Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(13):2389-2396. doi:10.7150/jca.24079 This issue Cite
Research Paper
PDE4a predicts poor prognosis and promotes metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma
1. Department of Radiology, the 309th Hospital of Chinese People's Liberation Army, Beijing 100091, P.R. China
2. Department of Gastroenterology, Xi'an Central Hospital, Xian 710010, P.R. China
3. Department of Gastroenterology, the 6th Affiliated Hospital of Xinjiang Medical University, Urumchi 830002, P.R. China
#Contributed equally
Received 2017-11-29; Accepted 2018-3-23; Published 2018-6-14
Abstract
Phosphodiesterases (PDEs) was found to be involved in a variety of cancer pathologies by modulating the degradation of levels of cAMP/cGMP. However, the prognostic significance and biological effect of PDE4a in hepatocellular carcinoma (HCC) have not been understood completely. In the present study, PDE4a expression was detected in a cohort of HCC and matched adjacent liver tissues (n = 210) by immunohistochemistry staining and Western immunoblotting assay, And in vitro experiments were conducted to determine the effect of PDE4a on metastatic capacity of HCC cells. The data here displayed that the majority of HCC patients had higher PDE4a expression in tumor tissues compared to matched adjacent liver tissues and enhanced PDE4a expression in tumor tissues was associated positively with HBV infection, liver cirrhosis, higher serum AFP level, advanced TNM stage, vascular embolus, intrahepatic metastases and portal vein tumor thrombus (PVTT). Survival analyses suggested that higher PDE4a was indicated the poor prognosis of HCCs after liver resection. Ectopic expression of PDE4a in Huh7 cells leaded to significant repression of E-cadherin and up-regulated the expression of N-cadherin and Vimentin, and facilitated migration and invasion abilities. Silencing PDE4a in MHCC97h cells acquired the opposite results. Taken together, PDE4a triggered EMT in HCC cells and acted as a predictive factor candidate and a potential therapeutic target for HCC.
Keywords: PDE4a, hepatocellular carcinoma, clinical outcomes, EMT
Introduction
Hepatocellular carcinoma (HCC) is one of the most fatal cancers which accountes for 745,500 deaths worldwide in 2012[1]. Although the great improvement has been achieved on the diagnosis and therapy for HCC, its prognosis remains unfavorable. Liver resection is still the most common curative treatment for HCC patients. However, 5-year post-surgical survival rate was approximately 30% after liver resection[2]. Frequent metastase smade theleading contribution to unfavorable prognosis of HCC after surgery[3]. Conventional predictive factors related to HCC patients including tumor size, vascular invasion, tumor-node-metastasis(TNM) stage, portal vein tumor thrombus(PVTT) and intrahepatic metastases have the limited clinical value. It is essential to find out the novel effective post-surgical predictive factors for HCCs. The accurate understanding of the underlying mechanism of HCC metastasis is critical to identify the efficacy prognostic biomarkers and therapeutic target.
Epithelial to mesenchymal transition (EMT) has been found been responsible for the diffusion of cancer cells and cancer progression, which was characterized that epithelial cells forfeited their polarity and cell-cell contacts because of supression of E-cadherin and up-regulated expression of mesenchymal markers such as N-cadherin and Vimentin[4]. Our previous investigation revealed that partial EMT process was often activated in HCC tissue[5-8], especially in the metastatic lesion, and facilitated HCC progression through mediating cell proliferation, migration, invasion and autophagy. However, it remains unclear about the concrete regulatory mechanism of EMT in HCC, up to now.
Disturbances of cyclic nucleotide signalings have been established as a composite of distrinct component pathways related with multiple aspects of cancer biology[9-11]. Phosphodiesterases (PDEs) was found to be involved in a variety of cancer pathologies by modulating the degradation of levels of cAMP/cGMP [12, 13]. PDE4 was a member of PDE family divided into 11 different families according to substrate preference and inhibitor sensitivity. And based on disparate splice variants and functional isoforms, PDE4 was also classified into 4 subfamily genes (A-D)[14]. PDE4a was found to be aberrantly up-regulated and promote cancer growth in various brain cancers including medulloblastoma and glioblastoma[15]. To date, there has been limited understanding about the role of PDE4a on hepatocarcinogenesis. This investigation revealed that PDE4a was aberrantly over-expressed in HCC tissues and predicted the worse post-surgical outcome, which could be caused by PDE4a-induced EMT.
