3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2018; 9(13):2302-2307. doi:10.7150/jca.24932 This issue Cite
Research Paper
The Tumor Mutational Burden of Chinese Advanced Cancer Patients Estimated by a 381-cancer-gene Panel
1. Department of Medical Thoracic Oncology, Fujian Cancer Hospital, Fujian Medical University Cancer Hospital, Fujian, P.R. China.
2. Department of Oncology, Chinese PLA General Hospital, Beijing, P.R. China.
3. Department of Interventional Radiology, the Second Clinical Medical College (Shenzhen People's Hospital), Jinan University, Shenzhen, P.R. China.
4. The medical department, 3D Medicine Inc. Shanghai, P.R. China.
5. The bioinformatics department, 3D Medicine Inc. Shanghai, P.R. China.
6. Department of Thoracic Disease Center, Zhejiang Rongjun Hospital, Zhejiang, P.R. China.
7. Department of Pathology, Fujian Cancer Hospital, Fujian Medical University Cancer Hospital, Fujian, P.R. China.
8. Transplantation Center, Department of Transplant Surgery, The First Affiliated Hospital, Sun Yet-sen University, Guangzhou, P. R. China.
9. Oncology Department, East Hospital Affiliated to Shanghai Tongji University, Shanghai, P.R. China.
10. Department of medical oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, P.R. China.
*Co-first authors
Received 2018-1-14; Accepted 2018-4-2; Published 2018-6-6
Abstract
Purpose: Tumor mutational burden (TMB) calculated by whole-exome sequencing (WES) is proved to be effective to predict the clinical benefit of immune checkpoint blockades. However, WES is not commonly used in China. We aimed to determine if a 381-caner-gene panel (CGP) could be used to estimate TMB, delineate the landscape of TMB of Chinese patients and identify mutated genes and pathways related to higher TMB.
Methods: We first evaluated the correlation between the TMB estimated by a 381-cancer-gene panel MasterView and WES using the data from the melanoma sample cohort. 3023 formalin fixed, paraffin-embedded tumor specimens from 2932 Chinese patients with advanced solid tumor were profiled for 381 gene sequencing, the baits of which covered 4,557 exons of 365 cancer-related genes and 47 introns of 25 genes frequently rearranged in cancer (All performed in a lab who achieved full marks five times in the external quality assessment by College of American Pathologists [CAP]). Using the sequencing data, we estimated the TMB of Chinese advanced solid tumor and identified mutated genes and pathways related to higher TMB level.
Results: 381-CGP-mutational burden was strongly associated with those calculated by WES (R2 = 0.978). The median TMB for each tumor type was 5.65 (colorectal cancer), 4.84 (lung cancer), 4.03 (hepatobiliary cancer), 4.03 (gastric carcinoma), 4.03 (breast cancer) mutations/mb respectively. No correlation was observed between TMB level and age (P = 0.577) or gender (P = 0.307). The TMB of patients with mismatch repair (MMR) or DNA repair response (DDR) pathway deficiency was significantly higher than that without MMR or DDR pathway deficiency (P < 0.001).
Conclusion: The 381-cancer gene panel is a clinical practicable method to assess tumor mutational burden compared with whole exome sequencing. MMR and DDR deficiency are correlated with higher tumor mutational burden of Chinese patients with advanced solid tumors.
Keywords: Tumor mutational burden, cancer-gene panel, mismatch repair, DNA damage response
Introduction
In recent years, great progress has been made in revolutionizing cancer treatment by targeting immune checkpoint to enhance anti-tumor immune response. Immune checkpoint blockades have been proven to be effective in the treatment of melanoma, Hodgkin lymphoma, urothelial carcinoma, renal cell carcinoma, non-small cell lung cancer, head and neck cancer, hepatocellular carcinoma, nasopharyngeal carcinoma, and tumors with mismatch repair deficiency [1-9]. However, not all patients can benefit from immunotherapy [10], which raises the question that which patients will be sensitive or resistant to checkpoint blockade. Therefore, to identify biomarkers that would enrich patients with responsiveness to checkpoint blockades has become a priority.
It has been demonstrated that tumor mutational burden (TMB), neoantigen burden, insertion and deletion burden, DNA mismatch repair (MMR) deficiency, the intensity of CD8+ T cell infiltrates, untratumoral PD-L1 expression and immune-regulatory mRNA expression signatures can be proposed as distinct biomarkers of response to immune checkpoint blockades [7, 11-15]. Among these biomarkers, higher TMB are demonstrated to be related with responsiveness to immune checkpoint blockades [16], supposing that higher tumor nonsynonymous somatic mutations would lead to higher diversity of neoantigens, thus enhancing immune response [17].
