, Zhejiang Cancer Hospital
3. Zhejiang Key Laboratory of Diagnosis and Treatment Technology of Thoracic Oncology
4. Zhejiang Key Laboratory of Radiation Oncology
Received 2017-12-18; Accepted 2018-6-6; Published 2018-7-30
Abstract
Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations are one of the most common observed genetic events in lung adenocarcinoma. The present study aimed to characterize treatment patterns and to estimate survival for patients in China with advanced lung adenocarcinoma and KRAS mutation. We identified KRAS-mutant lung adenocarcinoma between February 2013 and June 2017 in Zhejiang Cancer Hospital. Patients' characteristics and treatment outcomes were analyzed. A total of 159 lung adenocarcinoma were included, and 26 (16.4%) patients harbored KRAS mutations. Compared to KRAS-wild patients, patients with KRAS-mutant tumors were more likely to be smokers (76.9% vs. 51.9%, P = 0.029). Median tumor mutation burden (TMB) was significantly higher in the KRAS-mutant cohort than in the KRAS-wild cohort (5.4 vs. 4.2 mutations/megabases; P=0.041). Of the 93 patients receiving first-line chemotherapy, the median progression-free survival (PFS) in the KRAS-mutant group was significantly shorter than in the KRAS-wild group (1.5 vs. 7.2 months; P<0.001). The median overall survival (OS) in the KRAS-mutant group was also significantly shorter than in the KRAS-wild group (hazard ratio for progression or death for patients with KRAS mutation, 3.260; 95% CI, 1.516 to 7.013; P=0.001). In summary, our findings have several important implications for the molecular characterization and therapeutic outcome of lung adenocarcinoma initiated by oncogenic KRAS. Since the number of KRAS-mutant lung cancer is considerable, it should be taken seriously in clinical diagnosis and treatment. KRAS-mutant lung adenocarcinoma was not sensitive to chemotherapy, new and effective drugs targeting the KRAS pathway are in urgent need.
Keywords: KRAS mutation, lung adenocarcinoma, clinicopathologic characteristics, survival outcome.
Introduction
Lung cancer is the leading cause of cancer death in the People's Republic of China as well as worldwide [1]. Approximately 80% of lung cancers are non-small-cell lung carcinoma (NSCLC), the overall 5-year relative survival rate for this cohort was less than 20%, patients with advanced NSCLC has an extremely high mortality rate [2]. In the past decade, the advent of personalized medicine has seen the introduction of a number of targeted treatments for the NSCLC with druggable driver gene mutations [3]. Among these, the presence of activating mutations of the epidermal growth factor receptor (EGFR) and of chromosomic rearrangements in the anaplastic-lymphoma kinase (ALK) protooncogene, have been well characterized genetic alterations with approved inhibitors to be put into the clinical practice and resulted in improved outcomes for patients [4,5]. Beyond EGFR and ALK mutations, a series of other oncogenic drivers have been identified as novel molecular targets with potential therapeutic implications, such as mutations in the genes KRAS, BRAF, HER2, PI3KCA, MET, RET [6].
Kirsten rat sarcoma viral oncogene homolog (KRAS), one of the guanosine triphosphatases (GTPases), belongs to the RAS kinase family involved in cell survival, cell cycle progression, cell polarity and movement, actin cytoskeletal organization, as well as vesicular and nuclear transport [7,8]. When activated by mutations, KRAS protein activates downstream cytosolic effectors including the RAF/MEK/ERK pathway and the PI3K/AKT/mTOR pathway to promote cell growth, proliferation, and survival [9,10]. The transforming protein that results is implicated in various malignancies, including lung adenocarcinoma [11], colorectal cancer [12], pancreatic cancer [13], and leukemias [14].
KRAS is one of the most frequently mutated genes in NSCLC, which are found in approximately 30% of lung adenocarcinomas in western populations and 10% in Asian populations. Despite activating KRAS mutations are one of the most common observed genetic events in lung adenocarcinoma, little progress has been made during the past decades with no new agents being approved for this indication. Currently, platinum-containing chemotherapy remains the standard treatment regimen for advanced NSCLC patients with KRAS mutations.
In previous research, KRAS has been explored as a potential prognostic and predictive factor in patients with resected NSCLC receiving chemotherapy [15-17]. However, the results remain controversial. Until now, published data about treatment patterns and outcomes for advanced NSCLC with KRAS mutation in China have been limited. The present study aimed to characterize treatment patterns and to estimate survival for patients in China with advanced lung adenocarcinoma and KRAS mutation.
Methods
Patients Enrollment and Follow up procedures
Included were patients with pathologically confirmed locally advanced or metastatic lung adenocarcinoma who received targeted NGS analysis at Zhejiang Cancer Hospital during the period February 2013 and June 2017. Patients' data were collected including the following variables: gene mutation status, gender, age, smoking history, stage, involvement status, therapeutic regimens, efficacy of treatment.
