Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(14):2451-2459. doi:10.7150/jca.24907 This issue Cite
Research Paper
Effect of FAM196B in human lung adenocarcinoma
1. Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215000, China
2. Department of Pathology, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215000, China.
*These authors contributed equally to this work.
Received 2018-1-12; Accepted 2018-4-11; Published 2018-6-14
Abstract
Lung cancer is the leading cause of cancer-related mortality worldwide. The present study focused on the function of family with sequence similarity 196 member B (FAM196B) in lung adenocarcinoma (LUAD). We analyzed lung carcinoma patients' data from the Cancer Genome Atlas (TCGA), and verify the change of FAM196B expression in 30 LUAD tissues and matched adjacent non-tumor tissues (> 5 cm) by tissue microarray. To investigate the role of FAM196B in LUAD, we used lentivirus-mediated short hairpin RNA (shRNA) to knockdown FAM196B expression in human LUAD cell line A549 and H1299 and assessed it by RT-qPCR and western blot. Celigo Imaging Cytometry System, MTT assays and colony formation were conducted to evaluate cell proliferation. Cell apoptosis were assayed by Annexin V staining. We found that FAM196B was significantly upregulated (P=5.06E-06) in LUAD compared with adjacent non-tumor tissues. Cell proliferation was inhibited in FAM196B-silenced A549 and H1299 cells using Celigo Imaging Cytometry System, MTT assays and colony formation assays. Apoptosis rate was significantly increased in FAM196B-shRNA group than the control group. In conclusion, Knockdown of FAM196B can inhibit cell proliferation, cell colony formation capacity, and promote cell apoptosis in A549 and H1299 cell lines. FAM196B may be one novel potential targets for treating patients with LUAD.
Keywords: FAM196B, Lung adenocarcinoma, shRNA, Proliferation, Apoptosis
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. A total of 1.8 million people are diagnosed with lung cancer and 1.6 million die of the disease every year [1, 2]. Non-small cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancer cases [3]. Lung adenocarcinoma (LUAD) has become the most common subtype accounting for 75% of NSCLC [4]. Although great advances have been made with therapies, the 5-year survival rate of lung cancer is still below 15% related to distant metastasis and local recurrence of the tumors [5]. The carcinogenesis and progression of LUAD are involving many genetic and environmental factors, and are multistep processes [6-8]. Although the study of LUAD has made great progress to date, the underlying molecular mechanisms remains largely unclear yet [9].Thus, it is important to investigate the genes involved in tumor progression, invasion and metastasis as well as identify their molecular mechanisms for the treatment and prognostic prediction of LUAD.
Family with sequence similarity 196 member B (FAM196B) locates at human chromosome 5q35.1 [10]. It includes two transcript variants (NM_001129891, 5981 bp; NM_001346304, 5962 bp) [11]. Chen and colleagues reported that FAM196B expressed higher in the embryonic ovary than the testis during mouse gonad development [12]. More recently, it was also reported that FAM196B was up-regulated in clear cell renal cell cancer cell line than finite cell lines [13]. Emerging evidence suggests FAM196B may participate in cancer development. However, little is known about the role of FAM196B in LUAD.
In this study, we analyzed the expression of FAM196B in LUAD using human LUAD tissues and data from the Cancer Genome Atlas (TCGA). The role and function of FAM196B in LUAD were explored by a series of experiments. We found that FAM196B expression was significantly up-regulated in LUAD, and lentivirus-mediated short hairpin RNA (shRNA) targeting FAM196B was used in order to ascertain its function in cell proliferation, cell cycle, and apoptosis in human Lung adenocarcinoma cell lines A549 and H1299. Our study revealed, for the first time that FAM196B was up-regulated in LUAD tissues and performs its biological functions in LUAD. These findings may provide valuable information for elucidation the pathogenesis mechanism of LUAD, and suggest that FAM196B may serve as a promising molecular target for LUAD treatments.
Materials and Methods
Patients and tissue specimens
A total of 30 patients diagnosed with LUAD in the First Affiliated Hospital of Soochow University were incorporated in our study from June, 2015 to July, 2017. Ethical approval was obtained from the ethics committee of the First Affiliated Hospital of Soochow University and informed written consent was obtained from all of subjects.