In most studies, tumor mutational burden is calculated by whole-exome sequencing (WES). However, in China WES is not usually applied in clinic. Instead, cancer-gene panels (CGPs) are now commonly used in clinic to find precise targeted therapies for patients [18]. A previous study has shown that CGPs with a proper composition of cancer genes can estimate TMB to predict clinical benefit of immune checkpoint blockades [19]. Therefore, in present study we sought to evaluate whether a 381 cancer gene panel could be used to estimate the TMB. Then we estimated the TMB of Chinese progressed solid tumor patients using the 381 cancer gene panel and identified mutated genes and pathways related to TMB level. We found that the TMB estimated by the 381 cancer-gene panel was well correlated with that calculated by WES, thus proving the 381 gene-panel is a promising method to predict clinical benefit to immunotherapy. We also found that mismatch repair or DNA repair response pathway mutations was associated with higher TMB. To our knowledge, this is the first study to demonstrate the TMB of Chinese patients with advanced solid tumors.
Materials and methods
Study design
We downloaded the Melanoma sample cohort from the paper with 100 WES sequencing data [20] to evaluate the association between the TMB estimated by the 381 cancer gene panel and WES-TMB. We enrolled 3023 formalin fixed, paraffin-embedded (FFPE) tumor specimens from 2932 Chinese patients with advanced solid tumor. We calculated the tumor mutational burden for each specimen using the sequencing data of the 381 cancer-gene panel.
381 cancer-gene panel profiling
The MasterView 381 cancer-gene panel covered 4,557 exons of 365 cancer-related genes and 47 introns of 25 genes frequently rearranged in 381 cancer-related genes. The detailed genes for the 381 cancer-gene panel were provided in Supplemental Table S1.
Detailed experiment design of protocol was as previously described [21]. In brief, we enrolled 3023 FFPE tumor specimens. The pathological diagnosis of the specimens was confirmed by hematoxylin and eosin (HE) staining. Specimens with a volume <1 mm or tumor cells <20% were excluded from the following profiling. 50-200 ng of DNAs extracted from the FFPE tumor specimens were broke into ~200 bp fragments, which were then sequenced in next generation sequencing platform for base substitutions, indel, CNA and DNA arrangement analysis.
Tumor mutational burden estimation
TMB was defined by non-silent somatic mutation counts in coding region per megabase of genome examined. SNV include both synonymous and non-synonymous mutations, as well as stopgain, stoploss, and splicing variants. Indel variants include both the frameshift or non-frameshift insertions and deletions. Non-coding alterations were not counted. All germline variants were filtered by paired adjacent normal sample or blood controls. An in-house developed script was used to filter false positive variants. Panel size was defined by base number of the coding region of targeted genes.
Statistical analysis
The data was analyzed by R 3.4.1 and Graphpad prism v6. The correlation between the TMB estimated by cancer gene panel and WES was performed by linear regression. Mismatch repair deficiency is defined as any mutation in coding region of any of the four genes, MSH2, MSH6, MLH1, PMS2 according to a previous study [7]. DDR deficiency is defined as any mutation in coding region of any of the 381 genes related to base excision repair, homology recombination repair, mismatch repair, non-homologous end joining, and nucleotide excision repair pathway, including ATM, BRAD1, BRIP1, CHEK1, CHEK2, PALB2, FANCA, FANCC, FANCD2, FANCE, FANCF, ATR, BAP1, CDK12, BLM, RAD50, RAD51, STK11, BRCA1, BRCA2, CDH1, MRE11A, AURKA, MLH1, MSH2, MSH6, PMS2, ERCC1, POLE, POLD1 and PRKDC, which were defined by paper reviewing. Comparisons of tumor mutational burden between two groups were performed by unpaired t-test assuming the variances were not equal.
Results
Tumor mutational burden could be estimated by the 381 cancer-gene panel accurately
At first, we tried to determine whether the TMB measured by the 381 MasterView gene panel, would be associated with whole exome TMB. We used the melanoma sample cohort from the paper with 100 WES sequencing data [20].