Patients who received chemotherapy were evaluated for response every two treatment cycles during treatment and then every 2 months after treatment. Patients received targeted therapy were evaluated for response one month after the initial treatment and then every 2 months during treatment. The response evaluation of the tumor to therapy was based on computed tomography (CT) or magnetic resonance imaging (MRI) scan. The short-term efficacy was defined based on version 1.1 of the Response Evaluation Criteria in Solid Tumors (RECIST) [18]. The long-term efficacy was evaluated according to the progression-free survival (PFS) and overall survival (OS). PFS was defined as the time from the initiation of treatment to the radiological evidence of PD. OS was calculated from initiation of treatment to death. Patients who were still alive till the last follow-up were recorded as censored. Patients who did not relapse or metastases were censored on the day of last follow-up.
Molecular Analysis
All mutational analyses were conducted at the core facility of Nanjing Shihe Jiyin Biotechnology Inc. (Nanjing, China). 5-8 of 10μm tissue sections from tumor FFPE samples were used for genomic DNA extraction with QIAamp DNA FFPE Tissue Kit (QIAGEN) following the manufacturer's instructions. Extracted tumor genomic DNA was fragmented into 300~350bp using Covaris M220 instrument (Covaris). Sequencing libraries were prepared with KAPA Hyper Prep kit (KAPA Biosystems) with optimized protocols. Libraries were then subjected to PCR amplification and purification before targeted enrichment.
DNA libraries from different samples were marked with unique indices during library preparation and up to 2 μg of different libraries were pooled together for targeted enrichment. Human cot-1 DNA (Life Technologies) and xGen Universal blocking oligos (Integrated DNA Technologies) were added to block nonspecific binding of library DNA to targeted probes. Customized xGen lockdown probes panel (Integrated DNA Technologies) were used to targeted enrich for 416 predefined genes. The hybridization reaction was performed by using NimbleGen SeqCap EZ Hybridization and Wash Kit (Roche). Dynabeads M-270 (Life Technologies) was used to capture probe-bind fragments, followed by library amplification with Illumina p5 (5' AAT GAT ACG GCG ACC ACC GA 3') and p7 primers (5' CAA GCA GAA GAC GGC ATA CGA GAT 3') in KAPA HiFi HotStart ReadyMix (KAPA Biosystems), and purification by Agencourt AMPure XP beads. Library quantification was analyzed by KAPA Library Quantification kit (KAPA Biosystems). The size distribution of libraries was measured by Agilent Technologies 2100 Bioanalyzer (Agilent Technologies). The enriched libraries were sequenced on Hiseq 4000 NGS platforms (Illumina) to coverage depths of at least 300x after removing PCR duplicates for FFPE.
Statistical Analysis
Statistical analysis was performed using SPSS22.0 package. Statistical significance was defined as when P<0.05. The categorical variables were analyzed by chi-square tests, or Fisher's exact tests when needed. The continuous variable was compared using ANOVA and Tukey's multiple comparison tests. Survivals were analyzed using the Kaplan -Meier method and were compared using the log-rank test.
Results
An overall characterization of cancer-related mutations identified in all patients
A total of 159 advanced lung adenocarcinoma patients treated in Zhejiang Cancer Hospital who received targeted NGS analysis (416 genes) between February 2013 and June 2017 were enrolled in this study. Patients' characteristics are described in Table 1. The median age of diagnosis was 59.0 years (ranging from 21 to 86 years), and more than half of the patients were male (66.0%). Nineteen (11.9%) patients were diagnosed as stage IIIB, and others were stage IV.
Characteristics between patients with KRAS mutations and without KRAS mutations.
| Characteristic | All patients | KRAS mutation | Non-KRAS mutation | P value |
|---|---|---|---|---|
| Gender | 159 | 26 | 133 | |
| Female | 54 | 5 | 49 | |
| Male | 105 | 21 | 84 | 0.113 |
| Age | ||||
| <60 years | 82 | 10 | 72 | |
| ≥60 years | 77 | 16 | 61 | 0.198 |
| Stage | ||||
| IIIB | 19 | 1 | 18 | |
| IV | 140 | 25 | 115 | 0.317 |
| Smoking history | ||||
| Yes | 89 | 20 | 69 | |
| No | 70 | 6 | 64 | 0.029* |
| bone metastasis | ||||
| Yes | 70 | 11 | 59 | |
| No | 89 | 15 | 74 | 0.512 |
| Intracranial metastasis | ||||
| Yes | 20 | 4 | 16 | |
| No | 139 | 22 | 117 | 0.746 |
| Liver metastasis | ||||
| Yes | 19 | 5 | 14 | |
| No | 140 | 21 | 119 | 0.202 |
| Pleural effusion | ||||
| Yes | 27 | 5 | 22 | |
| No | 132 | 21 | 111 | 0.776 |
A total of 407 cancer-related genetic mutations were detected in these patients with a median of 2.6 mutations per patient and a range of 1-9 mutations per patient (Figure 1A). The most common driver genetic alterations were EGFR mutations in 72 (45.3%) patients, followed by ALK mutations in 16 (10.1%), KRAS mutations in 26 (16.4%), ROS1 mutations in 7 (4.4%), BRAF mutations in 6 (3.8%), RET mutations in 5 (3.1%), MET mutations in 5 (3.1%), HER2 mutations in 5 (3.1%), NF1 mutations in 4 (2.5%), PIK3CA mutations in 3 (1.9%), STK11 mutations in 2 (1.3%), FGFR mutations in 2 (1.3%) and others in 6 (3.8%) (Figure 1B).