Methods
Clinical Specimens
Written informed consent was achieved from all HCC patients involved in this study. The use and collection of the samples were reviewed and approved by the ethics committee of the 309th Hospital of Chinese People's Liberation Army according to the Helsinki Declaration of 2013 (No.20070120, 20 Jan 2007). There were 210 HCC patients receiving curative liver resection between 2007 and 2009 in the 309th Hospital of Chinese People's Liberation Army recruited in the present study. Before hepatectomy, no patients received any chemotherapy and/or radiotherapy. During surgery, HCC tissues and adjacent liver tissues (>2 cm distance to the resection margin) were collected and skept in liquid nitrogen. Clinical data were collected from the electronic medical records and the details were presented in Table 1. Follow-up visits for the survival analysis were conducted for up to 90 months after surgery.
Cell Lines
The immortalized human liver cell line (LO2) and HCC cell lines (MHCC97h, SK-Hep1, PLC/PRF/5, Hep3B, and Huh7) were grown with DMEM (Gibco, USA) with 10% FBS in a humidified 5% CO2 incubator at 37°C.
Immunohistochemistry Staining (IHC)
IHC assay against PDE4a was conducted as described previously[16]. The primary antibody targeting PDE4a was purchased from Abcam Co. (Catalog No.: ab14607, MA, USA). The score of IHC staining was evaluated by two experienced pathologists who observed all slides independently. Staining intensity was divided into four grades: 0 (negative), 1 (weakly positive), 2 (moderately positive), and 3 (strongly positive). The percentage of positive cells was classified into 5 grades including 0 for 0-5%, 1 for 6-25%, 2 for 26-50%, 3 for 51-75%, and 4 for > 75%. The ultimate score wasacquired by multiplying the staining intensity and the percentage of positive cells.
Plasmid Construction and Cell Transfections
The PDE4a cDNA was cloned into pCMV-Tag2B vector (Stratagene, USA) to be PDE4a expressing plasmid. The PDE4a expressing plasmid and control plasmid were transfected into Huh7 cells by FuGENE6 reagent respectively to be Huh7 PDE4a cells and PDE4a Vector cells. The siRNA sequence against PDE4a (Catalog No.: sc-41596) and relative srambled siRNA sequence (Catalog No.: sc-37007) were from Santa Cruz Biotechnology (CA, USA), which were transfected into MHCC97h cells by siRNA transfection reagent (Catalog No.:sc-29528, Santa Cruz Biotechnology, USA) respectively to be MHCC97h PDE4a siRNA cells and MHCC97h Scr siRNA cells.
RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA of HCC cells was isolated with TRIzol® reagent (Invitrogen, CA, USA) and then transcribed reversely into cDNA using Superscript III Reverse Transcriptase (Invitrogen, CA, USA). QRT-PCR was conducted by SYBR Green quantitative PCR protocol with the following primers: Forward GAAACGTGTCAGCAGTTGGC, Reverse TGTCACCATCGTGTCCACAG. There were six replicates in each measurement and the detection was repeated three times.
Immunoblot Analysis
Briefly speaking, Western immunoblotting was performed according to the protocol presented previously[17]. The primary antibodies were rabbit polyclonal to PDE4a (Catalog No.: ab14607, Abcam, USA), mouse monoclonal to E-cadherin (Catalog No.: ab1416, Abcam, USA), rabbit polyclonal to N-cadherin (Catalog No.: ab18203, Abcam, USA), mouse monoclonal to Vimentin (Catalog No.: ab18203, Abcam, USA), and mouse monoclonal to β-actin (Catalog No.: ab8226, Abcam, USA). After incubated with the primary antibodies, the blots were then incubated respectively with secondary antibodies conjugated with HRP (Abcam, USA). Protein levels were detected using the HyGLO HRP detection kit from Denville (Metuchen, NJ, USA). β-actin was used as the internal control.
Migration and Invasion Assays
Cell migration capacity of HCC cells was detected by wound healing assay and transwell assay was carried out to measure the cell invasion of HCC cells as introduced previously[6].
Cell Viability and Proliferation Assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed to detect cell viability. HCC cells were plateded in 96-well plates at 1 × 104 cells/well. After HCC cells were washed by PBS twice, 20 µl 5 mg/ml MTT was added to the culture medium. After incubation at 37°C for 4 h, HCC cells in each well were treated by 150 μl dimethyl sulfoxide from Sigma-Aldrich (MO, USA). The absorbance was examined at 570 mm. DNA uptake of 5-bromo-2 ' -deoxyuridine-5 ' -monophosphate (BrdU) was determined by ELISA assay. HCC cells were seeded into the 96-well plates at 5,000 cells per well. After 24h, ELISA assay was carried out using the BrdU ELISA kit (Roche, IN, USA). All tests were repeated at least six times.
