Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(15):3062-3069. doi:10.7150/jca.19619 This issue Cite
Research Paper
Identification of Nucleobindin-2 as a Potential Biomarker for Breast Cancer Metastasis Using iTRAQ-based Quantitative Proteomic Analysis
1. Department of Pathology, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou, Guangdong 510623, China;
2. Department of Pathology, Hunan Cancer Hospital & The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan 410013, China;
3. Department of Pathology, Hunan Normal University Medical College, Changsha, Hunan 410013, China;
4. Department of Social Medicine, Hunan Provincial People's Hospital & The Affiliated Hospital of Hunan Normal University, Changsha, Hunan 410007, China;
5. Department of Breast Internal Medicine, Hunan Cancer Hospital & The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan 410013, China;
6. Key Laboratory of Cancer of the Ministry of Health, Cancer Research Institute, Xiangya School of Medicine, Central South University, Changsha, Hunan 4100078, China.
* Liang Zeng and Jingmin Zhong contributed equally to this work.
Received 2017-2-12; Accepted 2017-3-17; Published 2017-9-2
Abstract
Metastasis is a lethal step in the progression of breast cancer. None of the metastasis-associated biomarkers identified up to now has a definite prognostic value in breast cancer patients. This study was designed to identify biomarkers for breast cancer metastasis and predictors of the prognosis of breast cancer patients. The differentially expressed proteins between 23 paired primary breast tumor and metastatic lymph nodes were identified by quantitative iTRAQ proteomic analysis. Immunohistochemistry was applied to locate and assess the expression of NUCB2 in paired primary breast tumor and metastatic lymph node tissues (n = 106). The relationship between NUCB2 expression and the clinicopathological characteristics of breast cancer patients (n = 189) were analyzed by χ2 test. Kaplan-Meier analysis and Cox hazard regression analysis were utilized to investigate the relationship between its expression and prognosis of breast cancer patients. The iTRAQ proteomic results showed that 4,837 confidential proteins were identified, 643 of which were differentially expressed in the primary breast cancer tissues and the paired metastatic lymph nodes. NUCB2 protein was found decreased in paired metastatic lymph nodes (P = 0.000), with the positive expression rate being 82% in primary breast cancer tissues and 47% in paired metastatic lymph nodes, respectively. According to Kaplan-Meier analysis, the overall survival time of patients with positive expression of NUCB2 protein were shorter than those with negative NUCB2 expression (P = 0.004). Cox regression model suggested that NUCB2 was a risk factor of breast cancer patients (P = 0.045, RR = 1.854). We conclude that NUCB2 can be used as a potential biomarker for breast cancer metastasis and a prognostic predictor of breast cancer patients.
Keywords: Breast cancer, Metastasis, Prognosis, iTRAQ, Nucleobindin-2.
Introduction
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer death among females worldwide, with an estimated 1.67 million new cases and 521,900 deaths in 2012. In developing countries, breast cancer incidence and mortality are rising, which have exceeded developed countries [1]. Metastasis is considered as a lethal step in the progression of breast cancer. Several risk factors have been identified to be related to breast cancer metastasis, such as tumor size, nodal status, histological grade, vascular invasion, etc. In addition, the expression of estrogen receptor (ER) and progesterone receptor (PR) and the amplification of HER2/c-erbB2 are closely linked with breast cancer metastasis, which provides the guidance to the adjuvant therapy for breast cancer [2]. Other molecules, such as miR-185-5p [3], cadherin-5 [4], Ku86 [5], and transforming growth factor β (TGF-β), are also associated with breast cancer metastasis. In addition, epithelial-to-mesenchymal transition (EMT) [6-8], circulating tumor cells [9], and the tumor microenvironment [10, 11] also contribute to the metastatic spread of breast cancer. While how these molecules and/or processes contribute to breast cancer metastasis is still not clearly understood, none of these has a definite prognostic value as a biomarker in breast cancer patients.
In recent years, innovations in high-throughput proteomic profiling approaches have allowed for highly sensitive, accurate, and quantitative identification of altered proteins in multiple samples at the same time. Isobaric tags for relative and absolute quantitation (iTRAQ) has been used successfully for the characterization of protein bioindicators of diverse effects [12]. Our goal in this study is to explore the differentially expressed proteins in matched primary breast tumor and metastatic lymph node tissue samples by the iTRAQ technique coupled with LC-MS/MS, aiming to identify reliable biomarkers for the prediction of breast cancer metastasis and prognosis.
Materials and Methods
Study subjects and clinical data
A total of 23 paired female fresh primary breast tumor and metastatic lymph node tissues were collected and used for iTRAQ followed by LC-MS/MS. All of these patients were diagnosed as breast cancer and received surgeries without any pre-operative chemotherapy or radiotherapy in Hunan Cancer Hospital & The Affiliated Cancer Hospital of Xiangya School of Medicine of Center South University from January to May, 2014. Histological type was confirmed by two pathologists in the Department of Pathology of Hunan Cancer Hospital. Each tissue sample was divided into two parts, one part used for routine pathological examination and the other stored in liquid nitrogen for iTRAQ analysis.
In addition, 106 paraffin-embedded tissue samples for immunohistochemistry were obtained from female patients with metastatic breast cancer who had received surgeries in Hunan Cancer Hospital between 1996 and 2008. These were paired primary breast tumors and lymph node metastases (n = 106). All of these patients had no pre-operative chemotherapy or radiotherapy and the pathological parameters of the patients together with patients without lymph node metastasis (n = 83) were summarized in Table 1. This study was approved by the Research Ethics Committee of Hunan Cancer Hospital, China and informed consent was obtained from all of the patients. The experiments performed in this study comply with the current laws of China relating to the use of human subjects.