mutation types and mutation number identified in 159 patients. A. The number of mutations identified in each patient was plotted to a histogram. B. Total mutations detected in 159 patients were classified according to the mutation genes.
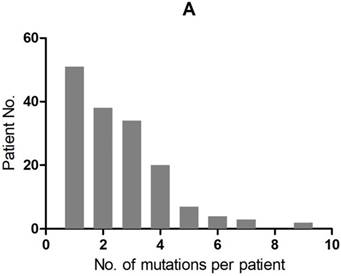
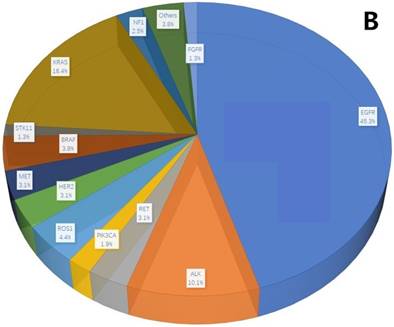
Clinicopathologic characteristics associated with KRAS-mutant NSCLC
KRAS mutations were present in 5 female and 21 male with an average age of 60 years (range, 48-73 years). Six patients (23.1%) were never smokers. Histopathologic stage varied and included IIIB (n = 1) and IV (n = 25). Compared to patients with non-KRAS mutation, patients with KRAS-mutant tumors were more likely to be smokers (76.9% vs. 51.9%, P = 0.029). There were no significant differences in the gender, age, stage, metastatic sites between patients with KRAS-mutant and KRAS wild-type tumors (Table 1). More details of the 26 patients' KRAS mutations are listed in Table 2.
Tumor mutation burden between KRAS-mutant patients and KRAS-wild patients
Of the 159 patients, the median tumor mutation burden (TMB) was 3.7 (range from 1.1 to 16.7) mutations/megabases. The median TMB was significantly higher in the KRAS-mutant group (5.4 mutations/megabases; range from 1.1 to 16.7) than in the KRAS-wild group (4.2 mutations/megabases; range from 1.1 to 11.0) (P=0.041). In addition, 13 of the 26 KRAS mutant patients have co-existence TP53 mutations. The median TMB was 5.9 (range from 2.2 to 16.7) mutations/megabases in this cohort.
Treatment efficacy and survival between KRAS-mutant and KRAS-wild patients
Efficacy of the first-line therapy for all 159 patients were listed in Table 3. In the KRAS-mutant group, all the 26 patients received chemotherapy as the first-line treatment. Among them, 13 patients received pemetrexed+cisplatin/carboplatin regimen, 4 patients received paclitaxel+cisplatin/carboplatin regimen, 4 patients received gemcitabine+cisplatin/carboplatin regimen, 1 patient received pemetrexed, 1 patient received docetaxel+carboplatin, 1 patient received docetaxel+bevacizumab, 1 patient received pemetrexed+cisplatin and bevacizumab, 1 patient was included in the clinical trial and received nivolumab+pemetrexed+carboplatin. In the KRAS-wild group, 66 patients received targeted therapy and 67 patients received chemotherapy as the first-line treatment. The objective response rate (ORR) (11.5% vs 43.6%, P=0.002) and the disease response rate (DCR) (50.0% vs 84.2%, P<0.001) were both significantly lower in the KRAS-mutant cohort than in the KRAS-wild cohort.
