Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(18):3903-3915. doi:10.7150/jca.20958 This issue Cite
Research Paper
Effects of Triple Effective RNA (teRNA) on the Inhibition of Hepatocellular Carcinoma Cells
1. Department of Pathological Anatomy, Nantong University, Nantong, China;
2. Small RNA Technology and Application Institute, Nantong University, Nantong, China;
3. Biomics Biotechnologies Co., Ltd., Nantong, China.
* These authors contributed equally.
Received 2017-5-10; Accepted 2017-8-31; Published 2017-10-23
Abstract
The occurrence and development of hepatocellular carcinoma (HCC) is a complicate process involved in genetic mutation and epigenetic regulation. Successful HCC therapy needs multi-targets be involved. The aim of this study was to provide a triple effective RNA (teRNA) which composed of the specific siRNAs targeting NET-1 and VEGF and dsRNA activating TLR3, and explored its anti-HCC roles and mechanism. Real-time quantitative PCR (RT-qPCR), Western blot, immunofluorescence staining, MTT, Annexin V-FITC flow cytometry, Transwell and in-vitro Angiogenesis assay were used to measure the cell biological functions and protein expression analysis. Furthermore in in-vivo mouse model, teRNA inhibited tumor growth were detected by immunohistochemistry and TUNEL assay. Results showed that the proliferation, migration and angiogenesis of HCC cells were inhibited by teRNA effectively, the cell apoptosis also was induced, and further tumor growth was suppressed in-vivo. The gene silencing mechanism of teRNA was in an Ago2-dependent manner with no interferon response. The study suggests that NET-1, VEGF and TLR3 might be better targets for HCC treatment and combined these targets in form of a multi-target small RNA, teRNA could be a stagey for the development of anti-HCC drugs.
Keywords: hepatocellular carcinoma, VEGF, NET-1, TLR3, siRNA, sliRNA, teRNA.
Introduction
Hepatocellular carcinoma (HCC) is a global and complex health problem with high recurrence rate and mortality, easy metastasis, which accounted for the fifth of global malignancies, and is the second cause of cancer death in China [1, 2]. Since HCC is a highly vascular and entity tumor which involves different genetic chromosomal aberrations, gene mutations and etc., so far overall effect of HCC treatment is not ideal, yet find an effective way to cure HCC [3, 4]. Therefore, multiple targeted key regulatory genes which related to HCC occurrence and development pathway has become a new strategy for the treatment of HCC, particularly the inhibition of tumor cell proliferation, migration, promote cell apoptosis and many more [5, 6] .With the finding of sorafenib (a multikinase inhibitor) [7], makes a great progress and proves that molecular therapies have an effect on HCC. There is an urgent need to find other more molecular agents of HCC pathways to improve patient survival.
NET-1 (TSPAN1) is a member of the transmembrane 4 superfamily (TM4SF). TM4SF is a group of proteins containing four hydrophobic transmembrane domains forming two extracellular loop structures with different size [8, 9]. In prior study of Chen et al. showed that NET-1 was high expressed in HCC patients and plays an important role [10]. Recently, NET-1 was tumor-related gene was reported, which promoted the proliferation, invasion and metastasis of tumor cells. Chen et al. had demonstrated that the most important role of NET-1 in HCC progression was a cell proliferation promoter [11, 12]. The over-expression of NET-1 in HCC was involved in cancer cell proliferation [13]. These reports suggest that NET-1 may be an optional target for HCC therapy.
HCC is a highly vascular and entity tumor even at very early stages of the disease was notoriously malignant blood [14]. VEGF (also referred to as VEGF-A, vascular permeability factor) was a critical pro-angiogenic factor in cancer. Tumor cells secret VEGF through paracrine effect via VEGF receptor 2 (VEGFR2) expressed on endothelial cells to pro-angiogenesis [15, 16]. Experimental studies demonstrated that inhibition of tumor angiogenesis was pivotal for HCC treatment [17, 18]. Wu et al. used dual gene targeted siRNA (DGT siRNA) conjugate composed of NET-1 and VEGF siRNA sequences could inhibit proliferation and angiogenesis of HCC [19]. Meanwhile, in our previous study showed that Toll like receptor 3 (TLR3) expressed in HCC was correlated with apoptosis, proliferation and angiogenesis [20], and activation of TLR3 by specific double-stranded RNA (dsRNA) was resulted in the inhibition of growth, angiogenesis and triggers apoptosis of HCC cells [21-23].
In the study, we attempt to combination of tumor specific genes NET-1 and VEGF inhibition, and TLR3 activation for HCC treatment and aim to develop a triple effective RNA (teRNA) which composes of NET-1 and VEGF targeted siRNAs and a TLR3 activated dsRNA. The combined effects for HCC inhibition and mechanism of teRNA were evaluated by cell biology functional assay both in in-vitro and in-vivo model.
Materials and Methods
siRNAs, small ligand RNAs (sliRNAs) and triple effective RNA (teRNA) design and construction
The siRNAs that targeting human NET-1 (NCBI reference number: NM005727.3) and VEGF (NCBI reference number: NM 001287044.1) genes were designed and screened by our previous studies [19, 24]. A specific double-stranded RNA (dsRNA) named sliRNA (BM-06) was screened for activating TLR3 provided by Biomics Biotechnologies Co. Ltd (Biomics Biotech) (China). Furthermore, the pre-screened siRNAs and BM-06 were constructed into a small double-stranded RNA molecule for NET-1/VEGF dual inhibition and TLR3 activation named teRNA. teRNA was designed accordingly with the sequences and structures listed in Table 1. A dsRNA had no-homology with human gene was used as a negative control (NC_siR). RNAi dependent protein Argonaute-2 (Ago2) targeted siRNA (Ago2_siR) was also designed and used. All the siRNAs, BM-06 and teRNA were chemically synthesized by Biomics Biotechnologies Co. Ltd (China).
Cell culture and transfection
Human HCC cell lines: SMCC-7721, MHCC-97H, MHCC-97L and HepG2, human normal liver cells LO2 were obtained from Shanghai Zhongshan Hospital Liver Cancer Research Institute (China). These cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, USA), 100 units/mL penicillin and 100 μg/mL streptomycin at 37 oC in 5% CO2 humidified atmosphere. Cells were transfected with siRNAs, BM-06 and teRNA using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer's instructions.