Clinicopathological characteristics of breast cancer patients
| Parameters | Numbers |
|---|---|
| Median age (years, range) | 45 (30-73) |
| Median follow up period for survivors (months, range) | 80 (8-210) |
| Histological type, n (%) | |
| Ductal | 154 (81.5) |
| Lobular | 35 (18.5) |
| Histological grade, n (%) | |
| I & II | 33 (17.5) |
| III | 156 (82.5) |
| Nodal status, n (%) | |
| Negative | 83 (43.9) |
| Positive | 106 (56.1) |
| Tumor size (d, cm), n (%) | |
| d ≤ 2 | 56 (29.6) |
| 2 < d ≤ 5 | 112 (59.3) |
| d > 5 | 20 (10.6) |
| Unknown | 1 (0.5) |
| Clinical stage, n (%) | |
| I | 29 (15.3) |
| II | 100 (52.9) |
| III | 59 (31.2) |
| Unknown | 1 (0.5) |
| ER, n (%) | |
| Negative | 90 (47.6) |
| Positive | 99 (52.4) |
| PR, n (%) | |
| Negative | 78 (41.3) |
| Positive | 111 (58.7) |
| C-erbB-2, n (%) | |
| Negative | 57 (30.2) |
| Positive | 122 (64.6) |
| Unknown | 10 (5.3) |
| Menstrual history, n (%) | |
| Premenopause | 123 (65.1) |
| Postmenopause | 66 (34.9) |
| Modus operandi, n (%) | |
| Radical mastectomy | 82 (43.4) |
| Modified radical mastectomy | 100 (52.9) |
| Other type | 7 (3.7) |
iTRAQ proteomics
The iTRAQ kit was purchased from AB Sciex (USA) and proteomic analysis performed essentially as described in the manufacturer's instructions. The prepared lysates were treated with 4 µl reducing reagent for 1 h at 60°C and then blocked by 2 µl Cysteine blocking reagent for 10 min at room temperature. After centrifugation, the supernatant was collected and incubated with trypsin and tetraethylammonium bromide (TEAB) overnight at 37°C. The next day, the prepared sample and iTRAQ reagents were mixed together. Afterwards, two-dimensional LC-MS/MS was used to analyze the labeled samples and Proteinpilot 4.0 (AB Sciex) protein retrieval software was used to retrieve the separated peptides. This process was repeated for three times and the average was accepted as the final results. A greater than 1.5-fold change in expression was considered different between the primary breast tumor tissue and the paired metastatic lymph node tissue. This research was assisted by the FitGene BioTechnology proteomics platform (http://www.fitgene.com).
Immunohistochemistry
Paraffin sections were baked in oven at 60°C for 2 h, and then dewaxed in two changes of xylene for 10 min each, followed by hydration in graded ethanol for 5 min each. For antigen retrieval, the slides were immerged into citric acid retrieval solution and heated in a microwave and cooled down at room temperature. Endogenous peroxidase activity was quenched by 3% H2O2 for 15 min. After blocking of nonspecific binding, the sections were incubated with the NUCB2 antibody (1:200, Abcam, UK) at 4°C overnight. Next, the sections were incubated with biotinylated secondary antibody followed by incubation with the streptavidin-peroxidase conjugate and color development using the DAB/H2O2 system. A negative control was included by replacing the primary antibody with PBS.
The immunostaining was evaluated by two independent experienced pathologists. The results of the two reviewers were compared and any discrepant scores were re-examined by both pathologists to reach a consensus score. A total immunostaining score (TIS) was calculated as the product of a proportion score and an intensity score. The proportion score described the estimated fraction of positive-stained tumor cells (0: 0-4%; 1: 5-25%; 2: 26-50%; 3: 51-75%; 4: >75%). The intensity score represented the estimated staining intensity (0: no or marginal staining; 1: weak; 2: moderate; 3: strong). The TIS ranged from 0 to 12. We defined 0 as negative, 1-4 as weak positive, 5-8 as moderate positive, and 9-12 as strong positive.
Statistical analysis
The difference of NUCB2 expression between paired primary breast tumor and metastatic lymph node was analyzed with McNemar test. In addition, the difference of NUCB2 expression between every other two different groups of patients with breast disease was evaluated with χ2 test. The relationship between NUCB2 expression and clinicopathologic characteristics was examined with χ2 test. The survival rate was calculated with the Kaplan-Meier method and examined with log-rank test. Multivariate Cox hazard analysis was used to analyze the role of NUCB2 in breast cancer prognosis.