Statistical Analysis
All data were expressed as mean ± standard deviation (SD). The statistically significant differences (P<0.05) were tested by Mann-Whitney U or Student's t test. Comparison of the Kaplan-Meier survival curves of HCCs with higher and lower PDE4a expression in HCC tissues compared with adjacent liver tissues was conducted by the log-rank test. Multivariate analysis was carried out by SPSS V17.0 software (SPSS Inc., Chicago, IL, USA).
Results
PDE4a was aberrantly up-regulated in HCC tissues
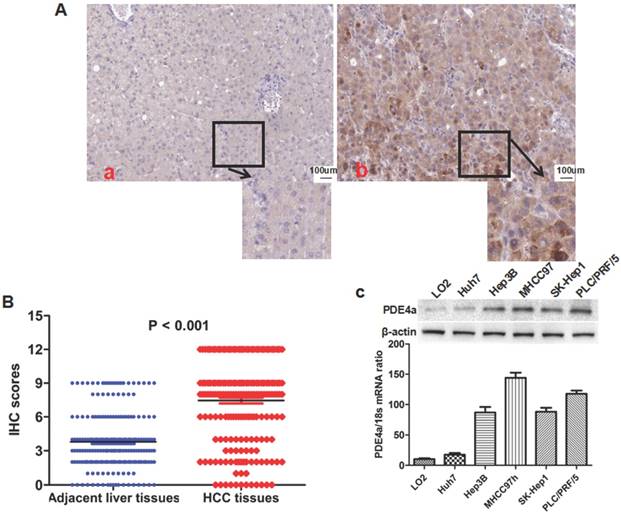
IHC staining assay showed that there was significantly more PDE4a expression in HCC tissues than adjacent liver tissus in 175 of 210 HCCs (83.3%). As shown in Fig.1A, PDE4a protein located predominantly in the cytoplasm. Mann-Whitney U test displayed that the IHC score of PDE4a protein in HCC tissues was notably higher than one in adjacent liver tissues (Fig.1B). After analysis of clinical features of HCCs, it was found that over-expression of PDE4a in HCC tissues was related positively with HBV infection, liver cirrhosis, higher serum AFP level, advanced TNM stage, microvascular invasion (MVI), intrahepatic metastases and portal vein tumor thrombus (PVTT) (Table 1). PDE4a expression was detected in human liver cell line LO2 and 5 kinds of human liver cancer cell lines including MHCC97h, SK-Hep1, PLC/PRF/5, Hep3B and Huh7 cells using both qRT-PCR and Western immunobloting assays. As shown in Fig.1C, there was the highest level of PDE4a expression in MHCC97h cells, whereas Huh7 cells had the least PDE4a expression.
Demographic information and clinical feature of 210 HCC patients
| Clinicopathological features | No. of Patients | X2 | P | ||
|---|---|---|---|---|---|
| Low PDE4a | High PDE4a | ||||
| Age (years) | < 50 | 15 | 77 | 0.015 | 0.901 |
| ≥ 50 | 20 | 98 | |||
| Gender | Male | 21 | 108 | 0.036 | 0.849 |
| Female | 14 | 67 | |||
| HBV infection | Present | 27 | 160 | 6.103 | 0.014* |
| Absent | 8 | 15 | |||
| Serum AFP level (ng/mL) | < 400 | 3 | 34 | 0.256 | 0.613 |
| ≥ 400 | 22 | 151 | |||
| Tumor diameter (cm) | < 5 | 21 | 69 | 5.040 | 0.025* |
| ≥ 5 | 14 | 106 | |||
| Liver cirrhosis | Present | 19 | 129 | 5.291 | 0.021* |
| Absent | 16 | 46 | |||
| Edmondson-Steiner Classification | I + II | 17 | 76 | 0.313 | 0.576 |
| III + IV | 18 | 99 | |||
| TNM stage | I + II | 21 | 62 | 7.367 | 0.007* |
| III + IV | 14 | 113 | |||
| PVTT | Present | 2 | 39 | 5.098 | 0.024* |
| Absent | 33 | 136 | |||
| Intra-hepatic metastases | Present | 2 | 39 | 54.84 | <0.001 |
| Absent | 33 | 136 | |||
| MVI | Present | 8 | 87 | 8.492 | 0.004* |
| Absent | 27 | 88 | |||
Over-expression of PDE4a in HCC tissues predicted unfavorable prognosis after liver resection
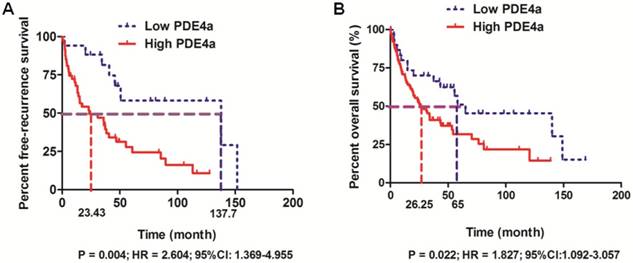