Sequences of siRNAs and teRNA
| Name | Sequences (5'-3') | |
|---|---|---|
| NET-1_siR | Sense | CCACAAUGGCUGAGCACUUdTdT |
| Antisense | AAGUGCUCAGCCAUUGUGGdTdT | |
| VEGF_siR | Sense | GAUCCGCAGACGUGUAAAUdTdT |
| Antisense | AUUUACACGUCUGCGGAUCdTdT | |
| BM-06 | Sense | UAGGUACUUCGAGAAGCCUAGUAdTdT |
| Antisense | UACCCUAUAGGCUUCUCGAAGUAdTdT | |
| teRNA | Sense | CCACAAUGGCUGAGCACUUGGGAGUACCCUGAUGAGAUCCUAGGUACUUCGAGAAGCCUAGUAUUdTdT |
| Antisense 1 | AAGUGCUCAGCCAUUGUGGdTdT | |
| Antisense 2 | GAUCUCAUCAGGGUACUCCdTdT | |
| Antisense 3 | UACUAGGCUUCUCGAAGUACCUAdTdT | |
| NC_siR | Sense | UUCUCCGAACGUGUCACGUdTdT |
| Antisense | ACGUGACACGUUCGGAGAAdTdT | |
| Ago2_siR | Sense | AAUCUCUUCUUGCCGAUCGdTdT |
| Antisense | CGAUCGGCAAGAAGAGAUUdTdT | |
Real-time quantitative PCR (RT-qPCR)
A RISO™ RNA isolation reagent (Biomics Biotech, China) was used to extract total RNA from cells with different treatments. The mRNA levels of target genes (VEGF, NET-1 and TLR3) were determined by RT-qPCR using EzQuickTM one-step qPCR kit (Biomics Biotech, China) according to the manufacturer's instructions. To exclude the influence of interferon (IFN) on gene silencing effects of teRNA, the mRNA levels of 2'-5'-oligoadenylate synthetase 1 (OAS1) and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) genes were measured. For a positive control, cells were transfected with 20 ng/mL polyinosinic-polycytidylic acid (polyI:C) (InvivoGen, USA). Specific primers for each gene were presented in Table 2. Samples were tested in triplicate. GAPDH expression was used as an internal control.
Sequences of RT-qPCR primers
| Gene Name | Sequences (5'-3') | |
|---|---|---|
| NET-1 | F | CTTCATCCTCCTCCTCATCTTC |
| R | CAGGCACTACCAGCAACG | |
| VEGF | F | AGGAGGAGGGCAGAATCATCA |
| R | CTCGATTGGATGGCAGTAGCT | |
| TLR3 | F | TGGTTGGGCCACCTAGAAGTA |
| R | TCTCCATTCCTGGCCTGTG | |
| Ago2 | F | GCGGGTTGGGAAAAGTGGAAAC |
| R | ATCTGGGACGGAAGGCATTCTG | |
| Caspase-3 | F | ACTGGACTGTGGCATTGAG |
| R | GCTTGTCGGCATACTGTTTC | |
| Caspase-8 | F | GAAGAAGTGAGCAGATCAGAATTG |
| R | ATCTTCAGCAGGCTCTTGTTG | |
| Caspase-9 | F | CATCTTTGTGTCCTACTCTAC |
| R | CACCGAAACAGCATTAGC | |
| OAS1 | F | GTGAGCTCCTGGATTCTGCT |
| R | TGTTCCAATGTAACCATATTTCTGA | |
| IFIT1 | F | AATAGACTGTGAGGAAGGATGG |
| R | TCCAGGCGATAGGCAGAG | |
| GAPDH | F | GAAGGTGAAGGTCGGAGTC |
| R | GAAGATGGTGATGGGATTTC | |
F: Forward primer; R: Reverse primer.
Western blot
After treated as described above, cells were washed three times with ice-cold PBS, then lysed using mammalian cell total protein lysis buffer (Sangon Biotech, China). 50 μg total proteins were separated in 12% SDS acrylamide gels and transferred to polyvinylidene fluoride membranes (Millipore, USA). The membranes were blocked in PBS containing 5% skim milk, and incubated overnight at 4 oC with the primary antibodies: anti-VEGF (1:1,000; Abcam, USA), anti-NET-1 (1:1000; Santa Cruz, USA), anti-TLR3 (1:2,000; Santa Cruz, USA) and anti-β-actin (1:1000; Santa Cruz, USA) respectively. The membranes were then washed in PBST, incubated with a 1:1000 dilution of goat anti-mouse or goat-anti-rabbit HRP-conjugated secondary antibody (Sigma-Aldrich, USA) at room temperature for 2 h. After incubation with ECL chemiluminescence reagent (Amersham Life Science, USA), the membranes were exposed to a film (Kodak, Japan) to quantitatively measure the intensity of protein bands.
Cell proliferation assay
Cells were seeded in 96-well plates at a density of about 5×104 cells/ml for 24 h before transfection. At scheduled time points after treated, cell proliferation capacity was measured in triplicate using MTT according to the manufacturer's specifications (Beyotime, China). The absorbance at 490 nm was determined using a Microplate Reader (Bio-Rad 680). Cell growth curve was drawn after measured.
Cell migration assay
2×104/well cells were seeded and incubated in a 24-well for 24 h. Transwell chambers (Corning, USA) were pretreated with DMEM for 30 min. Cells were treated with 2.5% trypsin and suspended in a serum-free medium at a concentration of 1×106/ml prior to added into each upper chamber (200 μl), and 600 μl DMEM with 20% FBS were added into each lower chamber. Subsequently, the transwell chambers were incubated in a 37 oC for 48 h. The cells on the inner surface of the filter membrane (8.0-μm pore size) were removed. The cells on the lower surface of the membrane were stained with crystal violet, and counted in five random fields under a light microscope (200×).
