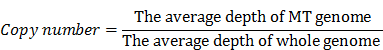Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(19):4011-4017. doi:10.7150/jca.21218 This issue Cite
Research Paper
Heteroplasmy and Copy Number Variations of Mitochondria in 88 Hepatocellular Carcinoma Individuals
1. Jining Medical University, Jining, Shandong 272067, China;
2. Collaborative Innovation Center for Birth Defect Research and Transformation of Shandong Province, Jining Medical University, Jining, Shandong 272067, China;
3. BGI-Shenzhen, Shenzhen, 518083, China.
* These authors contributed equally to this work
Received 2017-5-28; Accepted 2017-8-28; Published 2017-10-23
Abstract
Hepatocellular carcinoma (HCC) is the third leading cause of cancer mortality worldwide. In this study, we had analysed the copy number variations and heteroplasmic mutations of mitochondria (MT) in 88 HCC individuals. The average copy number of MT genome in normal samples was significantly greater than that in tumor samples. Overall, the number of heteroplasmic mutations in 88 tumor and their matched normal samples were 241 and 173, respectively. There was higher positive ratio of heteroplasmic mutations in tumor samples (86%) than normal samples (73%). Worthwhile mention, ND1 gene harbored greater mutation frequency and more nonsynonymous mutations in tumor samples. Interestingly, 202 tumor-specific heteroplasmic mutations were detected. Moreover, ND1, ND3, ND4, ND5 and ND6 genes had higher ratio of nonsynonymous versus synonymous mutations in tumor-specific heteroplasmic mutations. It might suggest that the disorder of NADH dehydrogenase (complex I) resulted by heteroplasmic mutations may have close relation with tumorigenesis of hepatocellular carcinoma. This study provided theoretical basis for further understanding mechanism of tumorigenesis from the perspective of mitochondrial heteroplasmic mutations.
Keywords: Heteroplasmy, Mitochondrial Genome, Copy Number, Hepatocellular carcinoma.
Introduction
Hepatocellular carcinoma (HCC) was the third leading cause of cancer mortality worldwide, responsible for more than half a million deaths each year [1]. The incidence and mortality in China were about 3 55,595 and 322,417 cases in 2011[2]. HCC had strong association with chronic hepatitis B virus (HBV) and chronic hepatitis C virus (HCV) infections, and additionally alcohol abuse [3]. Currently, HBV carriers and chronic B hepatitis patients were estimated around 93 million and 30 million in China [4].
Mitochondrion (MT) was an extremely important cellular organelle. It was best known for adenosine triphosphate (ATP) production, synthesizing ~95% of energy required for metabolism. MT genome encoded 13 polypeptides of the respiratory chain complexes, as well as 22 transfer RNAs and two ribosomal RNAs participating in mitochondrial protein synthesis [5]. Mitochondria also played a key role in the regulation of cellular function and cell apotosis [6-8]. Previous study found that mitochondria had the correlation with tumorigenesis by increasing production of reactive oxygen species (ROS), decreasing oxidative phosphorylation and increasing in glycolysis [9-11].
Oxidative stress was increased in individuals infected with HBV [12, 13], and the accompanied reactive oxygen was an important inducer of DNA mutations [14]. Because of lacking histones and DNA repair system, mutations of MT genome could accumulate to a greater extent [15, 16]. In the past few years, mutations of MT genome had been widely detected in several types of cancer [17-20]. For instance, mutations of DLOOP region had been linked to less differentiated hepatocellular carcinomas (HCC) and with stage progression and prognosis in non-small cell lung cancers [21, 22]. Many types of human malignancy, including colorectal cancer, liver cancer, breast cancer, lung cancer, prostate cancer, skin cancer and bladder cancer, were related to tumor-specific mutations of MT genome [10, 23].
Traditionally, the copy number and mutations of MT genome were detected by PCR-based methods. Significant bias could be caused by primer designing and the PCR process. With the rapid development of massively parallel sequencing (MPS) technology, whole genome sequencing (WGS) will bring new insight to the detection of copy number variations and mutations of MT genome.