The follow-up information was achieved in 158 among 210 HCC patients (75.2%). 158 HCCs with the follow-up information were divided into High PDE4a and Low PDE4a group using the ratio of PDE4a expression in HCC/adjacent liver tissuses as cut-off value. The cumulative recurrence rates at 1-, 3- and 5- year after surgery were 34.5%, 55.5% and 75.6% in High PDE4a group, while those in Low PDE4a group were 5.9%, 18.6% and 41.8%. Additionally, comparison of Kaplan-Meier survival revealed that HCC patients from High PDE4a group had apparently higher post-surgical recurrence rates (P=0.004; HR=2.604; 95%CI: 1.369-4.955), as presented in Fig.2A. The median survival time of patients from High PDE4a group was 26.25 months which was remarkably lower than one of patients from Low PDE4a group (65 months). The 1-, 3- and 5-year post-surgical survival rates in High PDE4a group were 70.8%, 40.9% and 31.7%, while those in Low PDE4a group were 80.0%, 70.0% and 31.7%. Analysis of Kaplan-Meier survival curves demonstrated that overall survival time of HCC patients from High PDE4a group was markedly worse than those from Low PDE4a group (Fig.2B, P=0.022; HR=1.827; 95%CI: 1.092-3.057). Moreover, univariate and multivariate analysis identified over-expression of PDE4a in HCC tissues as well as advanced TNM stage, MVI, intrahepatic metastases and PVTT as the independent predictive factors for the unfavorable post-surgical overall survival time, as shown in Table 2. Thus, these data supported strongly that PDE4a was an efficient biomarker to predict the worse post-surgical outcome of HCCs.
The PDE4a was up-regulated in HCC tissues and HCC cell lines. A. The representative picture of PDE4a IHC staining in adjacent liver tissues (a) and HCC tissues (b). There was significantly more PDE4a expression in HCC tissues in contrast to adjacent liver tissues; B. Mann-Whitney U test demonstrated that PDE4a expression in HCC tissues was apparently higher than one in adjacent liver tissues; C. Both qRT-PCR and Western immunoblotting showed that there was the higher level of PDE4a expression in 5 kinds of HCC cell line (Huh7, Hep3B, MHCC97h, SK-Hep1 and PLC/PRF/5) than human liver cell line LO2. Among 5 kinds of HCC cell lines, MHCC97h cells had the highest level of PDE4a expression, while there was the least PDE4a expression in Huh7 cells.
PDE4a predicted the unfavorable prognosis of HCC patients after liver resection. A. HCC patients with the higher PDE4a expression in tumor tissues had the magnificently higher post-surgical recurrence rates compared to those with the lower PDE4a expression in tumor tissues (P=0.004; HR=2.604; 95%CI:1.369-4.955); B. HCC patients from High PDE4a group had the shorter overall survival time than those from Low PDE4a group (P=0.022; HR=1.827; 95%CI: 1.092-3.057).