Immunohistochemistry (IHC)
For IHC assay, 5-mm sections from formalin-fixed-paraffinembedded tissue were transferred to polylysine-coated slides, and incubated with primary antibodies against FAM196B produced in rabbit (1:200 dilution; Santa Cruz, CA, USA). The sections were then incubated with secondary antibody conjugated by horseradish peroxidase (HRP). Examination was performed using 3, 3'- diaminobenzidine (DAB) substrate and hematoxylin. Finally, digital images were taken with a Leica Photo Microscope (Leica).
Cell lines and culture conditions
Human lung adenocarcinoma cell lines A549 and H1299 were originally purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Roswell Park Memorial Institute (RPMI)-1640 medium was purchased from (Thermo Fisher Scientific, Waltham, MA, USA). Fetal bovine serum (FBS) was purchased from Ausbian (Thermo Fisher Scientific). RPMI-1640 medium containing 10% FBS and 100U penicillin-streptomycin at 37°C in a humidified atmosphere supplemented with 5% CO2 was used to culture the human lung adenocarcinoma cell lines A549 and H1299.
Real-time Quantitative PCR (RT-PCR) for Endogenous FAM196B Expression Detection in LUAD Cell Lines
RNA extraction from theA549 and H1299 cell lines was performed with a TRIzol kit (Pufei Corp., Shanghai, China) and reverse transcribed into cDNA with random primers by M-MLV reverse transcriptase (Promega, Madison, WI) according to the manufacturer's instructions. Two sets of primers used for PCR were designed by Beacon Designer 2 software (Premier Biosoft International, Palo Alto, CA) as follows:
GAPDH-F, 5'-TGACTTCAACAGCGACACCCA -3',
GAPDH-R, 5'-CACCCTGTTGCTGTAGCCAAA -3';
FAM196B-F, 5'-TCCCCAAGGTGCAAAGCAAT -3',
FAM196B-R, 5'-GGACGTGTGTCATCCAAAGGG-3'.
The SYBR Green Real-Time PCR assay kit (Takara, Japan) was used for quantitative real-time PCR (RT-PCR) on Mx3000P qPCR system (Agilent, Santa Clara, CA USA) light cycler 480 (Roche, Indianapolis, IN, USA). The quantitative RT-PCR comprised an initial denaturation at 95°C for 45 s, then 40 cycles at 95°C for 5 s, and 60°C for 30 s. The PCR products from FAM196B and GAPDH were 202 bp and 121 bp, respectively. All samples were analyzed in triplicate. GAPDH was amplified as an internal control. For Real-Time PCR data analysis, cycle threshold (Ct) difference (ΔCt) was used for making comparisons between samples. The relative mRNA expression (FAM196B/ GAPDH) was determined by the 2 -ΔΔCt method.
Lentiviral shRNA vector construction
For knockdown of FAM196B, lentiviral shRNA vector targeting human FAM196B gene was constructed by Genechem (Shanghai, China). The sequences targeting of FAM196B were synthesized and cloned into a lentiviral vector pGVX115-GFP (Genechem, Shanghai, China), which was named pGV115-shFAM196B.The pGVX115-GFP was used as negative control and was named pGVX115-shCtrl.
Lentivirus vector production
The lentiviral particles were produced by co-transfection of pGV115-shFAM196B/pGV115-shCtrl with pHelper system (Genechem, Shanghai, China) into 293T cells according to the manufacturer's instruction. Lentiviruses were harvested 48 h post-transfection.
Cell Proliferation and Colony Formation Assays
FAM196B-shRNA and control cell cultures were trypsinized and resuspended , and then were seeded in five 96-well plates at a cell density of 2000 cells/100 μl/well. They were incubated at 37°C and 5% CO2 overnight. On the second day, we used Cellomics ArrayScan VTI (Thermo Fisher Scientific) to continuously measure the GFP expression in each well over a 5-d period. The data obtained were used for statistical data mapping and construction of cell proliferation curves.