In this study, published WGS data were used to investigate the heteroplasmic mutations and copy number variations of the mitochondrial genome in 88 pairs of tumor and matched normal samples [24]. The result indicated that there was lower copy number of mitochondrial DNA in tumor samples. Heteroplasmic mutations were detected by Mito-seek [25], and 241 and 173 heteroplasmic mutations were found in 88 pairs of tumor and matched normal samples. Interestingly, we found that ND1 gene harbored higher mutation frequency, and nonsynonymous mutations were more frequently detected in tumor samples. In particular 202 mutations were detected only in tumor samples. The data of copy number variations and heteroplasmic mutations could provide the basis for further understanding mechanism of tumorigenesis.
Materials and Methods
Samples and data preparation
We studied 88 Chinese individuals who were diagnosed with HCC at Queen Mary Hospital. Sample collection and whole genome sequencing had been described in the previous publication [24]. Written informed consents were obtained from each patient. The study protocol was approved by the Institutional Review Board of Queen Mary Hospital [26]. The data was anonymized before the whole genome sequence data was accessed. Sequence data had been deposited in the European Nucleotide Archive (ENA, http://www.ebi.ac.uk/ena/) under accession number ERP001196 and in GigaDB (http://dx.doi.org/10.5524/100034).
Heteroplasmic Mutations Calling and Copy number Calculation
Low-quality and adaptor-contamination reads were removed to obtain clean reads for subsequent analysis; meanwhile, MT genome (NC_012920) was used as reference. Then clean reads were aligned to this reference genome by using the BWA software [27]. A mapping quality score was assigned to each alignment by BWA. After removing the PCR duplicates and low mapping quality (Q<10) reads, we used the Mito-seek software to call heteroplasmic mutations [25]. In each sample, we calculated the copy number of MT genome according to the ratio of the average depth of MT genome vs whole human genome. The equation we used to calculate the copy number of MT genome was as followed:
Results
Copy number variations and heteroplasmic mutations between tumor and normal samples
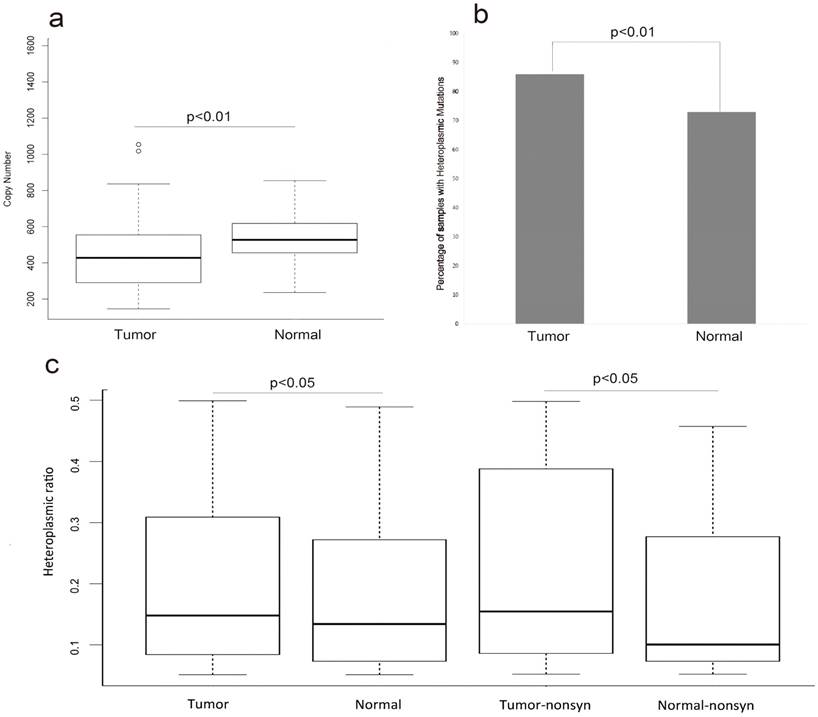
To evaluate whether or not any changes of MT genome within the HCC patients, we compared the copy number of MT genome between the tumor samples and their matched normal samples in the 88 HCC patients. The average copy number of MT genome in the normal samples was significantly higher than that in tumor samples (FIG.1a, p-value<0.01, T-test). The average copy number of MT genome was 453 and 532 in the tumor samples and their adjacent normal samples, respectively (Table S1).