More details of the 26 patients with KRAS mutations.
| Patient ID. | Gene Name | Mutation | Co-mutation genes | Chemotherapy regimen |
|---|---|---|---|---|
| 001 | KRAS | G12V | TP53, FLT4, PDGFRB, AKT2, CCND1, ERCC2, FGFR1, FGFR4 | Docetaxel + bevacizumab |
| 002 | KRAS | G12D | None | Pemetrexed + cisplatin |
| 003 | KRAS | G12C | TP53, ERCC2, PIK3CA | Paclitaxel + cisplatin |
| 004 | KRAS | G12A | TP53 | Pemetrexed + carboplatin |
| 005 | KRAS | G12S | TP53, IL7R, MYC, RECQL4, RICTOR, LKB1 | Pemetrexed + carboplatin |
| 006 | KRAS | G12C | TP53, CDKN2A | Pemetrexed |
| 007 | KRAS | G12S | TP53, PIK3CA, CDK4, RB1, SOX2 | Pemetrexed + cisplatin |
| 008 | KRAS | G12C | None | Gemcitabine + carboplatin |
| 009 | KRAS | G13D | CHEK1, SMARCA4 | Pemetrexed + carboplatin |
| 010 | KRAS | G12D | MEK2, CCND1, LKB1 | Gemcitabine + carboplatin |
| 011 | KRAS | G12D | NF2, CDKN2A, GNAS, KMT2A, MPL, SMAD4 | Pemetrexed + cisplatin + bevacizumab |
| 012 | KRAS | G12C | TP53, RB1 | Pemetrexed + carboplatin |
| 013 | KRAS | G12C | FGFR3, RB1, LKB1, NTRK1 | Pemetrexed + carboplatin |
| 014 | KRAS | G13D | TP53, HGF | Pemetrexed + carboplatin |
| 015 | KRAS | G13C | BRAF | Gemcitabine + carboplatin |
| 016 | KRAS | G12R | CDKN2A, SMARCA4, STK11 | Pemetrexed + cisplatin |
| 017 | KRAS | G12C | TP53, ERCC2, PIK3CA | Paclitaxel + cisplatin |
| 018 | KRAS | G12V | TP53, CDKN2A, GRM3 | Gemcitabine + cisplatin |
| 019 | KRAS | Q61H | TP53, DDR2 | Pemetrexed + cisplatin |
| 020 | KRAS | G12A | STK11 | Paclitaxel + cisplatin |
| 021 | KRAS | G12C | - | Pemetrexed + carboplatin |
| 022 | KRAS | G12C | NF2, STK11 | Paclitaxel + carboplatin |
| 023 | KRAS | G12V | TP53, CBL, IL7R, RICTOR | Docetaxel + carboplatin |
| 024 | KRAS | G13C | TP53 | Pemetrexed + carboplatin |
| 025 | KRAS | Q61K | - | Pemetrexed + carboplatin + Nivolumab (Clinical trial) |
| 026 | KRAS | G13C | BTK, SMARCA4, STK11 | Pemetrexed + carboplatin |
Comparison of efficacy between KRAS-mutant patients and KRAS-wild patients.
| Comparison of efficacy in the total 159 patients. | |||
| KRAS-mutant patients (n= 26) | KRAS-wild patients (n= 133) | P value | |
| CR | 0 | 0 | |
| PR | 3 | 58 | |
| SD | 10 | 54 | |
| PD | 13 | 21 | |
| ORR | 3 (11.5%) | 58 (43.6%) | 0.002 |
| DCR | 13 (50.0%) | 112 (84.2%) | <0.001 |
| Comparison of efficacy in the 93 patients receiving first-line chemotherapy. | |||
| KRAS-mutant patients (n= 26) | KRAS-wild patients (n= 67) | P value | |
| CR | 0 | 0 | |
| PR | 3 | 20 | |
| SD | 10 | 35 | |
| PD | 13 | 12 | |
| ORR | 3 (11.5%) | 20 (29.9%) | 0.106 |
| DCR | 13 (50.0%) | 55 (82.1%) | 0.003 |
Efficacy of the first-line chemotherapy for the 93 patients were also listed in Table 3. The DCR (50.0% vs 82.1%, P=0.003) was significantly lower in the KRAS-mutant cohort than in the KRAS-wild cohort. There were no significant differences in ORR between patients with KRAS mutation and non-KRAS mutation.
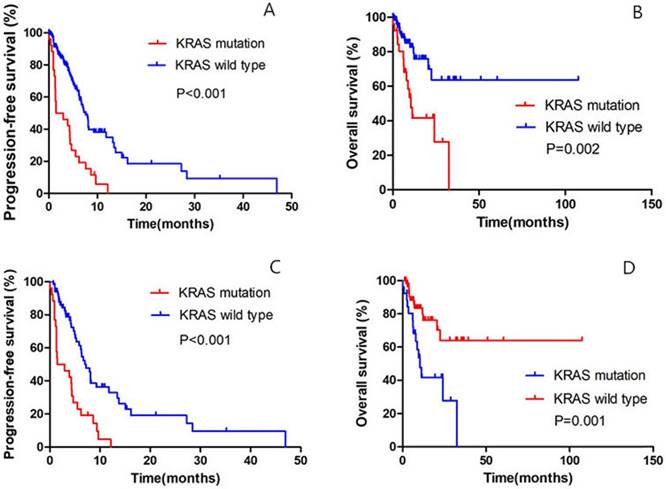
Until the last follow up, 25 patients in the KRAS-mutant group and 62 patients in the KRAS-wild group had disease progression. Of all the 159 patients, the median PFS was 1.5 months (95% confidence interval [CI], to 4.7) in the KRAS-mutant cohort, as compared with 7.6 months (95% CI, 6.5 to 8.7) among patients in the KRAS-wild cohort (hazard ratio for progression or death for patients with KRAS mutation, 3.131; 95% CI, 1.946 to 5.037; P<0.001) (Fig. 2A); the median OS in the KRAS-mutant cohort was significantly shorter than in the KRAS-wild cohort (hazard ratio for progression or death for patients with KRAS mutation, 3.182; 95% CI, 1.597 to 6.341; P=0.002) (Fig. 2B).