Univariate and multivariate analyses of predictive factors in HCC patients with liver resection
| Clinicopathological features | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| RR (95% CI) | P value | RR (95% CI) | P value | |
| Age, year (<50 versus ≥50) | 0.782 (0.318 - 1.726) | 0.415 | 0.651 (0.277-1.237) | 0.524 |
| Gender (Female versus Male) | 0.354 (0.075 - 1.352) | 0.361 | 0.432 (0.069 - 1.686) | 0.505 |
| HBV infection | 1.925 (0.998 - 3.399) | 0.095 | 1.729 (0.895 - 2.529) | 0.122 |
| Serum AFP level (ng/mL) (≥ 400 versus < 400) | 1.528 (0.579 - 1.981) | 0.155 | 1.338 (0.578 - 1.988) | 0.257 |
| Tumor diameter (cm) (≥ 5 versus < 5) | 1.752 (0.612 - 2.428) | 0.091 | 1.487 (0.524 - 2.197) | 0.129 |
| Liver cirrhosis | 2.110 (0.977 - 2.352) | 0.124 | 1.857 (0.769-3.272) | 0.110 |
| Edmondson-Steiner Classification (III + IV versus I + II) | 1.381 (0.485 - 1.420) | 0.095 | 1.128 (0.569 - 2.155) | 0.138 |
| Advanced TNM staging | 1.932 (1.188 - 4.460) | 0.037 | 1.799 (1.395 - 3.715) | 0.044 |
| PVTT | 2.695 (1.853 - 5.257) | 0.025 | 2.495 (1.572 - 4.913) | 0.033 |
| Intrahepatic metastases | 1.582 (1.089 - 3.228) | 0.008 | 1.399 (1.090 - 2.652) | 0.019 |
| Higher PDE4a in tumor tissue | 1.827 (1.092-3.057) | 0.008 | 1.558 (1.164 - 2.615) | 0.012 |
| MVI | 2.358 (1.398-5.037) | 0.002 | 1.988 (1.125-4.289) | 0.001 |
Enforced expression of PDE4a accelerated malignant behavior of Huh7 cells via inducing EMT phenotype
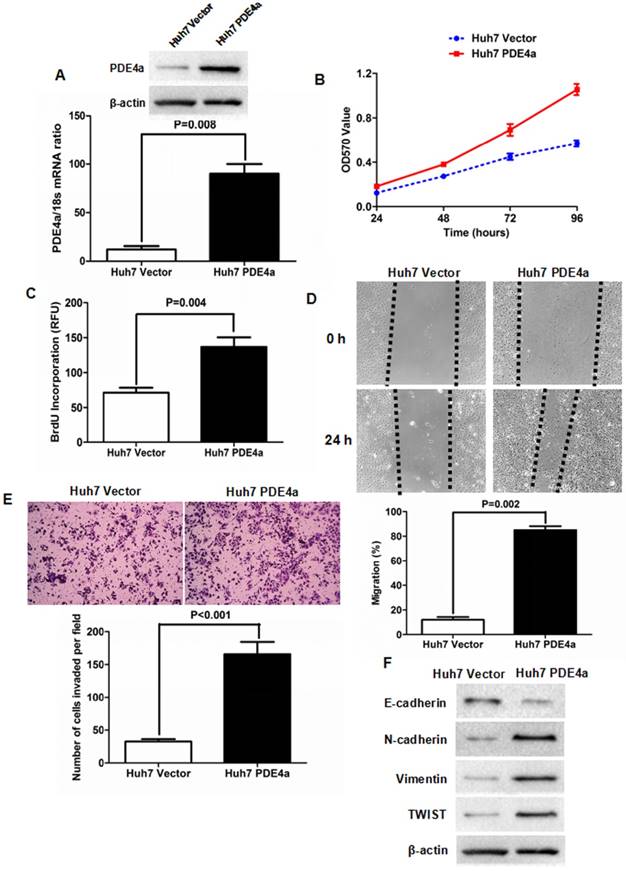
To figure out whether PDE4a was involved in the progression of HCC, PDE4a expressing plasmid was constructed and transfected into Huh7 cells. As shown in Fig.3A, both qRT-PCR and Western immunoblotting assays confirmed that there was more PDE4a expression in Huh7 PDE4a cells than Huh7 Vector cells at the levels of Mrna and protein. As assessed by MTT assay, cell viability of Huh7 cells was strengthened by ectopic expression of PDE4a (Fig.3B). BrdU incoporation detection showed that cell proliferation of Huh7 PDE4a cells was significantly up-regulated by enhancing PDE4a expression (Fig.3C). Scratch wound healing assay was conducted to evaluate the migration of Huh7 and found that Huh7 PDE4a cells had the higher migration capacity than Huh7 Vector cells (Fig.3D). Consistently, the invasion ability of Huh7 PDE4a cells was magnificently higher than one of Huh7 Vector cells (Fig.3E). And Western immunoblotting revealed that there were less E-cadherin expression and more expression of both N-cadherin and Vimentin in Huh7 PDE4a cells than Huh7 Vector cells (Fig.3F). And the expression of TWIST which has been well-known as EMT regulator was also increased by enhanced of PDE4a (Fig.3F). Therefore, these results demonstrated that PDE4a evoked EMT phenotype and consequently promoted cell viability, proliferation, migration and invasion of HCC cells.