There were 241 and 173 heteroplasmic mutations in tumor and normal samples, respectively (Table S1, Table S2). We found higher positive ratio of heteroplasmic mutation in tumor samples 86% (76/88) than that in normal samples 73%(65/88) (FIG.1b, p-value<0.01,Chi-Squared test). Next, we surveyed the heteroplasmic ratio of heteroplasmic mutations. There was significant higher heteroplasmic ratio in tumor samples than that in normal samples. Next we divided these heteroplasmic mutations of tumor and normal samples into synonymous and nonsynonymous mutations and surveyed the difference of heteroplasmic ratio. The results revealed that there was higher heteroplasmic ratio of nonsynonymous mutations in tumor samples (FIG.1c, p<0.05, T-test).
The copy number and heteroplasmic mutations of tumor and normal samples (a) represented that the difference of copy number variation in tumor and normal samples (T-test, p<0.01). (b) represented the difference of the ratio of samples with heteroplasmic mutations between tumor and normal samples (Chi-squared test, p<0.01). (c)represented that comparing the heteroplasmic ratio of heteroplasmic mutations in tumor and normal samples (T-test, p<0.05) and comparing the heteroplasmic ratio of nonsynonymous mutations in tumor and normal samples (T-test, p<0.05).
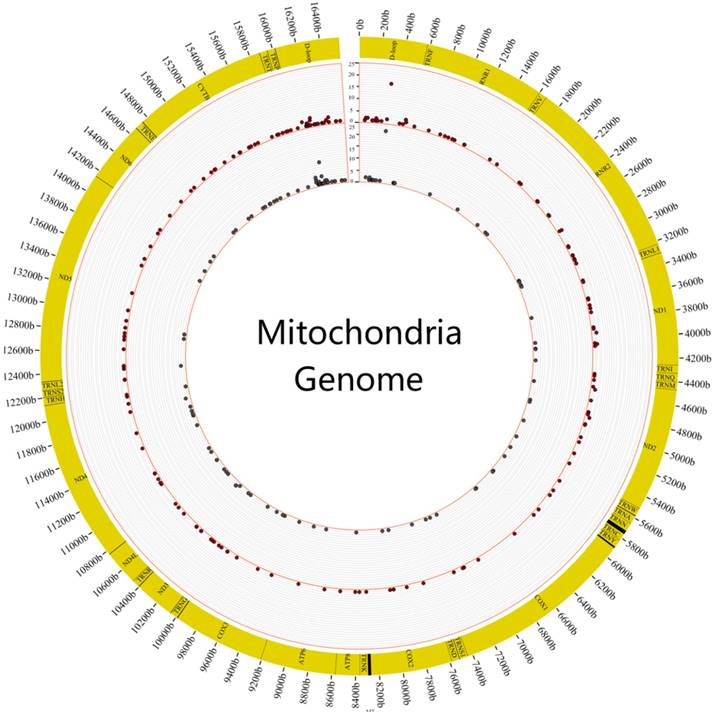
The Inclination of Heteroplasmic Mutations
We first surveyed the frequency of heteroplasmic mutations in these 88 individuals. It revealed that heteroplasmic mutation was prone to occur in the region of DLOOP (FIG.2). The highest frequency of heteroplasmic mutation was C312A of mitochondrial genome in the tumor and normal samples.