To test the effect of FAM196B-shRNA on A549 and H1299 cell viability, MTT[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide]- based assay was used. Cells were plated into 96-well plates at a density of 2,000 cells/well in 100 μl medium and incubated for 24 hours. Then we used shCtrl, FAM196B-shRNA to treat these cells for 1, 2, 3, 4 and 5 days, respectively. 20 μl of 5 mg/ml MTT solution (Sigma, St Louis, MO, USA) was added per well and then the cells were incubated for another 4 h. Cell viability was assessed at 490/570 nm using multi-microplate test system (POLARstar OPTIMA, BMG Labtechnologies, Germany). Each experiment was performed in triplicate.
For colony formation assays, cells were seeded in six-well plates at a density of 300 cells/well with each experimental group being plated in three wells. The cells were incubated over a period of 10 d, and half of the medium was replaced every 3 d. The cell colonies were fixed with 1 mL paraformaldehyde each well (Sinopharm Group, Beijing, China) for 30 min followed by Giemsa staining (500 μL/well; Shanghai Dingguo, Shanghai, China) for 20 min. Finally, the colonies were photographed and counted by fluorescence microscopy (Olympus, Japan). Each experiment was performed in triplicate.
Apoptosis Assay
FAM196B-shRNA and control cell cultures were trypsinized and resuspended in logarithmic growth phase. After washing the cells, Annexin V-APC Apoptosis Detection Kit (eBioscience, USA) was used to assess the apoptosis according to the manufacturer's instructions. The cell concentration was adjusted to 1*106/mL with staining buffer; 100 μL of this suspension was stained with 5 μL AnnexinV-APC. The cells were analyzed using FCM within 1 h. Each experiment was performed in triplicate.
Statistical Analysis
All data were presented as the mean ± SD. The IHC values were compared using the Mann- Whitney test. Results were analyzed using two-tailed Student's t-test to assess statistical significance. A p<0.05 was considered statistically significant. All statistical analyses were performed with the Statistical Package of the Social Sciences software v.17.0 (SPSS Inc.). * Indicated P < 0.05; ** indicated P < 0.01 and *** indicated P < 0.001.
Results
FAM196B is significantly up-regulated in LUAD tissues
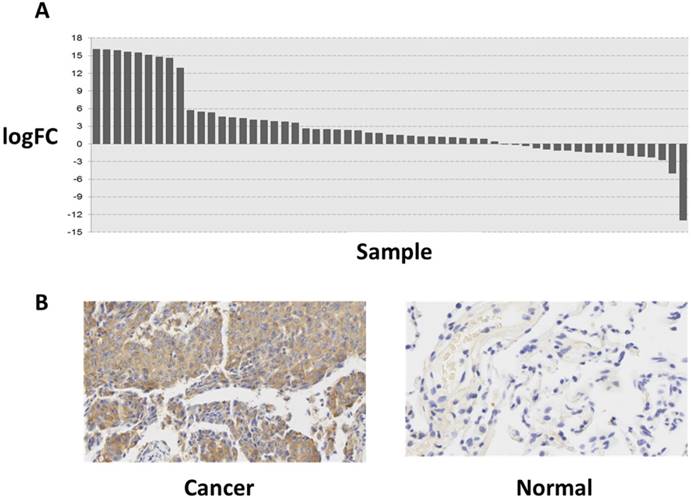
To explore the role of FAM196B in LUAD progression, the mRNA expression profiling of LUAD tissues (case group) and adjacent non-tumor tissues (control group) was retrieved from TCGA database. The difference of expression level of FAM196B between LUAD and adjacent non-tumor tissues was analyzed and was depicted using a histogram. As shown in Figure 1A, FAM196B was significantly upregulated (P = 5.06E-06) in LUAD compared with adjacent non-tumor tissues. To verify the change of FAM196B expression in protein level, we examined its expression levels in tissue microarray including 30 LUAD tissues and matched adjacent non-tumor tissues (>5 cm) by IHC. The results showed that FAM196B protein expression was significantly up-regulated in LUAD tissues compared with normal tissues (P < 0.01) (Figure 1B). Relationships between the expression of FAM196B and clinicopathologic features of LUAD patients are summarized in Table 1.