Results
Differentially expressed proteins between primary breast tumor and metastatic lymph node tissues
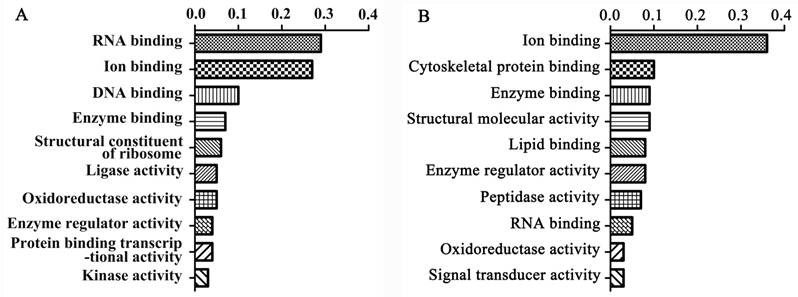
We first employed the iTRAQ technique to explore the differentially expressed proteins between 23 paired primary breast tumor and metastatic lymph node tissues. 4,837 confidential proteins were identified with a confidence interval (CI) ≥ 95%. Among these, 643 were differently expressed proteins, including 402 up-regulated and 241 down-regulated proteins in metastatic lymph nodes compared with the primary breast tumors. Gene ontology (GO) analysis revealed that the up-regulated proteins could be divided into 39 functional groups. The top ten up-regulated functional groups included RNA binding (29%), ion binding (27%), DNA binding (10%), enzyme binding (7%), structural constituent of ribosome (6%), ligase activity (5%), oxide reductase activity (5%), enzyme regulator activity (4%), protein binding transcriptional activity (4%), and kinase activity (3%) (Figure 1A). The down-regulated proteins could be divided into 31 functional groups. The top ten down-regulated functional groups included ion binding (36%), cytoskeletal protein binding (10%), enzyme binding (9%), structural molecular activity (9%), lipid binding (8%), enzyme regulator activity (8%), peptidase activity (7%), RNA binding (5%), oxide reductase activity (5%), and signal transducer activity (3%) (Figure 1B). The proteins up-regulated in the metastatic lymph nodes compared with the primary breast tumors were summarized in Table 2.
NUCB2 is differentially expressed in lymph node metastasis in breast cancer
The expression of NUCB2, which is one of the most up-regulated proteins associated with DNA binding, was 1.92-fold higher in metastatic lymph node tissues than primary breast tumor tissues. According to GO analysis, NUCB2 is located in various compartments of the cell and its molecular functions include ion binding and DNA binding (http://www.uniprot.org/uniprot/P80303). The basic biological information about NUCB2 was summarized in Table 3.
Functional groups of differentially expressed proteins between the primary breast tumors and metastatic lymph nodes determined by GO analysis. The top ten functional groups of proteins up-regulated (A) or down-regulated (B) in metastatic lymph nodes compared with primary breast tumors are shown.
Up-regulated proteins in metastatic lymph nodes compared to primary tumors
| Protein name | Accession number | Sequence coverage (%) | Molecular weight | Isoelectric point | Ratio |
|---|---|---|---|---|---|
| Transcriptional repressor protein YY1 | sp|P25490|TYY1_HUMAN | 20.05 | 44,713 | 6.22 | 2.31 |
| Golgin subfamily B member 1 | sp|Q14789|GOGB1_HUMAN | 38.45 | 376,019 | 4.66 | 1.93 |
| Dual specificity protein phosphatase 23 | sp|Q9BVJ7|DUS23_HUMAN | 63.33 | 16,588 | 8.15 | 1.93 |
| U1 small nuclear ribonucleoprotein C | sp|P09234|RU1C_HUMAN | 30.19 | 17,394 | 10.14 | 1.92 |
| Ribosome biogenesis protein BRX1 homolog | sp|Q8TDN6|BRX1_HUMAN | 28.05 | 41,401 | 10.60 | 1.92 |
| Pescadillo homolog | tr|B3KXD6|B3KXD6_HUMAN | 50.33 | 51,464 | 4.87 | 1.92 |
| WD repeat-containing protein 70 | tr|D6RIW8|D6RIW8_HUMAN | 27.39 | 43,240 | 4.69 | 1.92 |
| Translocation protein SEC62 | sp|Q99442|SEC62_HUMAN | 31.58 | 45,862 | 7.09 | 1.92 |
| 28S ribosomal protein S23, mitochondrial | sp|Q9Y3D9|RT23_HUMAN | 56.84 | 21,771 | 9.28 | 1.92 |
| Nucleobindin-2 | sp|P80303|NUCB2_HUMAN | 46.9 | 50,196 | 4.78 | 1.92 |
| Complement component 1 Q subcomponent-binding protein, mitochondrial | sp|Q07021|C1QBP_HUMAN | 57.45 | 31,362 | 4.47 | 1.92 |
| WD40 repeat-containing protein SMU1 | sp|Q2TAY7|SMU1_HUMAN | 31.77 | 57,544 | 7.15 | 1.92 |
| Chromatin target of PRMT1 protein | sp|Q9Y3Y2|CHTOP_HUMAN | 67.74 | 26,397 | 12.74 | 1.91 |
| Copper chaperone for superoxide dismutase | tr|J3KNF4|J3KNF4_HUMAN | 38.43 | 27,086 | 5.76 | 1.91 |
| Isoform 2 of Treacle protein | sp|Q13428-2|TCOF_HUMAN | 27.85 | 144,314 | 9.78 | 1.91 |
| Eukaryotic translation initiation factor 5B | sp|O60841|IF2P_HUMAN | 35.98 | 138,827 | 5.15 | 1.89 |
| DDRGK domain-containing protein 1 | sp|Q96HY6|DDRGK_HUMAN | 50.32 | 35,611 | 4.81 | 1.89 |
| 28S ribosomal protein S22, mitochondrial | sp|P82650|RT22_HUMAN | 47.49 | 41,280 | 7.85 | 1.89 |
| CAP-Gly domain-containing linker protein 1 | tr|J3KP58|J3KP58_HUMAN | 38.75 | 148,129 | 4.99 | 1.89 |