The TMB estimated by the 381-cancer-gene panel was highly correlated with WES TMB
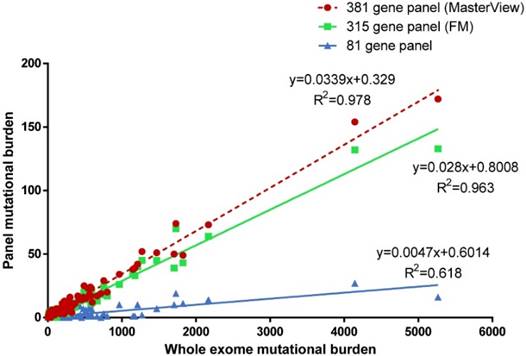
We sampled the 381 MasterView gene panel, Foundation Medicine 315 gene panel and 81 gene panel from this 100-sample cohort and calculated the correlation between panel sampling TMB and WES TMB (Figure 1). We found that the tumor mutational burden estimated by panel sampling was highly correlated with WES TMB (381 gene panel, P < 0.001, R2 = 0.978; 315 gene panel, P < 0.001, R2 = 0.963; 81 gene panel, P < 0.001, R2=0.618). However, this result indicated that 381 gene panel (R2 = 0.978) was better correlated with whole exome TMB than 81 gene panel (R2 = 0.618), which was consistent with a previous conclusion that it was suggested that cancer-gene panels comprised of more than 300 cancer-genes were used to estimate tumor mutational burden [19].
The tumor mutational burden across different cancer types
We then assessed the TMB of different cancer type in the patients profiled in our lab. 3023 specimens from 2923 patients were profiled for the 381 cancer-gene panel. As shown in Table 1, the patients contained 1704 male patients and 1228 female patients. The median age of the patients was 55 years old, with a range of 4-88. There were 16 patients under 18 years old. There were mainly 5 kinds of tumor for these patients: hepatobiliary cancer (738), lung cancer (582), breast cancer (196), colorectal cancer (243) and gastric cancer (169). The tumor types of the other 1004 patients were unknown.
Baseline characteristic of included patients
| Characteristic | |
|---|---|
| Age, median (range) | 55 (4-88) |
| Sex, n (%) | |
| male | 1704 (58.12%) |
| female | 1228 (41.88%) |
| Tumor type, n (%) | |
| hepatobiliary cancer | 738 (25.25%) |
| lung cancer | 582 (19.85%) |
| colorectal cancer | 243 (8.29%) |
| breast cancer | 196 (6.68%) |
| gastric cancer | 169 (5.76%) |
| undefined | 1004 (34.24%) |
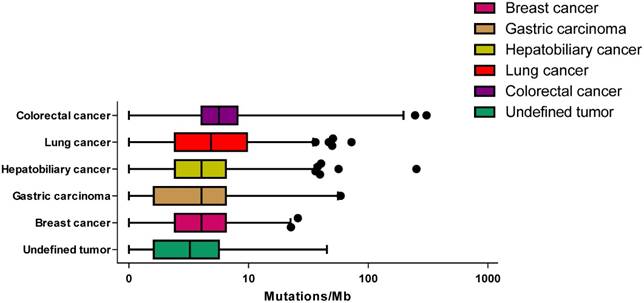
Across these specimens, the median TMB was 4.03, with a range of 0-608. Across these tumor types, the median TMB ranged from 4.03 mutations/mb in breast cancer to 5.65 mutations/mb in colorectal cancer (Figure 2). No statistically significant difference was observed between male and female patients (unpaired t-test P = 0.307) and there was no association between age and TMB (P = 0.577).
Identifying genes and pathways associated with higher TMB
We then explored the mutated genes or pathways that were associated with higher TMB. We identified 20 genes which were significantly associated with higher TMB with adjusted P value <0.001 (Supplemental Table S2), including TP53, which was consistent with a previous study [22].
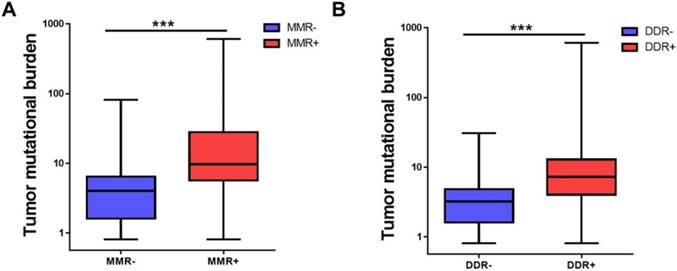
DNA damage response (DDR) including mismatch repair (MMR) is a mechanism to combat threats of DNA damage by detecting DNA lesions, signaling their presence and promoting their repair [23]. Defects in DDR may result in genomic instability, manifested as higher tumor mutational burden [18, 23]. It was demonstrated that DNA mismatch repair pathway was related to TMB level [7]. We then tried to identify whether MMR or DDR pathway was involved in the TMB level of Chinese advanced solid tumor. As shown in Figure 3, MMR deficiency (MMR+) patients had a higher TMB than MMR- patients (33.11 ± 78.20 vs 5.38 ± 6.34, adjusted P<0.001) and DDR deficiency (DDR+) patients had a higher TMB than DDR- patients (12.47 ± 29.93 vs 3.54 ± 3.21, adjusted P<0.001). MMR deficiency or DDR deficiency was also associated with higher TMB in the five tumor types (Supplemental Figure S1).