Knockdown Efficiency of FAM196B by shRNA Lentivirus Infection in A549 and H1299 cell line
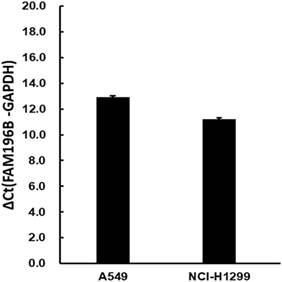
The moderate endogenous expression of FAM196B in A549 and H1299 cell lines were first detected using RT-PCR experiments. As shown in Figure 2, FAM196B was abundantly expressed moderately both in A549 and H1299 cell lines, indicating that A549 and H1299 were suitable for knocking down research.
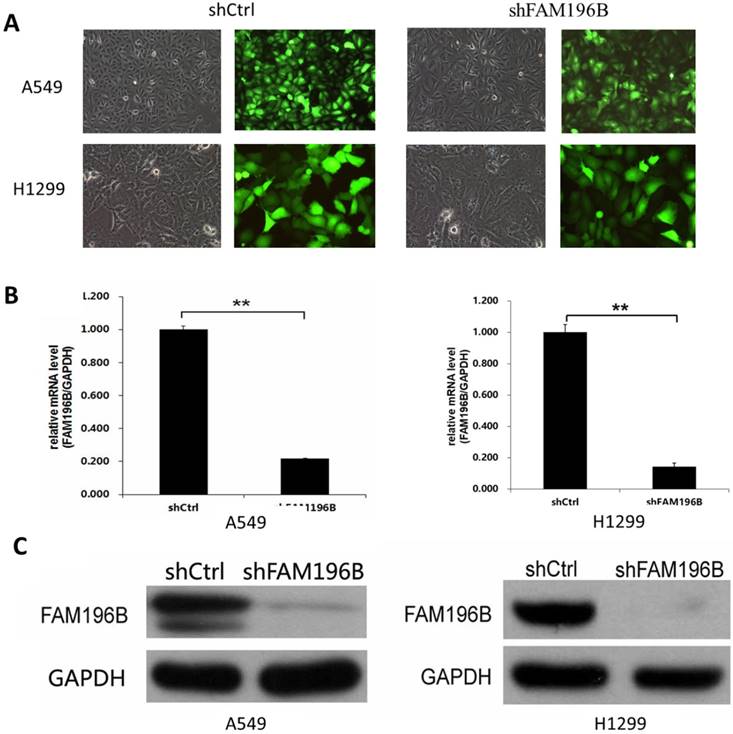
To investigate the role of FAM196B in A549and H1299 cell lines, shRNA targeting FAM196B was cloned into a lentiviral vector pGVX115-GFP. Then, FAM196B-shRNA lentivirus or shCtrl lentivirus were transfected into A549 and H1299 cell lines. As shown in Figure 3A, B, 3 days after infection, about 80% GFP-expressing cells were observed under fluorescence microscope, suggesting infection efficiency around 80%. RT-PCR assay suggested that FAM196B mRNA level was reduced by 80% in FAM196B-shRNA group. Western blot analysis demonstrated that FAM196B protein expression was downregulated post-infection with FAM196B-shRNA lentivirus as compared to cells infected with the control lentivirus in A549 and H1299 cell lines (Figure 3C).
Relationships between the expression of FAM196B and clinicopathologic features.
| FAM196B expression | ||||
|---|---|---|---|---|
| Low | High | P value | ||
| Gender | Male | 2 | 9 | 0.914 |
| Female | 3 | 16 | ||
| Age | ≤ 60 | 2 | 12 | 0.976 |
| > 60 | 2 | 13 | ||
| Pathology grade | I/II | 2 | 18 | 0.275 |
| III | 3 | 7 | ||
| Tumor size | ≤ 3cm | 3 | 13 | 0.787 |
| > 3cm | 2 | 12 | ||
| T classification | T1 | 3 | 12 | 0.706 |
| T2/T3 | 2 | 13 | ||
| Clinical classification | 1/2 | 1 | 3 | 0.583 |
| 3 | 2 | 21 | ||
The expression level of FAM196B in lung cancer was detected in TCGA database and LUAD tissue. (A) The logFC of the expression level of FAM196B in lung cancer compared to adjacent normal tissues based on TCGA dataset. (B) The expression level of FAM196B in LUAD tissues compared to adjacent normal tissues based on IHC.