| SAFB-like transcription modulator | sp|Q9NWH9|SLTM_HUMAN | 31.24 | 117,148 | 7.93 | 1.89 |
| Pentatricopeptide repeat domain-containing protein 3, mitochondrial | sp|Q96EY7|PTCD3_HUMAN | 25.54 | 78,550 | 6.37 | 1.88 |
| Ubiquitin-like modifier-activating enzyme 1 | sp|P22314|UBA1_HUMAN | 57.94 | 117,849 | 5.50 | 1.88 |
| Isoform 3 of UPF0696 protein C11orf68 | sp|Q9H3H3-3|CK068_HUMAN | 46.08 | 31,517 | 6.25 | 1.88 |
| Small nuclear ribonucleoprotein F | sp|P62306|RUXF_HUMAN | 66.28 | 9,725 | 4.34 | 1.87 |
| Isoform 3 of Cullin-4B | sp|Q13620-3|CUL4B_HUMAN | 41.84 | 84,017 | 6.74 | 1.87 |
| Epithelial cell adhesion molecule | sp|P16422|EPCAM_HUMAN | 41.72 | 34,932 | 7.40 | 1.85 |
| 26S protease regulatory subunit 8 | sp|P62195|PRS8_HUMAN | 81.77 | 45,626 | 7.48 | 1.78 |
| Protein S100-A8 | sp|P05109|S10A8_HUMAN | 67.74 | 10,835 | 6.98 | 1.72 |
| Isoform B of Nucleoporin SEH1 | sp|Q96EE3-1|SEH1_HUMAN | 45.84 | 46,578 | 7.87 | 1.68 |
| FAS-associated death domain protein | sp|Q13158|FADD_HUMAN | 45.19 | 23,279 | 5.40 | 1.51 |
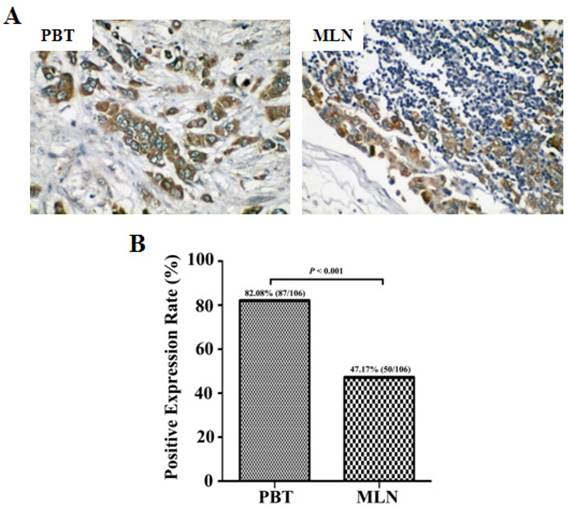
We further examined the expression of NUCB2 in primary breast tumors and paired metastatic lymph node tissues. NUCB2 staining was found predominantly in the cytoplasm of breast epithelial cells. The majority of the tissues examined showed weak staining of NUCB2 (TIS ≤ 4). NUCB2 expression was lower in metastatic lymph node than primary breast tumor tissues (P < 0.001). (Figure 2). The positive expression rate of NUCB2 was 82.1% (87/106) in primary breast tumors and 47.2% (50/106) in metastatic lymph nodes, respectively. The expression of NUCB2 in primary breast tumors and metastatic lymph nodes was summarized in Table 4. The results show that NUCB2 expression was down-regulated in metastatic lymph node tissues compared with primary breast tumors.
Basic biological information about NUCB2
| Name | Access number | Molecular weight | Isoelectric point | Cellular compartment | Molecular function |
|---|---|---|---|---|---|
| Nucleobinding-2 | sp|P80303|NUCB2-HUM | 50,196 | 4.80 | Nucleus | Ion binding |
| Cytoplasm | DNA binding | ||||
| Cytosol | |||||
| Extracellular region | |||||
| Plasma membrane | |||||
| Golgi apparatus | |||||
| Endoplasmic reticulum | |||||
| Nuclear envelope | |||||
| Extracellular space (secreted) |
Expression of NUCB2 protein in primary breast tumors and metastatic lymph node tissues. The paraffin-embedded tissue sections were immunohistochemically stained for NUCB2 expression and statistically evaluated by the χ2 test. (A) Representative immunohistochemical images showing the predominantly cytoplasmic staining of NUCB2 in breast tumor samples. Original magnification: 200×. (B) Summary of the positive expression rate of NUCB2 in breast tumor samples. PBT, primary breast tumor; MLN, metastatic lymph node.
Correlation between NUCB2 expression and clinicopathological parameters
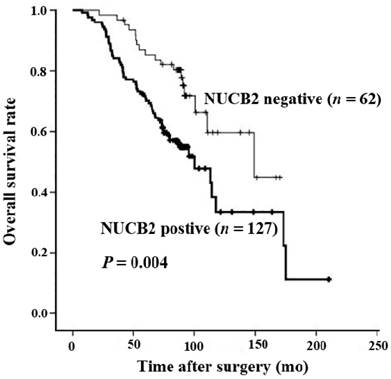
The correlation between NUCB2 expression and the clinicopathological parameters of breast cancer patients was analyzed by χ2. NUCB2 expression in breast tumors was significantly correlated with the extent of nodal invasion (P = 0.000), poor clinical stage (P = 0.001), and ER positivity (P = 0.020) (Table 5). On the other hand, no association was found between positive expression of NUCB2 and age, histological type, histological grade, tumor size, PR, HER2/c-erbB-2, menstrual history, or modus operandi. Kaplan-Meier analysis revealed that the overall survival time of patients with positive NUCB2 expression was significantly shorter than those with negative NUCB2 expression (P = 0.004, Figure 3). In addition, Cox regression analysis showed that among all the risk factors examined, only high expression of NUCB2 (P = 0.025, RR = 2.065) was a risk factor for short overall survival of breast cancer patients (Table 6). This means that high positive expression of NUCB2 is indeed associated with short overall survival time of breast cancer patients. Meanwhile, positive expression of NUCB2 in primary breast tumor is an independent risk factor for poor prognosis of breast cancer patients regardless of age, histological type, histological grade, nodal status, tumor size, clinical stage, ER, PR, HER2/c-erb-B2, menstrual history, and modus operandi.