Knockdown of PDE4a repressed the cell viability, proliferation, migration and invasion of MHCC97h cells
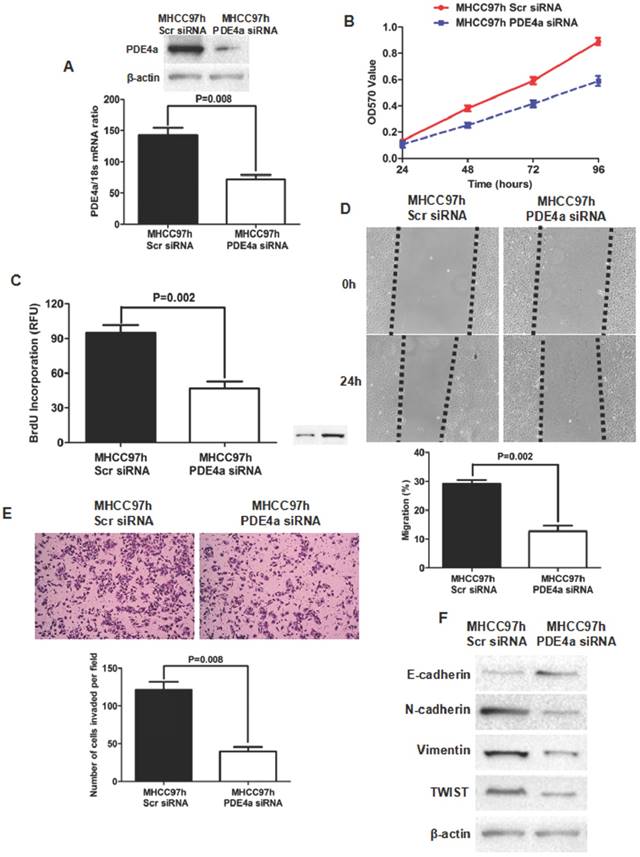
To further identify the oncogenic effect of PDE4a on HCC, siRNA sequences against PDE4a were transfected into MHCC97h cells and PDE4a expression was found inhibited dramatically by both qRT-PCR and Western immunoblotting (Fig.4A). MTT assay showed that silencing PDE4a leaded to significant repression of cell viability (Fig.4B). BrdU ELISA assay also revealed that cell proliferation of MHCC97h cells was restrained by repression of PDE4a (Fig.4C). Silencing PDE4a resulted in slower healing of scratch wounds inflicted on MHCC97h cells (Fig.4D). We carried out Transwell chamber (coated with the Matrigel) assay and found that the invasion capacity of MHCC97 cells was clearly impaired by knockdown of PDE4a (Fig.4E). In addition, knockdown of PDE4a increased E-cadherin expression and inhibited expression of N-cadherin and Vimentin (Fig.4F). TWIST expression was repressed by knockdown of PDE4a in MHCC97h cells (Fig.4F).
Discussion
Liver resection is the most common curative treatment and the post-surgical outcome remains unfavorable[18]. Recent investigations demonstrated that the 5-year rate of intrahepatic recurrence of HCC after liver resection was approximately 60%-100% at 5-years after resection[19]. Herein, it is urgent to develop the novel biomarkers to predict the post-surgical recurrence and survival of HCC. PDE4a is one of 4 members of PDE4 family (PDE4a, PDE4b, PDE4c, and PDE4d). It has been revealed that PDE4a modulated several critical biological processes, for instance chemotaxis, cytokine secretion, and proliferation[20-22] through controlling the degradation of cyclic 3'5'-adenosine monophosphate into 5-adenosine monophosphate[20, 23]. There have been few studied reported about the role of PDE4a on cancer progression. Garcia-Manero group revealed that PDE4 expression was apparently higher in patients with myelodysplastic syndromes (MDS) than in healthy individuals and PDE4 over-expression had the negative effect on survival[24]. PDE4 was established by Lerner group as the most important target for PDE inhbitor-driven apoptosis in chronic lymphocytic leukemia (CLL)[25]. Epstein group showed that T leukemic cell lines including CEM and Jurkat had aberrantly high level of PDE4 protein expression and PDE4 inhibitor notably strengthened glucocorticoid-induced apoptosis in CEM cells, and attenuated glucocorticoid resistance in a glucocorticoid-resistant CEM cell line[26].
Enforced expression of PDE4a induced EMT phenotype of Huh7 cell. A. Transfection of PDE4a expressing plasmid resulted in up-regulation of PDE4a in Huh7 cells; B. As assessed by MTT assay, enhanced expression of PDE4a increased the cell viability of Huh7 cells; C. ELISA assay displayed that there was magnificently more BrdU incorporation found in Huh7 PDE4a cells than Huh7 Vector cells; D. As examined by scratch wound healing assay, cell migration of Huh7 cells was strengthened by over-expression of PDE4a significantly; E. Transwell chamber coated with Matrigel matrix was used to measure the cell invasion ability and it was found that over-expression of PDE4a enhanced invasion capacity of Huh7 cells clearly; F. Enforced expression of PDE4a repressed E-cadherin expression and up-regulated the expression of N-cadherin, TWIST, Vimentin and TWIST in Huh7 cells. Supplementary fig. 1 presented the expression of E-cadherin, N-cadherin, Vimentin and TWIST examined by Western immunoblotting semiquantitatively.