Of all the 93 patients receiving chemotherapy as the first-line treatment, the median PFS was 1.5 months (95% confidence interval [CI], 0.3 to 4.7) in the KRAS-mutant cohort, as compared with 7.2 months (95% CI, 5.8 to 8.7) among patients in the KRAS-wild cohort (hazard ratio for progression or death for patients with KRAS mutation, 3.042; 95% CI, 1.801 to 5.137; P<0.001) (Fig. 2C); the median OS in the KRAS-mutant cohort was significantly shorter than in the KRAS-wild cohort (hazard ratio for progression or death for patients with KRAS mutation, 3.260; 95% CI, 1.516 to 7.013; P=0.001) (Fig. 2D).
Dramatic response to PD_1 inhibitor in patient with advanced NSCLC and KRAS mutation
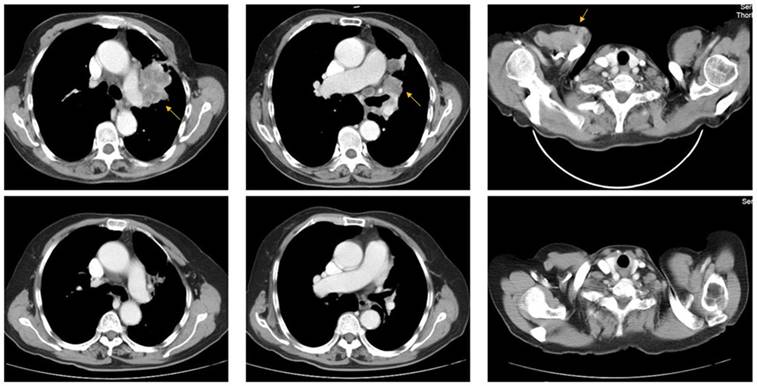
A 73-year-old woman (patient case 025) was diagnosed with lung adenocarcinoma with bilateral intrapulmonary, hilar lymph node, mediastinal lymph node, and subcutaneous metastasis (cT3N2M1b IV; AJCC 7th Edition). Expanded molecular testing revealed KRAS exon3 Q61K mutation. Then she was included in the clinical trial (NCT02477826) and received 4 cycles of nivolumab plus chemotherapy (pemetrexed and carboplatin) as first-line treatment from June 8, 2017 to Aug 10, 2017. After two cycles of treatment, the patient was with rapid dramatic clinical improvement, later confirmed as an excellent radiographic partial response by computed tomography scanning (Fig 3). After then, she received 5 cycles of nivolumab plus pemetrexed as maintenance therapy from Sep 1, 2017 to Nov 24, 2017. The PFS was 6.2 months, significantly better than the median PFS of 1.5 months in the KRAS-mutant cohort.
Comparison of PFS and OS between KRAS-mutant patients and KRAS-wild patients. A. Of the 159 patients, Kaplan-Meier PFS curves are shown between KRAS-mutant patients and KRAS-wild patients (P<0.001). B. Of the 159 patients, Kaplan-Meier OS curves are shown between KRAS-mutant patients and KRAS-wild patients (P=0.002). C. Of all the 93 patients receiving first-line chemotherapy, Kaplan-Meier PFS curves are shown between KRAS-mutant patients and KRAS-wild patients (P<0.001). D. Of all the 93 patients receiving first-line chemotherapy, Kaplan-Meier OS curves are shown between KRAS-mutant patients and KRAS-wild patients (P=0.001).
Representative pre- (upper row) and post-treatment (lower row) computed tomography (CT) images in a 73-year-old woman (patient case 025) diagnosed with advanced lung adenocarcinoma with KRAS exon3 Q61K mutation. After 2 cycles of treatment (Nivolumab + pemetrexed + carboplatin), metastatic lesions (yellow arrow) became markedly reduced compared with those in pre-treatment CT images.