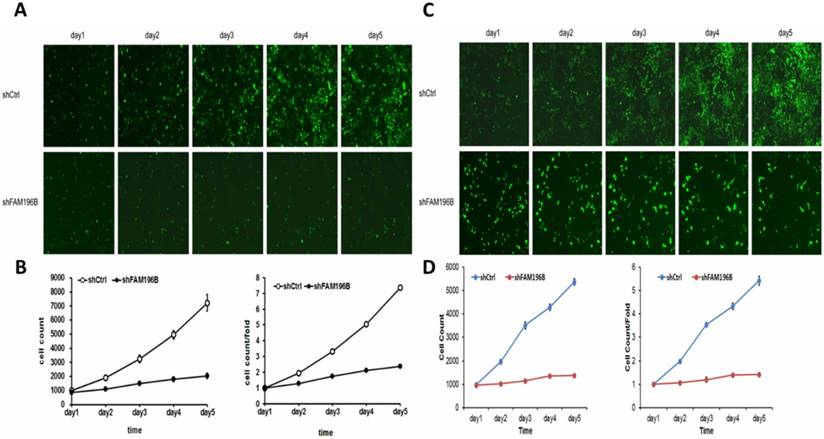
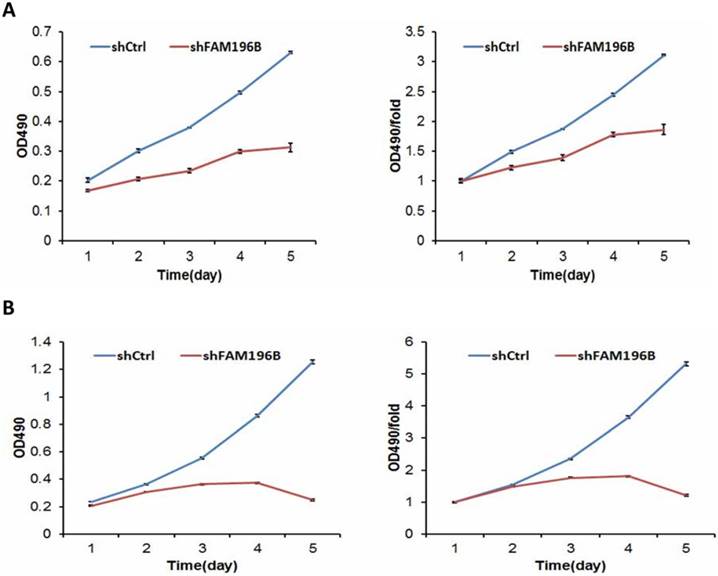
Knockdown of FAM196B Inhibited Cell Proliferation of A549and H1299 cell lines
To examine the effect of FAM196B on cell growth, A549 and H1299 cells were infected with lentivirus with FAM196B-shRNA and control group in a 5-day study, both Celigo Imaging Cytometry System and MTT assays were synchronously applied for deciphering cell growth curve. As illustrated in Figure 4, cell proliferation was significantly inhibited in FAM196B-shRNA cells relative to the control cells as assessed by Celigo Imaging Cytometry System after transfection.
The mRNA expression level of endogenous FAM196B in A549 and H1299 cell lines were detected by RT-PCR analysis. GAPDH was housekeeping gene for normalization.
MTT assay was performed to measure cell viability. FAM196B-shRNA and NC-siRNA were transfected into A549 and H1299 cell lines. We found that cell viability of FAM196B-shRNA group were significantly reduced from day 3 to day 5 compared with shCtrl group both in A549 and H1299 cell lines. (P< 0.001) (Figure 5).
The expression of FAM196B both in A549 and H1299 cell lines was detected through RT-PCR and Western Blot analysis after shFAM196B/shCtrl infection. (A)Transfection efficacy of lentivirus infection was observed through GFP; (B) the mRNA expression level of FAM196B in A549 and H1299 cell lines was detected through RT-PCR analysis; (C) the protein expression level of FAM196B in A549 and H1299 cell lines was detected through Western Blot analysis.