Discussion
Approximately 10-15% of patients with breast cancer have an aggressive disease and develop distant metastases within 3 years after the initial detection of the primary tumor. As it is not possible to accurately predict the risk of metastasis development in individual patients, 80% of the patients received adjuvant chemotherapy, among which 40% relapse and ultimately die of metastatic breast cancer [2]. Thus, we need to identify effective biomarkers or establish metastatic models to predict the occurrence of breast cancer metastasis to provide a better treatment for these patients. At present, many researches are focused on the different proteins of the primary tumor between breast cancer patients with or without lymph node metastases [13], or exploring the different proteins between cell lines with different metastatic potential [14-17]. However, no comparison has been made between the primary site and the metastatic site in patients with metastatic breast cancer.
Expression of NUCB2 in primary breast tumors and metastatic lymph nodes
| NUCB2 expression | ||||||
|---|---|---|---|---|---|---|
| TIS (0) | TIS (1-4) | TIS (5-8) | ||||
| n | % | n | % | n | % | |
| PBT | 19 | 17.9 | 73 | 68.9 | 14 | 13.2 |
| MLN | 56 | 52.8 | 47 | 44.3 | 3 | 2.8 |
TIS, total immunostaining score; PBT, primary breast tumor; MLN, metastatic lymph node.
Kaplan-Meier survival curves of patients with positive and negative NUCB2 expression. The overall survival rate of breast cancer patients that were positive or negative for NUCB2 expression was calculated with the Kaplan-Meier method and examined with log-rank test. There was significant difference between the two groups (log-rank P = 0.004).
Relationship between NUCB2 expression and clinicopathological parameters in breast cancer patients
| Characteristics | No. of patients | NUCB2 expression | P value* | |
|---|---|---|---|---|
| No | Yes | |||
| Age at diagnosis (years) | 0.304 | |||
| < 50 | 112 | 40 | 72 | |
| ≥50 | 77 | 22 | 55 | |
| Histological type | 0.555 | |||
| Ductal | 154 | 52 | 102 | |
| Lobular | 35 | 10 | 25 | |
| Histological grade | 0.632 | |||
| I- II | 33 | 12 | 21 | |
| III | 156 | 50 | 106 | |
| Nodal status | 0.000 | |||
| Negative | 83 | 43 | 40 | |
| Positive | 106 | 19 | 87 | |
| Tumor size (d, cm) | 0.277 | |||
| d ≤ 2 | 56 | 22 | 34 | |
| 2 < d ≤ 5 | 112 | 36 | 76 | |
| d > 5 | 20 | 4 | 16 | |
| Unknown # | 1 | |||
| Clinical stage | 0.001 | |||
| I | 29 | 15 | 14 | |
| II | 100 | 38 | 62 | |
| III | 59 | 9 | 50 | |
| Unknown # | 1 | |||
| ER | 0.020 | |||
| Negative | 90 | 37 | 53 | |
| Positive | 99 | 25 | 74 | |
| PR | 0.165 | |||
| Negative | 78 | 30 | 48 | |
| Positive | 111 | 32 | 79 | |
| C-erbB-2 | 0.095 | |||
| Negative | 57 | 23 | 34 | |
| Positive | 122 | 34 | 88 | |
| Unknown # | 10 | |||
| Menstrual history | 0.592 | |||
| Premenopause | 123 | 42 | 81 | |
| Postmenopause | 66 | 20 | 46 | |
| Modus operandi | 0.442 | |||
| Radical mastectomy | 82 | 29 | 53 | |
| Modified radical mastectomy | 100 | 30 | 70 | |
| Other type # | 7 | |||
* χ2 test.