Knockdown of PDE4a attentuated the malignant behaviors and reversed EMT phenotype in MHCC97h cells. A. As examined by both qRT-PCR and Western immunoblotting assays, PDE4a expression was found significantly decreased by transfection of PDE4a siRNA sequences; B. MTT assay showed that silencing PDE4a leaded to repression of cell viability in MHCC97h cells; C. BrdU ELISA assay displayed that knockdown of PDE4a restrained cell proliferation in MHCC97h cells significantly; D. Invasion capacity of MHCC97h cells was repressed clearly by knockdown of PDE4a; E. Silencing PDE4a was found to inhibit expression of both N-cadherin, Vimentin and TWIST, and increase E-cadherin expression by Western immunoblotting assay. Supplementary fig. 2 displayed the expression of E-cadherin, N-cadherin, Vimentin and TWIST examined by Western immunoblotting semiquantitatively.
However, no studies about the effect of PDE4a on HCC have been found yet. In this investigation, we detected the expression of PDE4a in HCC and adjacent liver tissues harvested during liver resection. PDE4a expression in HCC tissues was found clearly higher than that in adjacent liver tissues. In addition, higher PDE4a expression in HCC tissues was associated positively with the several unfavorable clinical characteristics including HBV infection, liver cirrhosis, higher serum AFP level, advanced TNM stage, microvascular invasion (MVI), intrahepatic metastases and portal vein tumor thrombus (PVTT) and further identified as the independent predictive factors for the poor overall survival time after surgery. PDE4a expression was also found up-regulated in five kinds of HCC cell lines compared with normal liver cell line. These data suggested strongly that PDE4a potentially mediated HCC progression positively. Pullamsetti group found that PDE4a contributed to EMT in alveolar epithelial cells and furthermore played a pivotal role in the TGFβ1-induced EMT involved in pulmonary fibrosis and lung cancer[27]. Therefore, we determined here whether PDE4a accelerated HCC progression via inducing EMT. Enforced expression of PDE4a enhanced the cell proliferation, viability, migration and invasion of Huh7 cells significantly. Besides it, ectopic expression of PDE4a repressed E-cadherin expression in Huh7 cells, while increased expression of TWIST, N-cadherin and Vimentin. Silencing PDE4a by siRNA sequences suppressed the cell proliferation, viability, migration and invasion of MHCC97h cells obviously. And knockdown of PDE4a leaded to up-regulation of E-cadherin and down-regulation of TWIST, N-cadherin and Vimentin in MHCC97h cells, as well. These data supported that PDE4a enhanced the malignant behaviors of HCC cells via driving EMT.
In summary, the present investigation revealed that PDE4a was aberrantly over-expressed in HCC tissues and predicted the poor post-surgical prognosis of HCCs. And PDE4a was initially found contributing to EMT of HCC cells, which could explain that PDE4a over-expression in HCC tissues was correlated positively with the unfavourable prognosis of HCCs after hepatectomy. Taken togather, PDE4a is involved closely in HCC progression by inducing EMT and could be the important biomarkers to predict the outcome of HCC after liver resection and therapeutic target.
Supplementary Material
Supplementary figures.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753-70
3. Xu M, Liu Z, Wang C, Yao B, Zheng X. EDG2 enhanced the progression of hepatocellular carcinoma by LPA/PI3K/AKT/ mTOR signaling. Oncotarget. 2017;8:66154-68
4. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178-96
5. Zheng X, Song T, Dou C, Jia Y, Liu Q. CtBP2 is an independent prognostic marker that promotes GLI1 induced epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget. 2015;6:3752-69
6. Gai X, Lu Z, Tu K, Liang Z, Zheng X. Caveolin-1 is up-regulated by GLI1 and contributes to GLI1-driven EMT in hepatocellular carcinoma. PLoS One. 2014;9:e84551
7. Zheng X, Vittar NB, Gai X, Fernandez-Barrena MG, Moser CD, Hu C. et al. The transcription factor GLI1 mediates TGFbeta1 driven EMT in hepatocellular carcinoma via a SNAI1-dependent mechanism. PLoS One. 2012;7:e49581