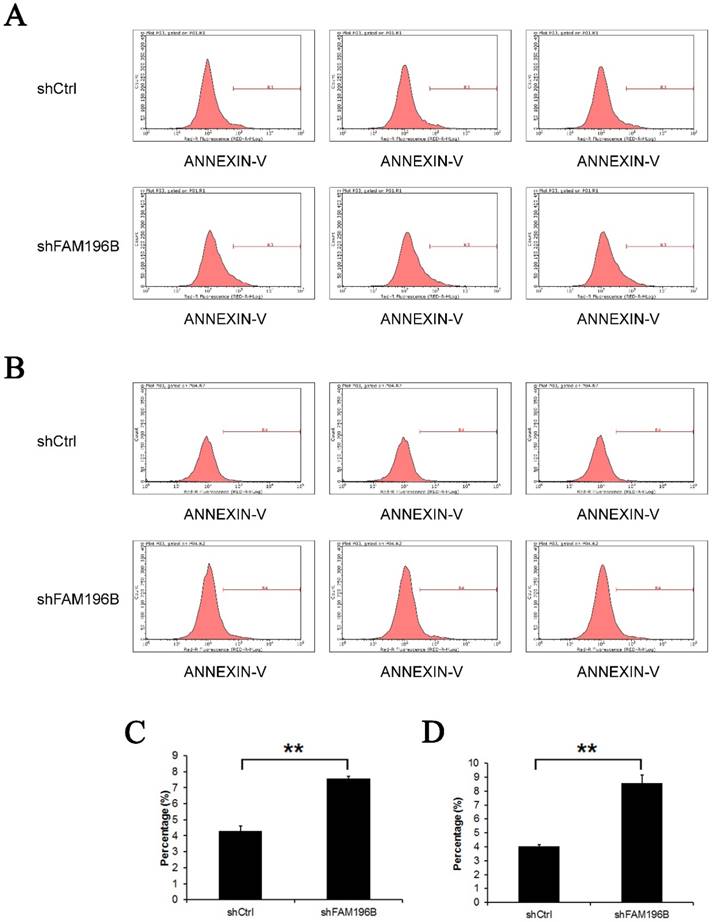
Knockdown of FAM196B induced the cell apoptosis in A549 and H1299 Cells
Annexin V staining was applied for detecting the effect of knockdown of FAM196B on cell apoptosisin by FACS. As shown in Figure 6,compared with the control, the percentage of cell apoptosis in FAM196B-shRNA group was markedly increased in A549 and H1299 cell lines (P< 0.001), suggesting that FAM196B-shRNA promoted A549 and H1299 cell apoptosis.
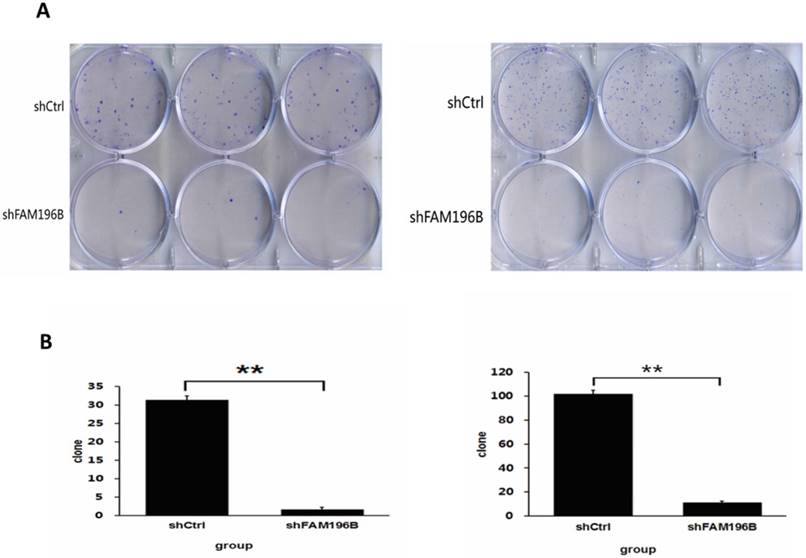
Knockdown of FAM196B inhibited colony formation capacity of A549 and H1299 cell lines
As revealed in Figure 7, in A549 and H1299 cell lines, the number of cell colony was significantly decreased after FAM196B-shRNA infection compared to shCtrl infection (P< 0.001).