# Unknown and other type cases are excluded from P-value calculation.
Cox regression analysis of risk factors for short overall survival time of breast cancer patients
| Variables | B | SE | Wald | Sig. | Exp (B) (95% CI) |
|---|---|---|---|---|---|
| Age | -0. 005 | 0. 020 | 0. 066 | 0. 797 | 0. 995 (0.956-1.035) |
| Histological type | 0. 423 | 0. 298 | 2. 017 | 0. 156 | 1. 526 (0.852-2.735) |
| Histological grade | 0. 654 | 0. 450 | 2. 056 | 0. 152 | 1. 906 (0.789-4.602) |
| Nodal status | 0. 338 | 0. 269 | 1. 575 | 0. 209 | 1. 402 (0.827-2.375) |
| Tumor size | 0. 093 | 0. 235 | 0. 156 | 0. 693 | 1. 097 (0.692-1.738) |
| Clinical stage | 0. 140 | 0. 413 | 0. 115 | 0. 734 | 1. 151 (0.512-2.587) |
| Estrogen Receptor | -0. 218 | 0. 334 | 0. 428 | 0. 513 | 0. 804 (0.418-1.546) |
| Progesterone Receptor | -0. 430 | 0. 342 | 1. 579 | 0. 209 | 0. 651 (0.333-1.272) |
| C-erbB-2 | -0. 70 | 0. 279 | 0. 062 | 0. 803 | 0. 933 (0.540-1.612) |
| Menstrual history | 0. 153 | 0. 406 | 0. 141 | 0. 707 | 1. 165 (0.526-2.581) |
| Modus operandi | 0. 281 | 0. 203 | 1. 921 | 0. 166 | 1. 324 (0.890-1.969) |
| NUCB2 expression | 0. 725 | 0. 323 | 5. 027 | 0. 025* | 2. 065 (1.096-3.891) |
95% CI: 95% confidence interval
*P < 0.05
In the present study, we employed the iTRAQ proteomic technique to screen the differentially expressed proteins between primary breast tumor and metastatic lymph node tissues. The initial search identified 4,837 confidential proteins and 643 differentially expressed proteins. Compared with primary breast tumors, 402 proteins were up-regulated and 241 proteins down-regulated in metastatic lymph nodes. The majority of these proteins are associated with RNA/DNA binding, enzyme binding and cytoskeleton protein binding, etc. These proteins are involved in lamellipodia formation [18, 19], angiogenesis [20, 21], circulating tumor cells [22], EMT [23], and tumor microenvironment [24] and may affect cell motility [25], cell proliferation [26, 27], invasion, migration, and metastasis [28, 29]. At the same time, several inflammatory factors were identified, such as complement C1r and complement component C8 gamma chain (C8G) etc. This might be an indication of a complex background including inflammatory response in the development of breast cancer metastasis [30, 31].
NUCB2 is a calcium-binding protein composed of 396 aa residues with a 24-aa signal peptide [23]. The human NUCB2 gene spans 55 kb and contains 14 exons and 13 introns [32]. It is highly conserved across mammalian and non-mammalian vertebrates, suggesting its physiological importance [33]. It is ubiquitously expressed in normal tissues, such as lymphoid tissue, pancreas, gut, and adipose [34, 35]. NUCB2 appears to play an important role in regulating energy homeostasis and food intake [36]. Adipose tissue plays important role in energy homeostasis and NUCB2 protein expression was significantly increased in high-fat-fed mice [37]. Li et al. reported that in high fat diet induced obese mice, gastric mTOR signaling and NUCB2 were increased [38]. Moreover, the activity of gastric mTOR is linked to the regulation of gastric NUCB2. A relation between obesity and ovarian cancer mortality was established and the N-terminal fragment of NUCB2 can inhibit the proliferation of human ovarian epithelial carcinoma HO-8910 cells through inducing apoptosis via the mTOR and RhoA/ROCK signaling pathway [39].
In addition, NUCB2 was also reported to be related to the development and progression of renal clear cell carcinoma [40] and prostate cancer [41-43]. In breast cancer, it has a positive association with lymph node metastasis and the ER status of the patients. Results of siRNA transfection experiments showed that NUCB2 was involved in cell proliferation, migration, and invasion in both ER-positive MCF-7 and ER-negative SK-BR-3 cells [44]. In our study, we analyzed the correlation between NUCB2 expression and clinicopathological features in 189 breast cancer patients. We demonstrated that NUCB2 has a significant positive correlation with the extent of nodal invasion, poorly clinical stage, and ER positivity. Obesity causes an increase in local estrogen production, which has a close correlation with breast cancer [45]. Suzuki et al. [23] demonstrated that NUCB2 was up-regulated by estrogen and our study showed that there was a positive correlation between NUCB2 expression and the ER status. Moreover, we found that breast cancer patients with high NUCB2 expression had a significantly poorer overall survival. Cox hazard regression analysis showed that NUCB2 expression and clinical stage were associated with overall survival of breast cancer. All these observations suggest that NUCB2 plays an important role in breast cancer progression and may be a potential prognostic biomarker for poor survival of breast cancer patients. Compared with the recently described serum biomarkers for breast cancer metastasis such as Y-box binding protein 1 (YB-1) [46] and HER2 [47], the immunohistochemical biomarker NUCB2 offers the advantages of increased sensitivity and higher specificity, because immunohistochemistry can reveal a biomarker change at an earlier stage and in a more specific location than a serum biomarker.