Discussion
In this study, we included 159 patients with advanced lung adenocarcinoma and 26 (16.4%) of them had KRAS mutations. The rate of KRAS mutations in our result was similar to those reported in Asian populations, but was lower than those reported in Caucasian populations [19, 20]. In our study, of the 26 KRAS-mutant patients, 8 was the G12C point mutation, 3 was G12V point mutation, 3 was the G12D point mutation, 3 was G13C point mutation, 2 was G13D point mutation, 2 was the G12S point mutation, 2 was the G12A point mutation, 1 was G12R point mutation, 1 was Q61H point mutation, and 1 was Q61K point mutation. Previous research also indicated that most of KRAS mutations in NSCLC occur at the G12C point mutation, followed by G12V and G12D [21]. In addition, our findings showed that compared to patients with non-KRAS mutation, patients with KRAS-mutant tumors were more likely to be smokers. This phenomenon was consistent with most of previous reports. In a study including 106 patients with lung adenocarcinoma, KRAS mutations were detected in 40 of 106 tumors (38%) and were significantly more common in smokers compared with nonsmokers (43% vs 0%; P=0.001) [22]. In a large French nationwide study only 6% of KRAS-mutant NSCLC patients were never smokers [23]. In another study, the frequency of KRAS mutation was not associated with smoking history, but the findings showed that never smokers were significantly more likely than former or current smokers to have a transition mutation (G→A) rather than the transversion mutations known to be smoking related (G→T or G→C; p<0.0001) [24]. Similar results have been reported by a research based on the cohort of lung adenocarcinomas patients in Korean [25].
Despite the fact that activating mutations of the KRAS gene are one of the most common recurring molecular aberrations in NSCLC, its utility as a direct treatment target remains disappointing [26]. In our study, all the 26 patients with KRAS mutations received chemotherapy as the first-line treatment. In the KRAS-wild group, 66 patients received targeted therapy and 67 patients received chemotherapy as the first-line treatment. The results demonstrated that both in the general cohort (n=159) and in the subgroup cohort (n=93) receiving first-line chemotherapy, KRAS mutations were associated with lower DCR, shorter PFS and OS. As far as we know, this is the first research from a single center in China, suggesting that the KRAS mutation is a poor predictive and prognostic indicator in advanced lung adenocarcinoma. Although the analysis of OS may be confounded by the long-periods tumor control of the KRAS-wild patients with actionable mutations who received targeted drugs, at least we can conclude that KRAS mutations have negative predictive and prognostic effect on efficacy and PFS for advanced lung adenocarcinoma receiving first-line chemotherapy.
Actually, conclusions about the prognostic value of KRAS mutations in NSCLC remain controversial. An initial research demonstrated that KRAS mutations were associated with poor disease-free survival (DFS, P=0.038) and OS (P=0.002) in resected NSCLC [27]. However, this result has not been reproduced at the E4592 trial in which KRAS mutations were not found to be prognostic for OS [28]. The results of a meta-analysis of resected NSCLC receiving adjuvant chemotherapy corroborate this finding, which showed that KRAS-mutant patients had the same prognosis as KRAS-wild patients [29]. The results of research exploring the prognostic value of KRAS mutations in advanced NSCLC seem to be more consistent. In a meta-analysis of 43 trials that included 5216 patients, KRAS mutations as an adverse prognostic factor for OS in advanced lung adenocarcinoma has been confirmed [30]. A recently published meta-analysis including more than 1000 III/IV TNM stage lung adenocarcinoma has also indicated that patients with KRAS mutations had a significantly shorter OS and PFS compared to wild-type lung cancer patients. However, in those studies authors did not collect the data of patients' treatment and the predictive role of KRAS mutations in such cohort receiving chemotherapy have not been analyzed [31].
The predictive value of KRAS mutations in NSCLC also remain uncertain. In a study analyzing the survival outcome according to KRAS mutation status in newly diagnosed patients with stage IV NSCLC treated with platinum doublet chemotherapy, KRAS mutations are not associated with inferior PFS (6.2m vs. 7.0m, p=0.51) and OS (15.6m vs. 19.0m, p=0.34) [32]. In another study analyzing the characterization of distinct types of KRAS mutation and its impact on first-line platinum-based chemotherapy in Chinese patients with advanced non-small cell lung cancer, KRAS mutation was a negative predictive factor of PFS, and patients with KRAS G12V mutations exhibited the poorest PFS compared with those with other KRAS mutant types [33]. In a meta-analysis including 41 studies, the results demonstrated that KRAS mutation is a weak, but valid predictor for chemotherapy in advanced NSCLC [34].