8. Jia YL, Xu M, Dou CW, Liu ZK, Xue YM, Yao BW. et al. P300/CBP-associated factor (PCAF) inhibits the growth of hepatocellular carcinoma by promoting cell autophagy. Cell Death Dis. 2016;7:e2400
9. Zhou M, Mok MT, Sun H, Chan AW, Huang Y, Cheng AS. et al. The anti-diabetic drug exenatide, a glucagon-like peptide-1 receptor agonist, counteracts hepatocarcinogenesis through cAMP-PKA-EGFR-STAT3 axis. Oncogene. 2017;36:4135-49
10. Riggle KM, Riehle KJ, Kenerson HL, Turnham R, Homma MK, Kazami M. et al. Enhanced cAMP-stimulated protein kinase A activity in human fibrolamellar hepatocellular carcinoma. Pediatr Res. 2016;80:110-8
11. Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;288:28893-9
12. Zhang L, Murray F, Zahno A, Kanter JR, Chou D, Suda R. et al. Cyclic nucleotide phosphodiesterase profiling reveals increased expression of phosphodiesterase 7B in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2008;105:19532-7
13. Marko D, Romanakis K, Zankl H, Furstenberger G, Steinbauer B, Eisenbrand G. Induction of apoptosis by an inhibitor of cAMP-specific PDE in malignant murine carcinoma cells overexpressing PDE activity in comparison to their nonmalignant counterparts. Cell Biochem Biophys. 1998;28:75-101
14. Tanaka M, Ishizuka K, Nekooki-Machida Y, Endo R, Takashima N, Sasaki H. et al. Aggregation of scaffolding protein DISC1 dysregulates phosphodiesterase 4 in Huntington's disease. J Clin Invest. 2017;127:1438-50
15. Goldhoff P, Warrington NM, Limbrick DD Jr, Hope A, Woerner BM, Jackson E. et al. Targeted inhibition of cyclic AMP phosphodiesterase-4 promotes brain tumor regression. Clin Cancer Res. 2008;14:7717-25
16. Yang W, Wu Y, Wang C, Liu Z, Xu M, Zheng X. FSTL1 contributes to tumor progression via attenuating apoptosis in a AKT/GSK-3beta - dependent manner in hepatocellular carcinoma. Cancer Biomark. 2017;20:75-85
17. Song T, Dou C, Jia Y, Tu K, Zheng X. TIMP-1 activated carcinoma-associated fibroblasts inhibit tumor apoptosis by activating SDF1/CXCR4 signaling in hepatocellular carcinoma. Oncotarget. 2015;6:12061-79
18. Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Med. 2014;11:e1001624
19. Dai WC, Cheung TT. Strategic overview on the best treatment option for intrahepaitc hepatocellular carcinoma recurrence. Expert Rev Anticancer Ther. 2016;16:1063-72
20. Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488-520
21. Srivani P, Usharani D, Jemmis ED, Sastry GN. Subtype selectivity in phosphodiesterase 4 (PDE4): a bottleneck in rational drug design. Curr Pharm Des. 2008;14:3854-72
22. Spina D. Phosphodiesterase-4 inhibitors in the treatment of inflammatory lung disease. Drugs. 2003;63:2575-94
23. Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC. Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov. 2014;13:290-314
24. Chamseddine AN, Cabrero M, Wei Y, Ganan-Gomez I, Colla S, Takahashi K. et al. PDE4 Differential Expression Is a Potential Prognostic Factor and Therapeutic Target in Patients With Myelodysplastic Syndrome and Chronic Myelomonocytic Leukemia. Clin Lymphoma Myeloma Leuk. 2016;16(Suppl):S67-73
25. Moon E, Lee R, Near R, Weintraub L, Wolda S, Lerner A. Inhibition of PDE3B augments PDE4 inhibitor-induced apoptosis in a subset of patients with chronic lymphocytic leukemia. Clin Cancer Res. 2002;8:589-95
26. Dong H, Zitt C, Auriga C, Hatzelmann A, Epstein PM. Inhibition of PDE3, PDE4 and PDE7 potentiates glucocorticoid-induced apoptosis and overcomes glucocorticoid resistance in CEM T leukemic cells. Biochem Pharmacol. 2010;79:321-9
27. Kolosionek E, Savai R, Ghofrani HA, Weissmann N, Guenther A, Grimminger F. et al. Expression and activity of phosphodiesterase isoforms during epithelial mesenchymal transition: the role of phosphodiesterase 4. Mol Biol Cell. 2009;20:4751-65
Author contact
![]() Corresponding author: Guokun Ao, paulzhengxjtu.edu.cn, Department of Radiology, the 309th Hospital of Chinese People's Liberation Army, 17 Heishanhu Rd, Haidian District, Beijing 100091, P.R. China
Corresponding author: Guokun Ao, paulzhengxjtu.edu.cn, Department of Radiology, the 309th Hospital of Chinese People's Liberation Army, 17 Heishanhu Rd, Haidian District, Beijing 100091, P.R. China

 Global reach, higher impact
Global reach, higher impact