In summary, several differentially expressed proteins involved in breast cancer metastasis were identified by iTRAQ. NUCB2 was one of the highest expressed proteins in the paired primary breast tumors and metastatic lymph nodes and showed positive correlation with poor survival of breast cancer patients. NUCB2 has the potential to be used as an effective and novel biomarker for the prognostic prediction of breast cancer.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81472496 to XD) and the Hunan Province Science and Technology Project (Grant No. 2014FJ6090 to LZ). We thank Proteomic technique platform by FitGene BioTechnology CO., Ltd (Guangzhou, P. R. China, http://www.fitgene.com).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108
2. Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591-602
3. Liu J, Chen Y, Shuai S, Ding D, Li R, Luo R. TRPM8 promotes aggressiveness of breast cancer cells by regulating EMT via activating AKT/GSK-3beta pathway. Tumour Biol. 2014;35:8969-77
4. Zeng Q, Zhang P, Wu Z, Xue P, Lu D, Ye Z. et al. Quantitative proteomics reveals ER-alpha involvement in CD146-induced epithelial-mesenchymal transition in breast cancer cells. J Proteomics. 2014;103:153-69
5. Zhao B, Xu B, Hu W, Song C, Wang F, Liu Z. et al. Comprehensive proteome quantification reveals NgBR as a new regulator for epithelial-mesenchymal transition of breast tumor cells. J Proteomics. 2015;112:38-52
6. Gilabert M, Launay S, Ginestier C, Bertucci F, Audebert S, Pophillat M. et al. Poly(ADP-ribose) polymerase 1 (PARP1) overexpression in human breast cancer stem cells and resistance to olaparib. PLoS One. 2014;9:e104302
7. Onishi T, Hayashi N, Theriault RL, Hortobagyi GN, Ueno NT. Future directions of bone-targeted therapy for metastatic breast cancer. Nat Rev Clin Oncol. 2010;7:641-51
8. Ma H, Liang C, Wang G, Jia S, Zhao Q, Xiang Z. et al. MicroRNA-mediated cancer metastasis regulation via heterotypic signals in the microenvironment. Curr Pharm Biotechnol. 2014;15:455-8
9. Wang B, Li J, Sun M, Sun L, Zhang X. miRNA expression in breast cancer varies with lymph node metastasis and other clinicopathologic features. IUBMB Life. 2014;66:371-7
10. Fry SA, Sinclair J, Timms JF, Leathem AJ, Dwek MV. A targeted glycoproteomic approach identifies cadherin-5 as a novel biomarker of metastatic breast cancer. Cancer Lett. 2013;328:335-44
11. Lagadec C, Romon R, Tastet C, Meignan S, Com E, Page A. et al. Ku86 is important for TrkA overexpression-induced breast cancer cell invasion. Proteomics Clin Appl. 2010;4:580-90
12. Putz SM, Boehm AM, Stiewe T, Sickmann A. iTRAQ analysis of a cell culture model for malignant transformation, including comparison with 2D-PAGE and SILAC. J Proteome Res. 2012;11:2140-53
13. Vergara D, Simeone P, del Boccio P, Toto C, Pieragostino D, Tinelli A. et al. Comparative proteome profiling of breast tumor cell lines by gel electrophoresis and mass spectrometry reveals an epithelial mesenchymal transition associated protein signature. Mol Biosyst. 2013;9:1127-38
14. Bouchal P, Roumeliotis T, Hrstka R, Nenutil R, Vojtesek B, Garbis SD. Biomarker discovery in low-grade breast cancer using isobaric stable isotope tags and two-dimensional liquid chromatography-tandem mass spectrometry (iTRAQ-2DLC-MS/MS) based quantitative proteomic analysis. J Proteome Res. 2009;8:362-73
15. Boutte AM, McDonald WH, Shyr Y, Yang L, Lin PC. Characterization of the MDSC proteome associated with metastatic murine mammary tumors using label-free mass spectrometry and shotgun proteomics. PLoS One. 2011;6:e22446
16. Jin L, Zhang Y, Li H, Yao L, Fu D, Yao X. et al. Differential secretome analysis reveals CST6 as a suppressor of breast cancer bone metastasis. Cell Res. 2012;22:1356-73
17. Naba A, Clauser KR, Lamar JM, Carr SA, Hynes RO. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. Elife. 2014;3:e01308
18. Pellegrino L, Krell J, Roca-Alonso L, Stebbing J, Castellano L. MicroRNA-23b regulates cellular architecture and impairs motogenic and invasive phenotypes during cancer progression. Bioarchitecture. 2013;3:119-24
19. Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D. et al. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev Cell. 2007;13:646-62
20. Wewer UM, Albrechtsen R. Tetranectin, a plasminogen kringle 4-binding protein. Cloning and gene expression pattern in human colon cancer. Lab Invest. 1992;67:253-62
21. Yang Y, Macleod V, Miao HQ, Theus A, Zhan F, Shaughnessy JD Jr. et al. Heparanase enhances syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282:13326-33
22. Kujawski R, Przybylowska-Sygut K, Mik M, Lewandowski M, Trzcinski R, Berut M. et al. Expression of the PLS3 Gene in Circulating Cells in Patients with Colorectal Cancer. Pol Przegl Chir. 2015;87:59-64
23. Oh IS, Shimizu H, Satoh T, Okada S, Adachi S, Inoue K. et al. Identification of nesfatin-1 as a satiety molecule in the hypothalamus. Nature. 2006;443:709-12
24. Fayard B, Bianchi F, Dey J, Moreno E, Djaffer S, Hynes NE. et al. The serine protease inhibitor protease nexin-1 controls mammary cancer metastasis through LRP-1-mediated MMP-9 expression. Cancer Res. 2009;69:5690-8