Apoptosis analysis
To observe cell apoptosis, cells were stained simultaneously with FITC-conjugated Annexin V and propidium iodide (PI) using Annexin V-FITC Apoptosis Detection Kit (KeyGen, China) and analyzed by FACScan flow cytometry (BD Biosciences, Germany) 48 h after siRNAs, BM-06 and teRNA treated. The apoptotic rate was determined by Cell Quest software.
In-vitro angiogenesis assay
Matrigel (BD Biosciences, Germany), as artificial extracellular matrix, was plated at 100 μl/well in 48-well plates and allowed to reach the solid phase after 30 min in a 37 oC incubator. Human umbilical vein endothelial cells (HUVECs) were then suspended in the supernatant of each group transfected cells or vehicle buffer (PBS) and plated on top of the Matrigel at a density of 3×104 cells/well. After 16 h incubation, the wells were imaged on an inverted phase contrast microscope. HUVECs normally form a branching plexus of tubes on Matrigel. A tube forming node was defined as one which had three or more branches coming from a common center. Quantification was blinded and carried out by counting each nodal branch point (≥3 branch of points referred to as a node). Branch point counts per image constituted the raw data for statistical analysis. There were four images per treated group. Experiments were run in triplicate.
Animal Tumor Model
Nude BALB/c mouse, female, 5-6 weeks old and weighing 18-20 g, were obtained from Experimental Animal Center of Nantong University (Experimental Animal Center production license number: SYXK (Su) 2012-0031) (China) and housed under pathogen free conditions according to the recommendations of animal guidelines for care and use of laboratory approved by the Animal Ethical Committee of Nantong University (China).
Nude mouse were inoculated subcutaneously at the anterior axilla with 1×107 cells in 200 μl PBS, The shortest axis (a) and the longest axis (b) of tumor were measured by caliper every day. The tumor volumes were calculated with the formula: volumes = 0.5×a×b2. When the tumor volume reached 100 mm3 at least, the tumor-bearing nude mouse models were established successfully.
After nude mice tumor formed, nude mouse were randomly assigned to three experimental groups: PBS treated group, liposome encapsulated teRNA treated group and sorafenib treated group. The liposome encapsulated teRNA was prepared and obtained from Biomics Biotechnologies Co. Ltd (China). Subcutaneous tumors were injected with liposome encapsulated teRNA (10 μg/50 μl/time), PBS (50 μl/time) or sorafenib (100 mg/kg) in 50 μl/ time by an intratumoral multi-point manner every three days for five times for longer than 30 days.
Hematoxylin-eosin (H&E) staining
After harvest, tumor tissues from mouse were formalin-fixed, paraffin-embedded, and sections (4 μm thick) were prepared for standard H&E staining. The changes in histology were assessed under a light microscope.
Immunohistochemical (IHC) staining
After deparaffinizing and rehydrating, the slides were treated with 3% H2O2 solution for 15 min at room temperature to block endogenous peroxidase. The slides were then soaked in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween-20, pH 6.0) at 96 oC/5 min for antigen retrieval. After blocking using 1% BSA (Sigma-Aldrich, USA), the following antibodies were used: anti-NET-1 (1:100 dilution; Santa Cruz, USA), anti-VEGF (1:100 dilution; Abcam, USA) , anti-Ki67 (1:100 dilution; Zymed Laboratories, USA), anti-cleaved-Caspase-3 (1:50 dilution; Cell Signaling, USA), anti-cleaved-Caspase-8 (1:50 dilution; Cell Signaling Technology, USA) and anti-TLR3 (1:100 dilution; Santa Cruz, USA). Antibodies were added to the slides, kept overnight at 4 oC and then incubated at room temperature with biotinylated secondary antibody (Santa Cruz, USA) for 30 min, followed with HRP-streptavidin for 15 min. After 3,3'-diaminobenzidine (DAB) (Sigma-Aldrich, USA) staining and hematoxylin counterstain. The expression of Ki67 was nuclear positive and others all were cytoplasm positive with yellow-brow. The sections were analyzed by two independent investigators with knowledge of histopathology. Five fields of view were captured on each section from different groups.
TUNEL Apoptosis Assay
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end-labeling (TUNEL) detection kit (Promega, USA) was employed for the detection of cell apoptosis. Briefly, paraffin-embedded sections were deparaffinized and dehydrated. Sections were treated with 20 μg/ml Proteinase K for 20 min, rinsed with 0.3% Triton X-100 for 10 min, incubated with TUNEL reaction mixture at 37 oC for 1 h, treated with HRP conjugated streptavidin (1:200; Beijing Zhongshan Biotech, China) at 37 oC for 30 min, treated with 0.04% DAB and 0.03% H2O2 at room temperature for visualization for 8-12 min, counter stained with hematoxylin. In the interval of experiment steps washing in PBS three times (3 min for each). In the negative control, TUNEL reaction mixture was replaced with PBS. The positive control sections were pre-treated with DNase I for 10 min followed by TUNEL staining. Cells with blue granules in the nucleus were regarded as positive for TUNEL.
Statistical methods
Experiments were performed independently at least three times. Statistical analysis was performed using SPSS17.0. The data were expressed as mean±SD. Differences between groups were evaluated with ANOVA or factorial design ANOVA. P<0.05 was considered to indicate a statistically significant result.
Results
The expression levels of VEGF, NET-1 and TLR3 in HCC cells
The mRNA and protein levels of VEGF, NET-1 and TLR3 in different HCC cells were higher than that in normal liver cell lines LO2 (P<0.05) detected by RT-qPCR and Western blot respectively. But the expression level of these genes were the highest in SMCC-7721 cells (P<0.05) (Figure 1).
Recently, the TLR3 ligand dsRNA has been reported to directly trigger apoptosis in HCC cells [22], however, little is known regarding the significance of TLR3 expression in cytoplasm or membrane of human HCC cells. Therefore, we furthermore compared the activation level of TLR3 and the effects on cell apoptosis after stimulated with BM-06 (10 μg/ml). The results showed that TLR3 was expressed both in membrane and cytoplasm of HCC cells post stimulation (incubated of BM-06 only with no transfection) or transfection of BM-06. The mRNA and protein levels of TLR3 in membrane were more higher than that in cytoplasm of MHCC-97H, MHCC-97L and HepG2 cells; while TLR3 expressed higher in cytoplasm of SMCC-7721 cell (P<0.05) (Figures 2A and 2B). Furthermore, the result of flow cytometry revealed that TLR3 high expressed in cytoplasm triggered apoptosis of SMCC-7721 cell than that in membrane, as well as other HCC cells (P<0.05) (Figure 2C).