The tumor mutational burden for different cancer type. The median mutation burden for different cancer type is plotted with points beyond 1-99 percentiles plotted individually.
MMR and DDR were associated with tumor mutational burden. (A) Comparison of tumor mutational burden in specimens of MMR+ and MMR-. (B) Comparison of tumor mutational burden in specimens of DDR+ and DDR-. TMB, tumor mutational burden, MMR, mismatch repair, DDR, DNA damage response. ***P<0.001.
Discussion
In the present study, we have demonstrated that tumor mutational burden (TMB) estimated by a 381 cancer-gene panel are highly correlated with the TMB calculated by whole exome sequencing. The detailed information of the TMB from 3023 specimens from Chinese patients with advanced solid tumors has been shown and no correlation has been found between TMB and age or gender. The related mutated genes or pathways have been analyzed and DDR and MMR mutation have been found to be related with higher TMB level. These data helps to delineate a picture of TMB of Chinese patients of different tumor types.
Our result suggested that tumor mutational burden estimated by the 381-cancer-gene panel was associated with that calculated by WES. There are several differences between cancer-gene panels and WES. It requires specimens to contain at least 40% tumor cells to perform WES, while 20% tumor cells would be applicable for cancer-gene panel profiling. Besides, WES covers 39 mb of human genome, while the 381 cancer-gene panel covers 2 mb, which suggests that WES cost much higher than cancer-gene panels. Considering the cost and requirement of specimen, cancer-gene panels may be more appropriate than WES to estimate TMB and predict the efficacy of immunotherapy. However, not all cancer-gene panels can be applied to estimate TMB. In this present study, we found that 381 cancer-gene panel associates with the TMB calculated by WES better than 81 cancer-gene panel. It requires at least a certain number of cancer genes to ensure the accuracy of TMB prediction.
This is the first study to describe the landscape of TMB of Chinese patients. We didn't observe any correlation between TMB and age, which was inconsistent with a previous study [18]. To be noted, our study only included 16 adolescent patients. It may be the reason why the correlation was not observed. Further, the median TMB for each tumor type ranged from 4.03 mutations/mb in breast cancer to 5.65 mutations/mb in colorectal cancer. There were patients with relatively high TMB in each tumor type, which suggested that there were patients of each tumor type who might respond well to immunotherapy.
Understanding the genes and pathways related to higher TMB is also important to guide immunotherapy. We identified 20 genes whose mutations are strongly associated with TMB including TP53. Previous studies have demonstrated that in lung adenocarcinoma, TP53 and KRAS mutations are associated with higher tumor mutational burden and predict the response to anti-PD-1 therapy [22]. Besides, our data demonstrated that deficiency in MMR or DDR pathway was related with higher TMB. Dysfunction in MMR or DDR pathway may result in new nonsynonymous somatic mutations, creating new epitopes for T cells to recognize. That's the reason why MMR or DDR mutation are associated with higher TMB. A recent study has demonstrated cancers with MMR deficiency respond well to immune checkpoint blockades, irrespective of cancer type [7].
Our study has several limitations. First, we didn't know the exact pathological classification for each patient, which might help to better understand the difference of TMB between different pathological classifications. Second, the number of patients is relatively small and the cancer type is limited. Further studies are needed to better delineate the TMB of Chinese patients.
Conclusion
These results show that the 381-cancer-gene panel can be applied to assess TMB compared with whole exome sequencing. We identify deficiency in mismatch repair and DNA repair response pathway may be associated with higher TMB.
Abbreviations
CAP: College of American Pathologists; CGP: cancer-gene panel; DDR: DNA repair response; FFPE: formalin fixed paraffin-embedded; HE: hematoxylin and eosin; MMR: mismatch repair; TMB: tumor mutational burden; WES: whole-exome sequencing.
Supplementary Material
Supplementary tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. PD-1 inhibitors effective in hodgkin lymphoma. Cancer Discov 2015, 5(2):102-103.
2. Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nature reviews Clinical oncology. 2017;14(8):463-482
3. Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC. et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet (London, England). 2017;389(10066):255-265
4. Three Drugs Approved for Urothelial Carcinoma by FDA. Cancer Discov 2017, 7(7):659-660.
5. Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER. et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. The New England journal of medicine. 2015;373(19):1803-1813
6. Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C. et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. The New England journal of medicine. 2016;375(19):1856-1867
7. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409-413
8. El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling THR. et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492-2502
9. Hsu C, Lee SH, Ejadi S, Even C, Cohen RB, Le Tourneau C, Mehnert JM, Algazi A, van Brummelen EMJ, Saraf S. et al. Safety and Antitumor Activity of Pembrolizumab in Patients With Programmed Death-Ligand 1-Positive Nasopharyngeal Carcinoma: Results of the KEYNOTE-028 Study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2017;35(36):4050-4056
10. Hsu C, Lee SH, Ejadi S, Even C, Cohen RB, Le Tourneau C, Mehnert JM, Algazi A, van Brummelen EMJ, Saraf S. et al. Safety and Antitumor Activity of Pembrolizumab in Patients With Programmed Death-Ligand 1-Positive Nasopharyngeal Carcinoma: Results of the KEYNOTE-028 Study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2017 JCO2017733675
11. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371(23):2189-2199
12. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568-571
13. Fang W, Chen Y, Sheng J, Zhou T, Zhang Y, Zhan J, Liu L, Huang J, Peng P, Zhang L. Association between PD-L1 Expression on Tumour-Infiltrating Lymphocytes and Overall Survival in Patients with Gastric Cancer. Journal of Cancer. 2017;8(9):1579-1585
14. Gridelli C, Ardizzoni A, Barberis M, Cappuzzo F, Casaluce F, Danesi R, Troncone G, De Marinis F. Predictive biomarkers of immunotherapy for non-small cell lung cancer: results from an Experts Panel Meeting of the Italian Association of Thoracic Oncology. Translational lung cancer research. 2017;6(3):373-386
15. Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, Wong YNS, Rowan A, Kanu N, Al Bakir M. et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. The Lancet Oncology. 2017;18(8):1009-1021
16. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY). 2015;348(6230):124-128
17. Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nature reviews Cancer. 2017;17(9):569
18. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J. et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome medicine. 2017;9(1):34
19. Campesato LF, Barroso-Sousa R, Jimenez L, Correa BR, Sabbaga J, Hoff PM, Reis LF, Galante PA, Camargo AA. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to PD-1 blockade in clinical practice. Oncotarget. 2015;6(33):34221-34227
20. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (New York, NY). 2015;350(6257):207-211
21. Su D, Zhang D, Chen K, Lu J, Wu J, Cao X, Ying L, Jin Q, Ye Y, Xie Z. et al. High performance of targeted next generation sequencing on variance detection in clinical tumor specimens in comparison with current conventional methods. Journal of experimental & clinical cancer research: CR. 2017;36(1):121
22. Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, Tu HY, Chen HJ, Sun YL, Zhou Q. et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23(12):3012-3024
23. Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071-1078
Author contact
![]() Corresponding authors: Xiaofeng Zhu, Transplantation Center, Department of Transplant Surgery, the First Affiliated Hospital, Sun Yet-sen University, Guangzhou, P. R. China. Tel: (+86) 20 87332200-8752, Email: 13602820518com. Hong Jiang, Oncology Department, East Hospital Affiliated to Shanghai Tongji University, 150 Jimo road, Pudong new area, Shanghai, P.R. China, 200120. Tel: (+86) 02138804518; E-mail: Jianghong2046com. Zhijie Wang, Department of medical oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Chaoyang District Panjiayuan nail No.17, Beijing, P.R. China, 100021. Tel: (+86)1087788029, Email: jie_969com
Corresponding authors: Xiaofeng Zhu, Transplantation Center, Department of Transplant Surgery, the First Affiliated Hospital, Sun Yet-sen University, Guangzhou, P. R. China. Tel: (+86) 20 87332200-8752, Email: 13602820518com. Hong Jiang, Oncology Department, East Hospital Affiliated to Shanghai Tongji University, 150 Jimo road, Pudong new area, Shanghai, P.R. China, 200120. Tel: (+86) 02138804518; E-mail: Jianghong2046com. Zhijie Wang, Department of medical oncology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Chaoyang District Panjiayuan nail No.17, Beijing, P.R. China, 100021. Tel: (+86)1087788029, Email: jie_969com

 Global reach, higher impact
Global reach, higher impact