Discussion
The main aim of this study was to explore the biological function of FAM196B in human LUAD cells and to elucidate the role in lung carcinogenesis. Lung carcinogenesis is a multifactorial process, involving cell adhesion, dissemination, motility, degradations of basal membrane and ECM and angiogenesis [9, 14, 15]. As we know, dysregulation of oncogenes and tumor suppressor gene (TSG) has been shown to play an important role in controlling cell survival, proliferation, migration, invasion, cell cycle, and apoptosis in cancer [16, 17].
Cell growth of A549 and H1299 after shFAM196B/shCtl infection was detected by Celigo calculation. (A) Cell growth of A549 after shFAM196B /shCtrl infection from day 1 to day 5; (B) cell growth curves of A549 was depicted according to Celigo calculation; (C) cell growth of H1299 after shFAM196B /shCtrl infection from day 1 to day 5; (D) cell growth curves of H1299 was depicted according to Celigo calculation.
Cell growth of A549 and H1299 cell lines after shFAM196B and shCtrl infection were examined through MTT assay. (A) Cell growth of A549 after shFAM196B /shCtrl infection from day 1 to day 5; (B) cell growth of H1299 after shFAM196B /shCtrl infection from day 1 to day 5.
Cell apoptosis of A549 and H1299 cell lines after shFAM196B and shCtrl infection were examined through flow cytometry. (A) Cell apoptosis analysis in A549 cells with shFAM196B or shCtl infection; (B) cell apoptosis analysis in H1299 cells with shFAM196B or shCtl infection; (C) the percentage of cell apoptosis in A549 cells; (D) the percentage of cell apoptosis in H1299 cells.
FAM196B is a protein coding gene and locates at 5q35.1. At present, it was only found that FAM196B was up-regulated in renal carcinoma [13].DNA methylation of FAM196B showed high differences in breast cancer [18].Recent studies showed that FAM196B expression was up-regulated in gastric cancer (GC) tissues and associated with poor tumor histology and T stage of GC. FAM196B also could promote gastric cancer cell proliferation through activating AKT signaling pathway [19]. At the same time, it was reported that oncogene MeCP2 could bind to the methylated CpG islands in the promoter region of FAM196B in GC [20].
Colony formation efficiency of A549 and H1299 cells were observed after shFAM196B/shCtrl infection for 11 days. (A) Cell colony of A549 and H1299 cells was stained by Giemsa. (B) The number of cell colony of A549 and H1299 cells in shFAM196B and shCtrl group.
In the present study, we found that FAM196B is significantly up-regulated in LUAD tissues (P=5.06E-06) compared with adjacent non-tumor tissues through TCGA database. Due to the limitation of sample size,we did not found that FAM196B protein expression was correlated with age, gender, pathologic grade, tumar size, T stages,clinical stages, or lifetime. In the further study, more samples will be collected for indicating the correlation between FAM196B expression and LUAD clinical feature. We also found that FAM196B was abundantly expressed moderately both in LUAD A549 and H1299 cell lines. Therefore, we construct lentivirus-mediated FAM196B shRNA and transfect into A549 and H1299 cell lines.Our results showed that downregulation of FAM196B decreasing colony formation capacity and cell proliferation, and inducing the cell apoptosis (P<0.001, FAM196B-shRNA group vs. control group). Based on the above research results, FAM196B could regulate lung adenocarcinoma cells proliferation in vitro. Its in vivo role is necessary to be elucidated in the further study.