The expression levels of VEGF, NET-1 and TLR3 in HCC cells. A. The mRNA levels of VEGF, NET-1 and TLR3 in different HCC cells compared with normal liver cell LO2 detected by RT-qPCR; B. The protein levels of VEGF, NET-1 and TLR3 in different HCC cells compared with normal liver cell LO2 detected by Western blot. *P<0.05, vs. LO2 cell.
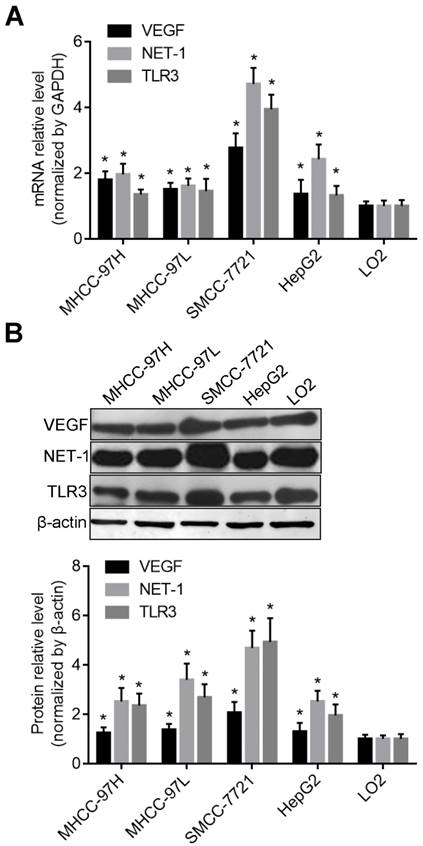
The expression levels of TLR3 in the membrane or cytoplasm of HCC cells. A. The mRNA levels of TLR3 in the membrane or cytoplasm of HCC cells detected by RT-qPCR; B. The protein levels of TLR3 in the membrane or cytoplasm of HCC cells detected by Western blot; C. The apoptosis of HCC cells were triggered by up-regulation of TLR3 by BM-06 in membrane or cytoplasm detected by flow cytometry.
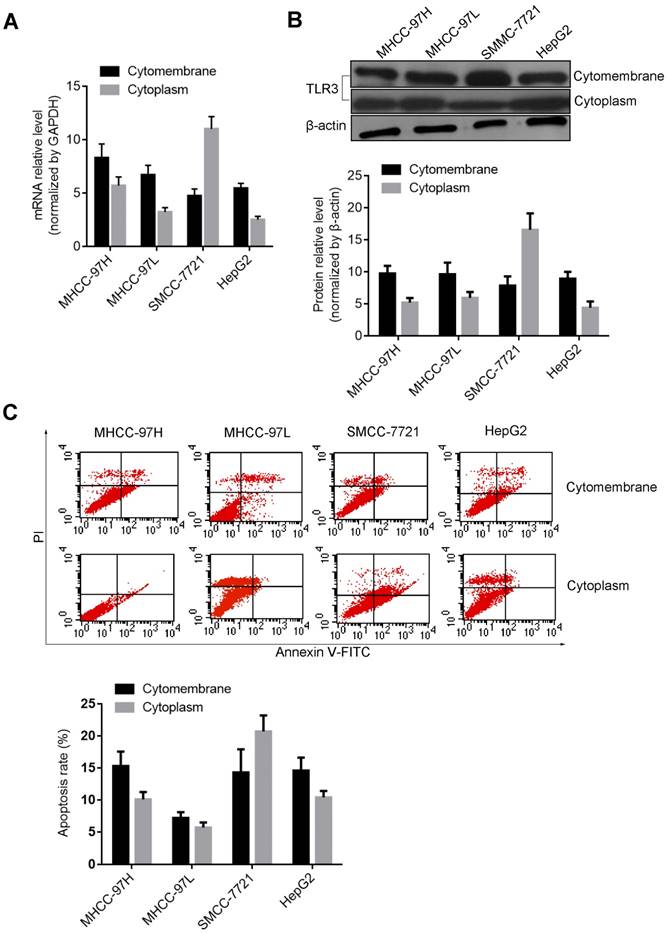
Effect of teRNA on the expression levels of VEGF, NET-1 and TLR3 in HCC cells
The mRNA and protein levels of VEGF, NET-1 and TLR3 regulated by teRNA were detected by RT-qPCR and Western blot. Compared with NC_siR treated cells, post treatment of teRNA, VEGF_siR, NET-1_siR or BM-06, the mRNA and protein levels of VEGF or NET-1 were decreased, but TLR3 were increased up to more than 2 fold (P<0.05) (Figure 3). There was no significant difference between NC_siR and untreated cells (P>0.05).
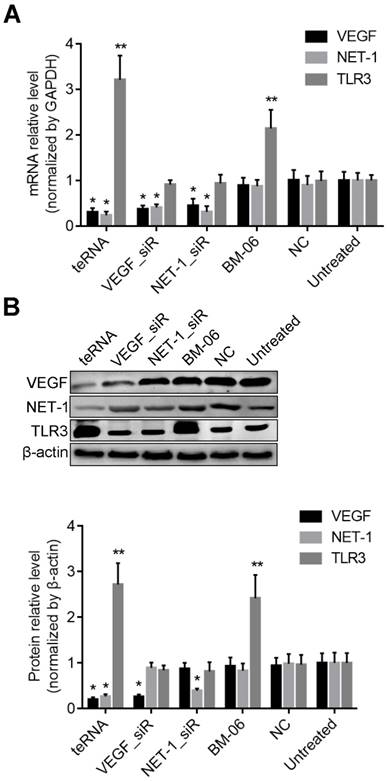
Effect of teRNA on VEGF and NET-1 in HCC cells through Ago2-dependent pathway
To study whether the teRNA silence genes via the Ago2-dependent pathway as conventional siRNAs, Ago2 was first knocked down by an Ago2_siR for 24 h (Figures 4A), then the cells were transfected with teRNA and single target siRNAs (VEGF_siR and NET-1_siR). After 24 h, silence effects of VEGF and NET-1 gene were measured by RT-qPCR, and showed that knockdown of individual siRNAs (VEGF_siR and NET-1_siR) and teRNA were effectively blocked with pre-treated of Ago2_siR (Figures 4B and 4C). The result revealed that teRNA trigger gene silencing through the same RNAi pathway as conventional siRNAs.