The co-existence of TP53 mutations was observed in 13 (50.0%) of patients with KRAS mutations in our results, which was consistent with previous report. Recently, lung adenocarcinoma with the co-existence of TP53 mutations and KRAS mutations were shown to have higher levels of TMB and inflammation markers, which may be helpful to select patients who will benefit from immune checkpoint blockade and other novel immunotherapy approaches [35, 36]. In our study, we found the similar result: the median TMB was significantly higher in the KRAS-mutant group than in the KRAS-wild group. In addition, in our current study, one patient with KRAS mutation (patient case 025) was included in the clinical trial (NCT02477826) and received 4 cycles of nivolumab plus chemotherapy (pemetrexed and carboplatin) as first-line treatment and later confirmed as an excellent radiographic PR as well as a relative long PFS. This case confirms the previous finding that NSCLC patients with KRAS mutation may benefit from checkpoint inhibitors. In a previous research investigating the potential relevant gene expression signatures that predict efficacy of checkpoint blockade, the results demonstrated TP53 and KRAS mutation had remarkable effect on increasing PD_L1 expression, facilitating T cell infiltration and augmenting tumor immunogenicity. And patients with TP53 and/or KRAS mutation showed sensitivity to PD_1 blockade [37]. Due to failure of therapeutic targeting drugs and unsatisfactory response to conventional chemotherapy in KRAS-mutant lung adenocarcinoma, it is worth exploring the role of immune checkpoint inhibitor in such cohort. In our future studies, we plan to collect more data from KRAS-mutant patients to analyze the TMB information and the value of immunotherapy.
Conclusions
In summary, our findings have several important implications for the molecular characterization and therapeutic outcome of lung adenocarcinoma initiated by oncogenic KRAS. First, this study identified KRAS mutations in about 16.4% of Chinese patients with lung adenocarcinoma. Due to the large population base and the first incidence of lung cancer in China, it can be anticipated that the number of KRAS-mutant lung cancer is considerable, which should be taken seriously in clinical diagnosis and treatment. Second, KRAS-mutant patients were associated with lower DCR, shorter PFS and OS than KRAS-wild patients receiving first-line chemotherapy. This indicates that new and effective drugs targeting the KRAS pathway are in urgent need. Third, the co-existence of TP53 mutations was observed in half of patients with KRAS mutations and the median TMB was significantly higher in the KRAS-mutant group than in the KRAS-wild group. Therefore, clinical trials enrolling patients with KRAS-mutant NSCLC should take into account the co-mutation status of individual tumors.
This emphasizes the significance of comprehensive genomic profiling in assessing patients with NSCLC.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No.81702248).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1-19
2. Chen WQ, Zheng RS, Baade PD. et al. Cancer Statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132
3. Boolell V, Alamgeer M, Watkins DN, Ganju V. The Evolution of Therapies in Non-Small Cell Lung Cancer. Cancers. 2015;7(3):1815-1846
4. Liu TC, Jin X, Wang Y, Wang K. Role of epidermal growth factor receptor in lung cancer and targeted therapies. Am J Cancer Res. 2017;7(2):187-202
5. Roskoski RJ. Anaplastic lymphoma kinase (ALK) inhibitors in the treatment of ALK-driven lung cancers. Pharmacol Res. 2017;117:343-356
6. Califano R, Abidin A, Tariq NU, Economopoulou P, Metro G, Mountzios G. Beyond EGFR and ALK inhibition: unravelling and exploiting novel genetic alterations in advanced non small-cell lung cancer. Cancer Treat Rev. 2015;41(5):401-11
7. Tsuchida N, Ryder T, Ohtsubo E. Nucleotide sequence of the oncogene encoding the p21 transforming protein of Kirsten murine sarcoma virus. Science. 1982;217(4563):937-9
8. Welman A, Burger MM, Hagmann J. Structure and function of the C-terminal hypervariable region of K-Ras4B in plasma membrane targeting and transformation. Oncogene. 2000;19(40):4582-91
9. Castellano E, Molina AM, Krygowska AA. et al. Ras signalling through PI3-Kinase controls cell migration via modulation of Reelin expression. Nat Commun. 2016;7:11245
10. Kuracha MR, Thomas P, Loggie BW, Govindarajan V. Bilateral blockade of MEK- and PI3K-mediated pahtways downstream of mutant KRAS as a treatment approach for peritoneal mucinous malignancies. Plos One. 2017;12(6):e0179510
11. Renaud S, Beau-Faller M, Massard G. KRAS in non-small-cell lung cancer: oncogenic addiction and epidermal growth factor receptor tyrosine kinase inhibitors. JAMA Oncol. 2016;2(10):1373
12. Pek M, Yatim S, Chen Y. et al. Oncogenic KRAS-associated gene signature defines co-targeting of CDK4/6 and MEK as a viable therapeutic strategy in colorectal cancer. Oncogene. 2017;36(35):4975-4986