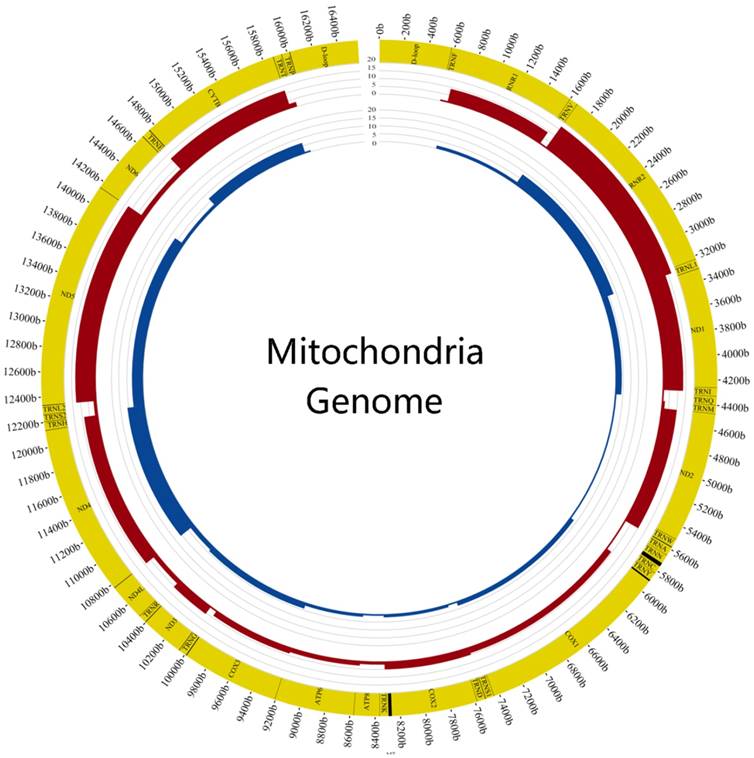
We also calculated the mutation frequency of genes and found the top 6 genes of tumor samples were DLOOP(41), RNR2(17), ND5(13), ND1(13), CYTB(12), ND2(10) and the normal samples were DLOOP(50), ND4(11), RNR2(8), ND5(7), CYTB(7), ND1(4) (FIG.3). The results of comparing mutation frequency of genes between tumor and normal samples showed that ND1 and ND2 have higher mutation frequency in tumor samples (Table S3). Meanwhile, the ratio of nonsynonymous versus synonymous heteroplasmic mutations in ND1 genes was greater in tumor samples (p<0.01, Chi-squared test) (Table 1).
The specific heteroplasmic mutations of tumor
In order to analyse the characteristics of tumor-specific heteroplasmic mutations in 88 HCC patients, heteroplasmic mutations shared by tumor and normal samples were filtered out, and consequently obtained 202 tumor-specific mutations in 68 tumor samples (Table S4). Overall, 32 synonymous and 67 nonsynonymous mutations were found in 68 samples (Table S5). Moreover, ND1, ND3, ND4, ND5 and ND6 genes had higher ratio of nonsynonymous versus synonymous mutations in tumor-specific heteroplasmic mutations. Among those 202 tumor-specific mutations, it is noted that six genes (DLOOP(28), RNR2(15), ND1(13), CYTB(11), ND5(11) and ND2(10)) were more frequently mutated (≥10 samples) than others (Table S6).
Discussion
In this study, we had studied the copy number of MT genome in 88 pairs of HCC tumor and matched normal samples, revealing the tumor samples possessed significantly lower copy number than the matched normal samples. This is consistent with previous studies in HCC [28]. The reduced copy number in tumor samples had high possibility to be link to the functional defects of mitochondria [29, 30].
Tumor samples not only had more heteroplasmic mutations, but also contained higher heteroplasmic ratio of heteroplasmic mutations than those normal samples. The phenomenon might indicate that there is higher cumulative mutation frequency in tumor samples. Accompany with the development of tumorigenesis, these heteroplasmic mutations of tumor samples might be accumulated effectively. Interestingly, there was also higher heteroplasmic ratio among these nonsynonymous mutations in tumor samples than in normal samples (p<0.05, T-Test), however no significant difference among these synonymous mutations. It might suggest that these nonsynonymous mutations might have a certain role in promoting tumorigenesis.
The distribution of mutation frequency of heteroplasmic sites in tumor and normal samples. The figure showed the distribution of mutation frequency in tumor and normal samples. The inner circle revealed the distribution of mutation frequency in normal samples and the outer circle revealed the distribution of mutation frequency in tumor samples.