Effect of teRNA on HCC cells with no interferon response
To prove the influence of interferon (IFN) on gene regulation effects of teRNA, the mRNA level of IFN related genes OAS1 and IFIT1 were analyzed at 48 h after treated by teRNA, conventional siRNAs: VEGF_siR and NET-1_siR served as controls. Poly(I:C), an acknowledged IFN inducer, was also transfected into cells as a positive control. The expression of OAS1 and IFIT1 were not induced by VEGF_siR, NET-1_siR, and teRNA, while poly(I:C) (10 ng/mL) transfection induced significant expression of OAS1 and IFIT1, respectively, compared to NC_siR treated cells (Figure 4D).
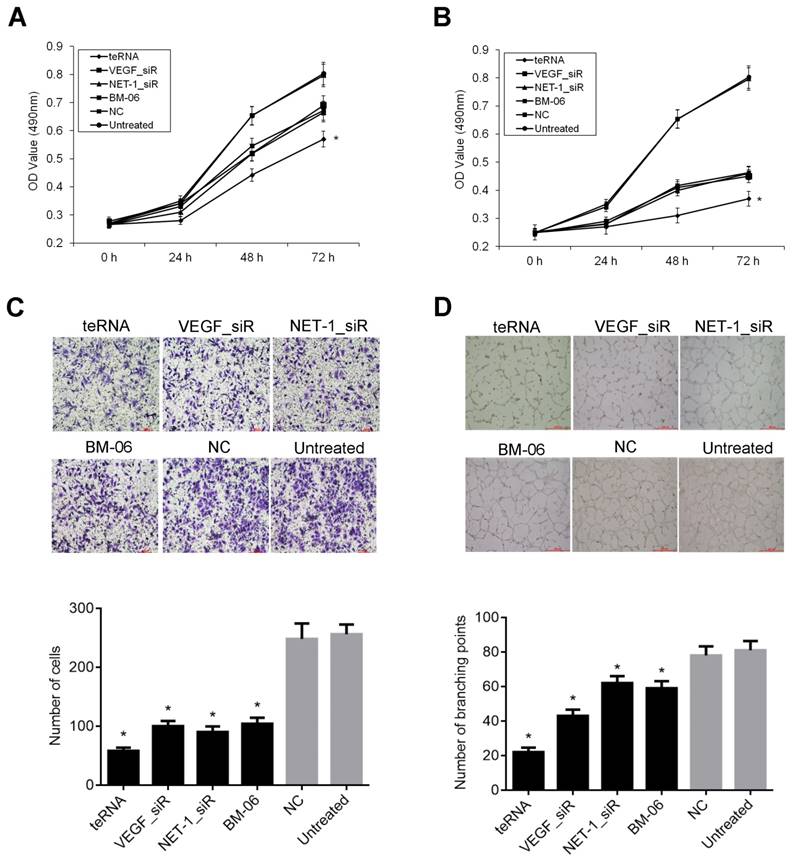
Effect of teRNA on the proliferation, migration and angiogenesis of HCC cells
The cells proliferation with different treatments were determined by MTT assay. After transfection for 24 h, 48 h and 72 h respectively, inhibition of cell proliferation was observed. After cells were transfected with 10 or 20 nM of teRNA, VEGF_siR, NET-1_siR and BM-06 result in decrease of proliferation significantly (P<0.05), compared to NC_siR treated cells. The effects of RNAs on growth inhibition were in a dose-dependent and time-dependent manner. The cell proliferation inhibited by teRNA was higher than either single target siRNAs (VEGF_siR, NET-1_siR and BM-06) treat cells at 48 h and 72 h (P<0.05) (Figures 5A and 5B). There was no significant difference between NC_siR and untreated cells (P>0.05).
The migration abilities of HCC cells inhibited by teRNA were analyzed by Transwell assay. After treated for 48 h, the cell numbers of migration were decreased significantly in teRNA, NET-1_siR, VEGF_siR and BM-06 treated cells, especially in teRNA treated ones (P<0.05) (Figure 5C). There was no significant difference between NC_siR and untreated cells (P>0.05).
The expression levels of VEGF, NET-1 and TLR3 were regulated by teRNA in HCC cells. A. The mRNA levels of VEGF and NET-1 were inhibited by teRNA, but TLR3 was activated detected by RT-qPCR; B. The protein levels of VEGF and NET-1 were inhibited by teRNA, but TLR3 was activated detected by Western blot. NC was the NC_siR treated cells as a negative control. *P<0.05, the expression levels of VEGF and NET-1 inhibited by teRNA or siRNAs vs. NC group; **P<0.05, the expression levels of TLR3 activated by teRNA or BM06, vs. NC group.
According to the ability of cancer cells can secrete pro-angiogenic factor [27], HUVEC tube formation model was used to observe the effect of teRNA on angiogenesis induce in HCC cells. The results shown that, after treated for 18 h, untreated cells formed an organized network of endothelial tubes with more tube formation nodes (85±15.35), but in teRNA (22±2.65), VEGF_siR (47±7.61), NET-1_siR (75±10.02) and BM-06 (44±9.11) treated cells were all markedly decreased (P<0.05) (Figures 5D). The tube formation nodes in teRNA treated cells were significantly less than that others (P<0.05). There was no significant difference between NC_siR and untreated cells (P>0.05).