25. Zhang H, Ma Y, Zhang S, Liu H, He H, Li N. et al. Involvement of Ras GTPase-activating protein SH3 domain-binding protein 1 in the epithelial-to-mesenchymal transition-induced metastasis of breast cancer cells via the Smad signaling pathway. Oncotarget. 2015;6:17039-53
26. Best A, Dagliesh C, Ehrmann I, Kheirollahi-Kouhestani M, Tyson-Capper A, Elliott DJ. Expression of Tra2 beta in Cancer Cells as a Potential Contributory Factor to Neoplasia and Metastasis. Int J Cell Biol. 2013;2013:843781
27. Kim MJ, Yoo YA, Kim HJ, Kang S, Kim YG, Kim JS. et al. Mitochondrial ribosomal protein L41 mediates serum starvation-induced cell-cycle arrest through an increase of p21(WAF1/CIP1). Biochem Biophys Res Commun. 2005;338:1179-84
28. Hua KT, Tan CT, Johansson G, Lee JM, Yang PW, Lu HY. et al. N-alpha-acetyltransferase 10 protein suppresses cancer cell metastasis by binding PIX proteins and inhibiting Cdc42/Rac1 activity. Cancer Cell. 2011;19:218-31
29. Zeng Y, Min L, Han Y, Meng L, Liu C, Xie Y. et al. Inhibition of STAT5a by Naa10p contributes to decreased breast cancer metastasis. Carcinogenesis. 2014;35:2244-53
30. van Kilsdonk JW, Takahashi N, Weidle U, Burtscher H, Jarry J, Daha MR. et al. Modulation of activated leukocyte cell adhesion molecule-mediated invasion triggers an innate immune gene response in melanoma. J Invest Dermatol. 2012;132:1462-70
31. Xu S, Venge P. Lipocalins as biochemical markers of disease. Biochim Biophys Acta. 2000;1482:298-307
32. Yamada M, Horiguchi K, Umezawa R, Hashimoto K, Satoh T, Ozawa A. et al. Troglitazone, a ligand of peroxisome proliferator-activated receptor-{gamma}, stabilizes NUCB2 (Nesfatin) mRNA by activating the ERK1/2 pathway: isolation and characterization of the human NUCB2 gene. Endocrinology. 2010;151:2494-503
33. Stengel A, Tache Y. Role of NUCB2/Nesfatin-1 in the hypothalamic control of energy homeostasis. Horm Metab Res. 2013;45:975-9
34. Kalnina Z, Silina K, Bruvere R, Gabruseva N, Stengrevics A, Barnikol-Watanabe S. et al. Molecular characterisation and expression analysis of SEREX-defined antigen NUCB2 in gastric epithelium, gastritis and gastric cancer. Eur J Histochem. 2009;53:7-18
35. Garcia-Galiano D, Pineda R, Ilhan T, Castellano JM, Ruiz-Pino F, Sanchez-Garrido MA. et al. Cellular distribution, regulated expression, and functional role of the anorexigenic peptide, NUCB2/nesfatin-1, in the testis. Endocrinology. 2012;153:1959-71
36. Zegers D, Beckers S, de Freitas F, Jennes K, Van Camp JK, Mertens IL. et al. Identification of mutations in the NUCB2/nesfatin gene in children with severe obesity. Mol Genet Metab. 2012;107:729-34
37. Ramanjaneya M, Chen J, Brown JE, Tripathi G, Hallschmid M, Patel S. et al. Identification of nesfatin-1 in human and murine adipose tissue: a novel depot-specific adipokine with increased levels in obesity. Endocrinology. 2010;151:3169-80
38. Li Z, Xu G, Li Y, Zhao J, Mulholland MW, Zhang W. mTOR-dependent modulation of gastric nesfatin-1/NUCB2. Cell Physiol Biochem. 2012;29:493-500
39. Xu Y, Pang X, Dong M, Wen F, Zhang Y. Nesfatin-1 inhibits ovarian epithelial carcinoma cell proliferation in vitro. Biochem Biophys Res Commun. 2013;440:467-72
40. Qi C, Ma H, Zhang HT, Gao JD, Xu Y. Nucleobindin 2 expression is an independent prognostic factor for clear cell renal cell carcinoma. Histopathology. 2015;66:650-7
41. Zhang H, Qi C, Wang A, Li L, Xu Y. High expression of nucleobindin 2 mRNA: an independent prognostic factor for overall survival of patients with prostate cancer. Tumour Biol. 2014;35:2025-8
42. Zhang H, Qi C, Wang A, Yao B, Li L, Wang Y. et al. Prognostication of prostate cancer based on NUCB2 protein assessment: NUCB2 in prostate cancer. J Exp Clin Cancer Res. 2013;32:77
43. Zhang H, Qi C, Li L, Luo F, Xu Y. Clinical significance of NUCB2 mRNA expression in prostate cancer. J Exp Clin Cancer Res. 2013;32:56
44. Suzuki S, Takagi K, Miki Y, Onodera Y, Akahira J, Ebata A. et al. Nucleobindin 2 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 2012;103:136-43
45. Boonyaratanakornkit V, Pateetin P. The role of ovarian sex steroids in metabolic homeostasis, obesity, and postmenopausal breast cancer: molecular mechanisms and therapeutic implications. Biomed Res Int. 2015;2015:140196
46. Ferreira AR, Bettencourt M, Alho I, Costa AL, Sousa AR, Mansinho A. et al. Serum YB-1 (Y-box binding protein 1) as a biomarker of bone disease progression in patients with breast cancer and bone metastases. J Bone Oncol. 2017;6:16-21
47. Yang H, Chen L, Zhou L, Liu C, Tang L, Zhang J. et al. Serum Human Epidermal Growth Factor 2 is a Novel Biomarker for Recurrence and Metastasis in Triple Negative Breast Cancer. Clin Lab. 2017;63:53-8
Author contact
![]() Corresponding author: E-mail: dengxiyunmededu.cn; Tel: +86-731-88912426; Fax: +86-731-88872260
Corresponding author: E-mail: dengxiyunmededu.cn; Tel: +86-731-88912426; Fax: +86-731-88872260

 Global reach, higher impact
Global reach, higher impact