Comparison of mutation frequency between tumor and normal samples The figure showed the difference of mutation frequency in tumor and normal samples. The inner circle revealed the distribution of gene frequency in normal samples and the outer circle revealed the distribution of gene frequency in tumor samples.
The number of synonymous and nonsynonymous mutations
| Gene | The number of tumor synonymous mutation | The number of tumor nonsynonymous mutation | The number of normal synonymous mutation | The number of normal nonsynonymous mutation |
|---|---|---|---|---|
| ND4 | 5 | 6 | 7 | 5 |
| ND1 | 3 | 12 | 4 | 1 |
| ATP8 | 0 | 2 | 0 | 1 |
| CYTB | 5 | 11 | 2 | 9 |
| ND5 | 4 | 12 | 1 | 6 |
| ND2 | 7 | 8 | 3 | 2 |
| ATP6 | 0 | 3 | 0 | 2 |
| COX3 | 2 | 2 | 1 | 4 |
| ND3 | 3 | 5 | 4 | 3 |
| COX1 | 4 | 3 | 3 | 4 |
| ND6 | 1 | 3 | 2 | 1 |
| COX2 | 1 | 4 | 1 | 1 |
| ND4L | 0 | 3 | 0 | 3 |
The table showed that the number of synonymous and nonsynonymous mutations in tumor and normal samples.
Furthermore, comparing the mutation frequency between tumor and normal samples, ND1 and ND2 had been found to have higher mutation frequency in tumor than normal samples. Additionally, ND1 gene included more nonsynonymous sties than synonymous mutations in tumor samples. Therefore, suggesting heteroplasmic mutations in ND1 might have an important contribution to tumorigenesis. In addition, it was observed that ND1, ND3, ND4, ND5, ND6 genes harbored more nonsynonymous than synonymous mutations in tumor-specific mutations. Bear in mind, these genes had been known to be part of complex I. Therefore, it suggested that nonsynonymous mutations of Complex I might be closely related to tumorigenesis. Moreover, nonsynonymous mutations could result in amino acid substitutions and disrupt the function of mitochondrial genes. The accumulation of certain mutations in the mitochondrial genome had been found to lead to deficiency in mitochondrial respiration and ATP generation [19, 28, 31]. For instance, mutations and dysfunction of ND5 are linked to promote the tumorigenesis through ROS alteration and AKT activation. Cell line carrying the heteroplasmic mutation in complex I manifested tumor growth enhancement, because dysfunctional Complex I could alter redox ratio (NADH/NAD) and activation of AKT pathway [32, 33]. Hereby, this study supplied evidence further suggesting nonsynonymous mutations of Complex I might have close relationship with tumorigenesis. Moreover, the copy number variation and heteroplasmic mutations in HCC samples could provide the basis for further understanding mechanism of tumorigenesis.
Supplementary Material
Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.
Acknowledgements
The study is sponsored by Doctoral Setup Foundation of Jining Medical University (No. 600491001), the Natural Science Foundation of Shandong Province (No. ZR2016CB49, No. ZR2016HM55). We are grateful to Asian Cancer Research Group, Inc. (ACRG).