In summary, our study revealed that FAM196B expression was up-regulated in lung cancer tissues. Knockdown of FAM196B can inhibit cell proliferation and cell colony formation capacity, promote cell apoptosis in A549 and H1299 cell lines. These findings may provide new insights for the tumorigenesis and tumor progression in lung adenocarcinoma. Therefore, FAM196B may be one novel potential target for treating patients with lung adenocarcinoma.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No.81672281), and Natural Science Foundation of Jiangsu Province (No.BE2015638).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Siegel RL, Ward EM, Jemal A. Global Cancer Incidence and Mortality Rates and Trends-An Update. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2016;25:16-27
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359-86
3. Kidd L, Driscoll DJ, Gersony WM, Hayes CJ, Keane JF, O'Fallon WM. et al. Second natural history study of congenital heart defects. Results of treatment of patients with ventricular septal defects. Circulation. 1993;87:I38-I51
4. Noone AM, Cronin KA, Altekruse SF, Howlader N, Lewis DR, Petkov VI. et al. Cancer Incidence and Survival Trends by Subtype Using Data from the Surveillance Epidemiology and End Results Program, 1992-2013. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2017;26:632-41
5. Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL. et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299-311
6. Li Y, Xiao X, Han Y, Gorlova O, Qian D, Leighl N. et al. Genome-wide interaction study of smoking behavior and non-small cell lung cancer risk in Caucasian population. Carcinogenesis. 2017
7. Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Translational lung cancer research. 2016;5:288-300
8. Wei R, DeVilbiss FT, Liu W. Genetic Polymorphism, Telomere Biology and Non-Small Lung Cancer Risk. Journal of genetics and genomics = Yi chuan xue bao. 2015;42:549-61
9. Chalela R, Curull V, Enriquez C, Pijuan L, Bellosillo B, Gea J. Lung adenocarcinoma: from molecular basis to genome-guided therapy and immunotherapy. Journal of thoracic disease. 2017;9:2142-58
10. Schmutz J, Martin J, Terry A, Couronne O, Grimwood J, Lowry S. et al. The DNA sequence and comparative analysis of human chromosome 5. Nature. 2004;431:268-74
11. Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R. et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nature genetics. 2004;36:40-5
12. Chen H, Palmer JS, Thiagarajan RD, Dinger ME, Lesieur E, Chiu H. et al. Identification of novel markers of mouse fetal ovary development. PloS one. 2012;7:e41683
13. Feng H, Zhang Y, Liu K, Zhu Y, Yang Z, Zhang X. et al. Intrinsic gene changes determine the successful establishment of stable renal cancer cell lines from tumor tissue. International journal of cancer. 2017;140:2526-34
14. Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Seminars in cancer biology. 2017
15. Zarogoulidis P, Tsakiridis K, Karapantzou C, Lampaki S, Kioumis I, Pitsiou G. et al. Use of proteins as biomarkers and their role in carcinogenesis. Journal of Cancer. 2015;6:9-18
16. He L, Zhang Y, Sun H, Jiang F, Yang H, Wu H. et al. Targeting DNA Flap Endonuclease 1 to Impede Breast Cancer Progression. EBioMedicine. 2016;14:32-43
17. Thomas C, Ji Y, Lodhi N, Kotova E, Pinnola AD, Golovine K. et al. Non-NAD-Like poly(ADP-Ribose) Polymerase-1 Inhibitors effectively Eliminate Cancer in vivo. EBioMedicine. 2016;13:90-8
18. Heyn H, Carmona FJ, Gomez A, Ferreira HJ, Bell JT, Sayols S. et al. DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis. 2013;34:102-8
19. Zhang J, Tong DD, Xue M, Jiang QY, Wang XF, Yang PB. et al. FAM196B acts as oncogene and promotes proliferation of gastric cancer cells through AKT signaling pathway. Cellular and molecular biology. 2017;63:18-23
20. Zhao L, Liu Y, Tong D, Qin Y, Yang J, Xue M. et al. MeCP2 Promotes Gastric Cancer Progression Through Regulating FOXF1/Wnt5a/beta-Catenin and MYOD1/Caspase-3 Signaling Pathways. EBioMedicine. 2017;16:87-100
Author contact
![]() Corresponding author: Haitao Ma, Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215000, China. E-mail: mht7403com
Corresponding author: Haitao Ma, Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215000, China. E-mail: mht7403com

 Global reach, higher impact
Global reach, higher impact