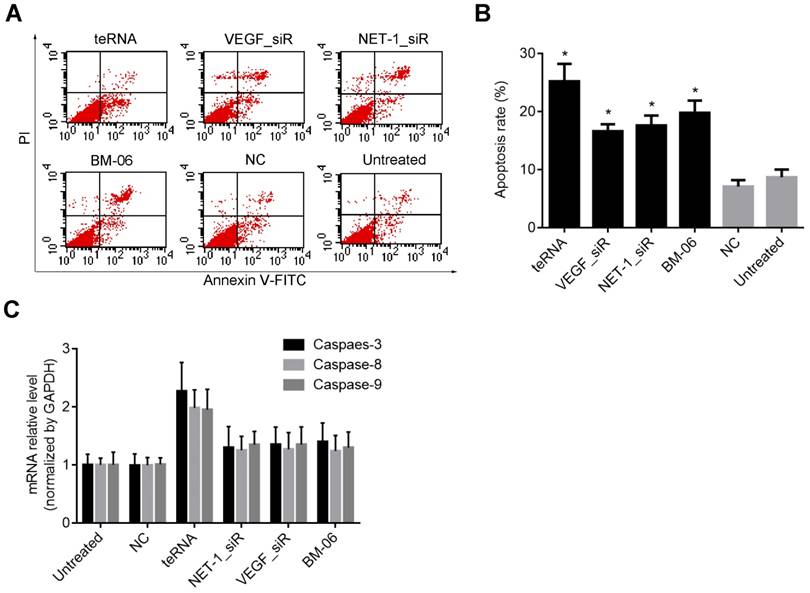
Effect of teRNA on the apoptosis and apoptosis-related genes inhibition in HCC cells
Cell apoptosis induced by teRNA was measured by FCM assay after Annexin V and PI staining. The result showed that cell apoptosis rate increased significantly in each group after treated with teRNA (25.2%), NET-1_siR (15.6%), VEGF_siR (14.4%), and BM-06 (20.8%), respectively, compared to the NC_siR treated cells (P<0.05), and the apoptosis of teRNA treated cell was higher than single target groups treated ones (P<0.05) (Figures 6A and 6B). There was no significant difference between NC_siR and untreated cells (P>0.05).
It was known that apoptosis involves a cascade of proteolytic reactions affected chiefly by the caspase family. Caspase-8 and Caspase-9 were the pro-apoptotic genes in either exogenous or endogenous apoptosis pathway; Caspase-3 was thought to be the major executor of apoptosis [25, 26]. Therefore, the expression levels of Caspase-3, Caspase-8 and Caspase-9 were analyzed after treatment. In teRNA, NET-1_siR, VEGF_siR and BM-06 treated cells, Caspase-3, Caspase-8 and Caspase-9 were all increased (P<0.05), compared to the NC_siR treated ones. And the expression level of Caspase-3, Caspase-8 and Caspase-9 in teRNA treated cells were the highest than others (P<0.05) (Figure 6C). There was no significant difference between NC_siR and untreated cells (P>0.05).
Inhibition of VEGF and NET-1 by teRNA in HCC cells via Ago2-dependent RNAi pathway in HCC cells detected by RT-qPCR. A. The mRNA levels of Ago2 inhibited by Ago2_siR in HCC cells, *P<0.05 vs. untreated cells; B. The mRNA levels of VEGF inhibited by teRNA via Ago2-dependent pathway, *P<0.05 vs. Ago2_siR treated cells; C. The mRNA levels of NET-1 inhibited by teRNA via Ago2-dependent RNAi pathway, *P<0.05 vs. Ago2_siR treated cells; D. No interferon response induced by teRNA in HCC cells, *P<0.05, IFIT1 expression levels; **P<0.05, OAS1 expression levels, vs. NC group.
Effects of teRNA on the proliferation, migration and angiogenesis of HCC cells. A. The proliferation of HCC cells inhibited by teRNA in the concentration of 10 nM was determined by MTT assay, *P<0.05 vs. NC group at 48 h and 72 h; B. The proliferation of HCC cells inhibited by teRNA in the concentration of 20 nM was determined by MTT assay, *P<0.05 vs. NC group at 48 h and 72 h; C. The migration abilities of HCC cells inhibited by teRNA detected by Transwell assay; D. The abilities of tube formation were inhibited by teRNA observed in HUVEC angiogenesis model.
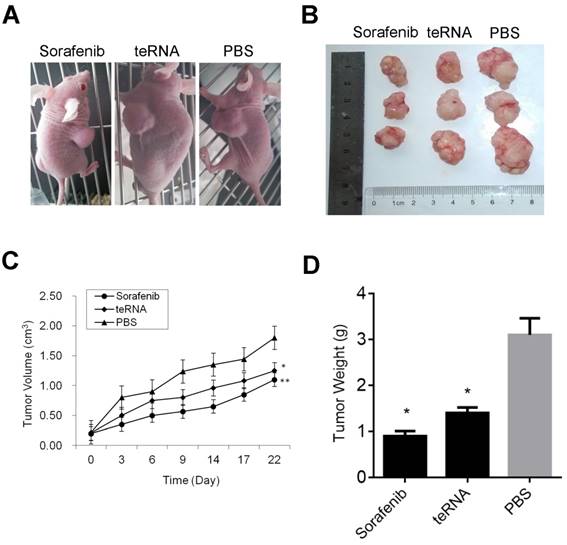
Effect of teRNA on tumor growth in-vivo of mouse model
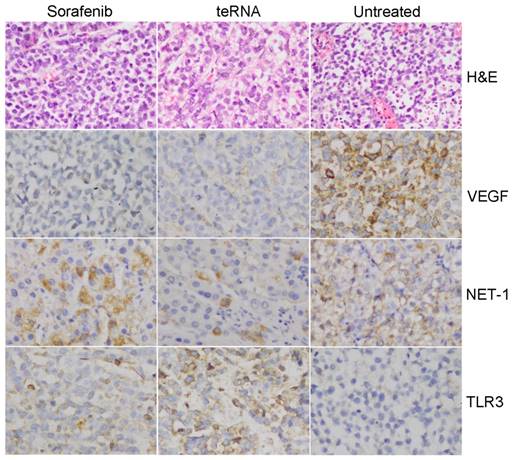
After injection of HCC cells into nude mice subcutaneously, tumors were formed at the tenth day. After tumors formed, the mouse models were randomly assigned to three experimental groups. After treated by liposome encapsulated teRNA and sorafenib, and PBS injection as negative control for 30 days, the size of tumors were significantly smaller in liposome encapsulated teRNA and sorafenib treated group than that in PBS treated group (P<0.05) (Figure 7). The result of H&E staining showed that the tumor cells grow nest path or stripe shaped, presented with obvious atypical and mitosis or massive necrosis in PBS treated group especially, whereas in teRNA and sorafenib treated group, tumor mitotic relatively rate. The result of immunohistochemistry showed that the expression level of NET-1 and VEGF were decreased and TLR3 was increased in teRNA treated group, and VEGF was only decreased in sorafenib treated group (Figure 8). The expression level of Caspase-3 and Caspase-8 were significantly increased and Ki67 was decreased in teRNA and sorafenib treated groups. Meanwhile, the result of TUNEL apoptosis assay also validated the results (Figure 9).