Author Contributions
Conceived and designed the experiments: W.Y.L and X.F.C. Analyzed the data: Y.W.Q, Y.H.S, M.H.T. Wrote the paper: W.Y.L, X.F.C, Q.H. Provided good advice: H.L.Z, C.X.L, M.Z, Q.S.K, S.Y.D.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gomaa AI, Khan SA, Toledano MB. et al. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World journal of gastroenterology: WJG. 2008;14:4300-8
2. Chen W, Zheng R, Zeng H. et al. The updated incidences and mortalities of major cancers in China, 2011. Chinese journal of cancer. 2015;34:502-7
3. Caldwell S, Park SH. The epidemiology of hepatocellular cancer: from the perspectives of public health problem to tumor biology. J Gastroenterol. 2009;44(Suppl19):96-101
4. Lu FM, Zhuang H. Management of hepatitis B in China. Chinese medical journal. 2009;122:3-4
5. Wei YH. Mitochondrial DNA mutations and oxidative damage in aging and diseases: an emerging paradigm of gerontology and medicine. Proceedings of the National Science Council, Republic of China Part B, Life sciences. 1998;22:55-67
6. Chen JZ, Gokden N, Greene GF. et al. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer research. 2002;62:6470-4
7. Polyak K, Li Y, Zhu H. et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nature genetics. 1998;20:291-293
8. Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004;4:755-62
9. Navaglia F, Basso D, Fogar P. et al. Mitochondrial DNA D-loop in pancreatic cancer: somatic mutations are epiphenomena while the germline 16519 T variant worsens metabolism and outcome. American journal of clinical pathology. 2006;126:593-601
10. Petros JA, Baumann AK, Ruiz-Pesini E. et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:719-24
11. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nature reviews Genetics. 2005;6:389-402
12. Kane JM 3rd, Shears LL 2nd, Hierholzer C. et al. Chronic hepatitis C virus infection in humans: induction of hepatic nitric oxide synthase and proposed mechanisms for carcinogenesis. The Journal of surgical research. 1997;69:321-324
13. Schwarz KB. Oxidative stress during viral infection: a review. Free radical biology & medicine. 1996;21:641-49
14. Beckman KB, Ames BN. Oxidative decay of DNA. The Journal of biological chemistry. 1997;272:19633-36
15. Croteau DL, Bohr VA. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. The Journal of biological chemistry. 1997;272:25409-12
16. Birch-Machin MA. Using mitochondrial DNA as a biosensor of early cancer development. British journal of cancer. 2005;93:271-272
17. Fliss MS, Usadel H, Caballero OL. et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017-19
18. Van Trappen PO, Cullup T, Troke R. et al. Somatic mitochondrial DNA mutations in primary and metastatic ovarian cancer. Gynecologic oncology. 2007;104:129-133
19. Wu CW, Yin PH, Hung WY. et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes, chromosomes & cancer. 2005;44:19-28
20. Zaragoza MV, Fass J, Diegoli M. et al. Mitochondrial DNA variant discovery and evaluation in human Cardiomyopathies through next-generation sequencing. PloS one. 2010;5:e12295
21. Matsuyama W, Nakagawa M, Wakimoto J. et al. Mitochondrial DNA mutation correlates with stage progression and prognosis in non-small cell lung cancer. Human mutation. 2003;21:441-3
22. Tamori A, Nishiguchi S, Nishikawa M. et al. Correlation between clinical characteristics and mitochondrial D-loop DNA mutations in hepatocellular carcinoma. Journal of gastroenterology. 2004;39:1063-68
23. Durham SE, Krishnan KJ, Betts J. et al. Mitochondrial DNA damage in non-melanoma skin cancer. Br J Cancer. 2003;88:90-5
24. Sung WK, Zheng H, Li S. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nature genetics. 2012;44:765-9
25. Guo Y, Li J, Li CI. et al. MitoSeek: extracting mitochondria information and performing high-throughput mitochondria sequencing analysis. Bioinformatics. 2013;29:1210-1
26. Hao K, Luk JM, Lee NP. et al. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC cancer. 2009;9:389
27. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754-1760
28. Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663-74
29. Isidoro A, Martinez M, Fernandez PL. et al. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem J. 2004;378:17-20
30. Vaupel P, Thews O, Kelleher DK. et al. Current status of knowledge and critical issues in tumor oxygenation. Results from 25 years research in tumor pathophysiology. Adv Exp Med Biol. 1998;454:591-602
31. Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647-62
32. Sharma LK, Fang H, Liu J. et al. Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum Mol Genet. 2011;20:4605-16
33. Park JS, Sharma LK, Li H. et al. A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis. Hum Mol Genet. 2009;18:1578-89
Author contact
![]() Corresponding authors: Weiyang Li (163.lwycom) or Xiaofang Cui (scucxfcom); Phone: 0086-0537-3616298
Corresponding authors: Weiyang Li (163.lwycom) or Xiaofang Cui (scucxfcom); Phone: 0086-0537-3616298

 Global reach, higher impact
Global reach, higher impact