13. Kamerkar S, LeBieu VS, Sugimoto H. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498-503
14. Fedders H, Alsadeq A, Schmäh J. et al. The role of constitutive activation of FMS-related tyrosine kinase-3 and NRas/KRas mutational status in infants with KMT2A-rearranged acute lymphoblastic leukemia. Haematologica. 2017:169870
15. Schiller JH, Adak S, Feins RH. et al. Lack of prognostic significance of p53 and K-ras mutations in primary resected non-small-cell lung cancer on E4592: A Laboratory Ancillary Study on an Eastern Cooperative Oncology Group Prospective Randomized Trial of Postoperative Adjuvant Therapy. J Clin Oncol. 2001;19:448-457
16. Shepherd FA, Domerg C, Hainaut P. et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173-2181
17. Tomasini P, Jao K, Kamel-Reid S. et al. Predictive value of coexisting KRAS and TP53 mutations on response to chemotherapy in non-small cell lung cancer (NSCLC). J Clin Oncol. 2015;33(suppl):11060a
18. Eisenhauer EA, Therasse P, Bogaerts J. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-47
19. Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72(10):2457-2467
20. Cancer Genome Atlas Research. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543-550
21. Wood K, Hensing T, Malik R, Salgia R. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol. 2016;2(6):805-812
22. Ahrendt SA, Decker PA, Alawi EA. et al. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer. 2001;92(6):1525-1530
23. Barlesi F, Mazieres J, Merlio JP. et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet. 2016;387(10026):1415-1426
24. Riely GJ, Kris MG, Rosenbaum D. et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14(18):5731-5734
25. Kim HR, Ahn JR, Lee JG. et al. The impact of cigarette smoking on the frequency of and qualitative differences in KRAS mutations in Korean patients with lung adenocarcinoma. Yonsei Med J. 2013;54(4):865-74
26. Manchado E, Weissmueller S, Morris J, Chen C, Wullenkord R, Lujambio A. A combinatorial strategy for treating KRAS mutant lung cancer. Nature. 2016;534(7609):647-651
27. Slebos RJ, Kibbelaar RE, Dalesio O. et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323(9):561-565
28. Wakelee HA, Stephenson P, Keller SM. et al. Post-operative rediotherapy (PORT) or chemoradiotherapy (CPORT) following resection of stage II and IIIA non-small cell lung cancer (NSCLC) does not increase the expected risk of death from intercurrent disease (DID) in Eastern Cooperative Oncology Group (ECOG) trial E3590. Lung Cancer. 2005;48(3):389-397
29. Shepherd FA, Domerg C, Hainaut P. et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31(17):2173-2181
30. Mascaux C, Lannino N, Martin B. et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92(1):131-139
31. Shen HC, Che KY, Cong L. et al. Diagnostic and prognostic value of blood samples for KRAS mutation identificatin in lung cancer: a meta-analysis. Oncotarget. 2017;8(22):36812-36823
32. Brady AK, McNeill JD, Judy B. et al. Survival outcome according to KRAS mutation status in newly diagnosed patients with stage IV non-small cell lung cancer treated with platinum doublet chemotherapy. Oncotarget. 2015;6(30):30287-30294
33. Jia YJ, Jiang T, Li XF. et al. Characterization of distinct types of KRAS mutation and its impact on first-line platinum-based chemotherapy in Chinese patients with advanced non-small cell lung cacner. Oncol Lett. 2017;14(6):6525-6532
34. Pan W, Yang Y, Zhu H, Zhang Y, Zhou RP, Sun XC. KRAS mutation is a weak, but valid predictor for poor prognosis and treatment outcomes in NSCLC: A meta-analysis of 41 studies. Oncotarget. 2016;7(7):8373-8388
35. Schabath MB, Welsh EA, Fulp WJ. et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35(24):3209-3216
36. Skoulidis F, Byers LA, Diao L. et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860-877
37. Dong ZY, Zhong WZ, Zhang XC. et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012-3024
Author contact
![]() Corresponding author: Xun Shi, Department of Medical Oncology, Zhejiang Cancer Hospital, 1 Banshan East Road, Hangzhou, China 310022. Tel: 86-571-88122082; Fax: 86-571-88122082; Email address: shi_xunxcn and Ying Jin, Department of Medical Oncology, Zhejiang Cancer Hospital, 1 Banshan East Road, Hangzhou, China 310022. Tel: 86-571-88122082; Fax: 86-571-88122082; Email address: jinyingorg.cn
Corresponding author: Xun Shi, Department of Medical Oncology, Zhejiang Cancer Hospital, 1 Banshan East Road, Hangzhou, China 310022. Tel: 86-571-88122082; Fax: 86-571-88122082; Email address: shi_xunxcn and Ying Jin, Department of Medical Oncology, Zhejiang Cancer Hospital, 1 Banshan East Road, Hangzhou, China 310022. Tel: 86-571-88122082; Fax: 86-571-88122082; Email address: jinyingorg.cn

 Global reach, higher impact
Global reach, higher impact