Discussion
Due to the occurrence of cancer development is a multi-step, complex biological process and multiple factors involved, thus the intervention of a single oncogene has some limitations. It is necessary to develop new multiple-target strategy for cancer treatment.
Although the molecular mechanism of HCC was not well known, the important events of HCC progression were evading apoptosis, limitless replicative potential and sustained angiogenesis [28]. Previous studies and our observations have suggested that NET-1, VEGF and TLR3 may be optional targets for HCC gene therapy. NET-1, as a marker of cell proliferation, was closely correlated with HCC progression. VEGF, as a critical pro-angiogenic factor, was closely correlated with HCC angiogenesis. In HCC cells, TLR3 agonists can directly inhibit cell proliferation and induce tumor cell apoptosis. And, the expression level of NET-1 and VEGF were high expressed in HCC cells and TLR3 was expressed in HCC cells (Figure 1). Additionally, the result of apoptosis assay showed that sliRNA (BM06) could activate the expression of TLR3 both in cytomembrane and cytoplasm of HCC cells, further inducing apoptosis of HCC cells (Figure 2).
In the study, we designed a new dsRNA molecule with triple functions based on our previous studies [19, 24], named triple effective RNA (teRNA), the designed teRNA combined the functions of NET-1/VEGF dual inhibition and TLR3 activation, and teRNA composed of the specific siRNAs targeting NET-1 and VEGF and dsRNA activating TLR3. The results proved that teRNA had functions of silencing both target genes (VEGF and NET-1) and activating TLR3 effectively both in mRNA and protein level (Figure 3).
Inhibition effects of teRNA on the apoptosis and apoptosis-related genes inhibition in HCC cells. A. Cell apoptosis induced by teRNA was measured by FCM assay after Annexin V and PI staining; B. Apoptosis rate of HCC cells; C. The expression levels of pro-apoptotic genes: Caspase-3, Caspase-8 and Caspase-9 in HCC cells were detected by RT-qPCR.
Inhibition effects of teRNA on tumor growth in HCC mouse model. A. The mouse models were randomly assigned to three experimental groups; B. The size of tumors were significantly decreased in liposome encapsulated teRNA and sorafenib treated group than that in PBS treated group; C. The volumes of tumors were significantly decreased in liposome encapsulated teRNA and sorafenib treated group than that in PBS treated group, *P<0.05 teRNA vs. PBS treated group, **P<0.05 sorafenib vs. PBS treated group; D. The weight of tumors were significantly decreased in liposome encapsulated teRNA and sorafenib treated group than that in PBS treated group, *P<0.05 vs. PBS treated group.
Pathological observation in HCC mouse model tumor tissues. A. HCC mouse model tumor tissues were detected by H&E staining (×200). B. The expression of NET-1, VEGF and TLR3 in HCC mouse model tumor tissues with different treatments were detected by immunohistochemistry staining (×200).
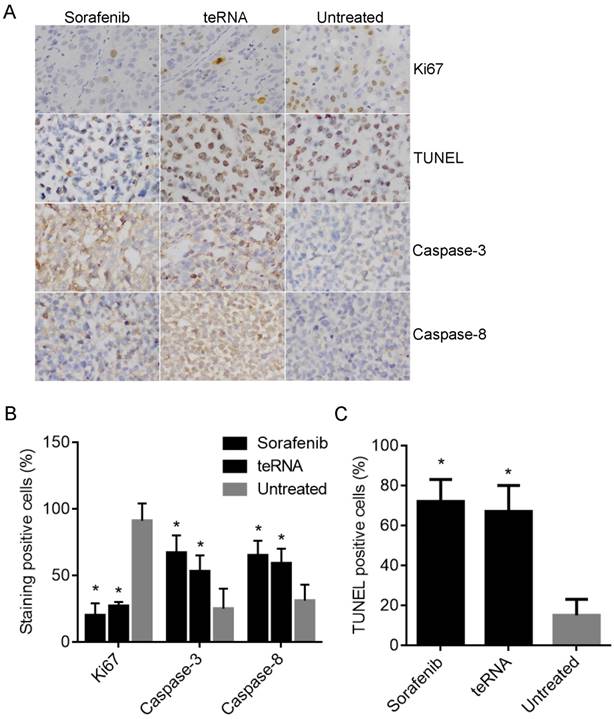
The expression of Ki67, Caspase-3 and Caspase-8 in HCC mouse model tumor tissues were detected by immunohistochemistry staining, and the cell apoptosis in tumor tissues were also detected by TUNEL Apoptosis assay. A. Representative micrographs of IHC staining and TUNEL Apoptosis assay (×200). B. The staining positive rate of Ki67, Caspase-3 and Caspase-8 by IHC staining. C. The cell apoptotic rate with TUNEL Apoptosis assay.
The previous study reported that dsRNA longer than 30 bp could induce interferon responses after being transfected into mammalian cells [29]. Therefore, interferon response genes OAS1 and IFIT1 were used in this study as sensitive biomarkers to evaluate immune responses resulted from teRNA. The result showed that there was no significant interferon response induced by teRNA, the RNAi function was in an Ago2-dependent manner (Figure 4).
Furthermore, teRNA had activities significantly of the proliferation, migration and angiogenesis suppression, and induction of apoptosis of HCC cells, as well as inhibition tumor growth in-vivo, the activity of teRNA was higher than either single targeting siRNAs (NET-1_siR and VEGF_siR) or BM-06 (Figures 5-9). Therefore, this new type of teRNA may have a great potential therapeutic application. The results suggest that NET-1, VEGF and TLR3 may be better combination targets for HCC treatment and combined functions of these targets in form of a triple effective RNA (teRNA) molecule might be a preferred stagey for the development of HCC therapeutic drugs.
Acknowledgements
This study was supported by a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, and from the Science Foundation of Nantong City, Jiangsu Province, China (No. BK2018001) and the Post-graduation Innovation Project Program of Nantong University (No. YKC16070).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-1255
2. Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844-855
3. Forner A, Gilabert M, Bruix J. et al. Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol. 2014;11:525-535
4. Sherman M. Hepatocellular carcinoma: epidemiology, risk factors, and screening. Semin Liver Dis. 2005;25:143-154
5. Chen J, Zhu S, Tong L, Li J, Chen F, Han Y, Zhao M, Xiong W. Superparamagnetic iron oxide nanoparticles mediated (131)I-hVEGF siRNA inhibits hepatocellular carcinoma tumor growth in nude mice. BMC Cancer. 2014;14:114
6. Chen J, Yang L, Chen H, Yuan T, Liu M, Chen P. Recombinant adenovirus encoding FAT10 small interfering RNA inhibits HCC growth in vitro and in vivo. Exp Mol Pathol. 2014;96:207-211
7. Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, Xu J, Sun Y, Liang H, Liu J, Wang J, Tak WY, Pan H, Burock K, Zou J, Voliotis D, Guan Z. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncology. 2009;10:25-34
8. Chen L, Li X, Wang GL, Wang Y, Zhu YY, Zhu J. Clinicopathological significance of overexpression of TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 2008;94:531-538
9. Yauch RL, Hemler ME. Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphoinositide 4-kinase. Biochem J. 2000;351(Pt 3):629-637
10. Chen L, Wang Z, Zhan X. et al. Association of NET-1 gene expression with human hepatocellular carcinoma. Int J Surg Pathol. 2007;15:346-353
11. Chen L, Yuan D, Zhao R, Li H, Zhu J. Suppression of TSPAN1 by RNA interference inhibits proliferation and invasion of colon cancer cells in vitro. Tumori. 2010;96:744-750
12. Chen L, Zhu Y, Li H, Wang GL, Wu YY, Lu YX, Qin J, Tuo J, Wang JL, Zhu J. Knockdown of TSPAN1 by RNA silencing and antisense technique inhibits proliferation and infiltration of human skin squamous carcinoma cells. Tumori. 2010;96:289-295
13. Chen L, Yuan D, Wang GL. et al. Clinicopathological significance of expression of tspan-1, Jab1 and p27 in human hepatocellular carcinoma. J Korean Med Sci. 2010;25:1438-1442
14. Schoenleber SJ, Kurtz DM, Talwalkar JA, Roberts LR, Gores GJ. Prognostic role of vascular endothelial growth factor in hepatocellular carcinoma: systematic review and meta-analysis. Br J Cancer. 2009;100:1385-92
15. Luo X, Feng GS. VEGFA genomic amplification tailors treatment of HCCs with sorafenib. Cancer Discov. 2014;4:640-641
16. Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2:795-803
17. Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011-1127
18. Llovet JM. Focal gains of VEGFA: candidate predictors of sorafenib response in hepatocellular carcinoma. Cancer Cell. 2014;25:560-562
19. Wu YY, Chen L, Wang GL, Zhang YX, Zhou JM, He S, Qin J, Zhu YY. Inhibition of hepatocellular carcinoma growth and angiogenesis by dual silencing of NET-1 and VEGF. J Mol Hist. 2013;44:433-445
20. Yuan MM, Xu YY, Chen L, Li XY, Qin J, Shen Y. TLR3 expression correlates with apoptosis, proliferation and angiogenesis in hepatocellular carcinoma and predicts prognosis. BMC Cancer. 2015;15:245
21. Chen L, Xu YY, Zhou JM, Wu YY, E Q, Zhu YY. TLR3 dsRNA agonist inhibits growth and invasion of HepG2.2.15 HCC cells. Oncol Rep. 2012;28:200-6
22. Guo Z, Chen L, Zhu Y, Zhang Y, He S, Qin J, Tang X, Zhou J, Wei Y. Double-stranded RNA-induced TLR3 activation inhibits angiogenesis and triggers apoptosis of human hepatocellular carcinoma cells. Oncol Rep. 2012;27:396-402
23. Wang G, Zhang M, Li Y, Zhou J, Chen L. Studying the Effect of Downregulating Autophagy-Related Gene LC3 on TLR3 Apoptotic Pathway Mediated by dsRNA in Hepatocellular Carcinoma Cells. Cancer Res Treat. 2017;49:230-245
24. Li T, Xue Y, Wang G, Gu T, Li Y, Zhu YY, Chen L. Multi-target siRNA: Therapeutic Strategy for Hepatocellular Carcinoma. J Cancer. 2016Jun25;7(10):1317-27
25. Degterev M. Boyce, J. Yuan, A decade of caspases. Oncogene. 2003;22(53):8543-8567
26. Ghavami S, Hashemi M, Ande SR, Yeganeh B, Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ, Los M. Apoptosis and cancer: mutations within caspase genes. J Med Genet. 2009;46(8):497-510
27. Niu G, Chen X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr Drug Targets. 2010;11:1000-17
28. Kudo M. Molecular targeted therapy for hepatocellular carcinoma: bench to bedside. Dig Dis. 2011;29:273-7
29. Elbashir SM, Harborth J, Lendeckel W. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498
Author contact
![]() Corresponding author: Li Chen, Professor, Department of Pathological Anatomy, Nantong University, 19 Qixiu Road, Nantong 226001, Jiangsu, China. Email: bl1edu.cn. Tel: +86-513-85051729; Fax: +86-513-85051729.
Corresponding author: Li Chen, Professor, Department of Pathological Anatomy, Nantong University, 19 Qixiu Road, Nantong 226001, Jiangsu, China. Email: bl1edu.cn. Tel: +86-513-85051729; Fax: +86-513-85051729.

 Global reach, higher impact
Global reach, higher impact