Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(15):3014-3027. doi:10.7150/jca.21169 This issue Cite
Research Paper
The epigenetic regulation of CXCL14 plays a role in the pathobiology of oral cancers
1. Department of Preventive and Public Oral Health, Nihon University School of Dentistry at Matsudo, 2-870-1 Sakae-cho Nishi, Matsudo, Chiba 271-8587, Japan
2. Research Institute of Oral Health, Nihon University School of Dentistry at Matsudo, 2-870-1 Sakae-cho Nishi, Matsudo, Chiba 271-8587, Japan
3. Department of Oral Health, Graduate School of Dentistry, Kanagawa Dental University, 82 Inaoka-cho, Yokosuka, Kanagawa 238-8580, Japan
4. Department of Biochemistry and Molecular Biology, Nihon University School of Dentistry at Matsudo, 2-870-1 Sakae-cho Nishi, Matsudo, Chiba 271-8587, Japan
Received 2017-5-24; Accepted 2017-7-9; Published 2017-8-25
Abstract
Background: Chemokines selectively attract and activate leukocytes and play roles in a variety of homeostatic and disease processes. Explore the biological properties of CXCL14 seems complicated due to unknown functional characteristics of CXCL14 in cancer.
Methods: To study the multistep process of oral cancer development, we analyzed oral samples spanning normalcy, dysplasia and cancer from multiple perspectives, revealing a cascade of progressive changes.
Results: CXCL14 protein was expressed in the cytoplasm adjacent to tumors. T classification (P<0.001), clinical stage (P=0.0013) and nodal metastasis (P=0.0035) were significantly associated with CXCL14 in relationships between CXCL14 expression levels and tumor and patient characteristics. Compared with non-tumor tissue, expression of the epidermal growth factor receptor (EGFR) gene was increased in dysplasia and was further sustained in cancer. Our data show an inverse relationship between CXCL14 and EGFR expression levels in tumor cells indicating that CXCL14 expression is beneficial for tumor suppression. To explore epigenetic regulation and the impact of CXCL14 on oral cancer, analysis of CpG islands methylation in the CXCL14 promoter region indicated that the abnormal hypermethylation of that promoter region in tumor cells and tissues is one of the mechanisms causing the reduced expression. Restoration of CXCL14 expression was induced by treatment with 5-aza-2′-deoxycytidine. Using in vivo mouse models, we demonstrate that the restoration of CXCL14 expression in irradiation-induced oral carcinoma cells induces the expression of Late Cornified Envelope (LCE) genes.
Conclusions: Our data suggest that LCE genes are a novel target of CXCL14 and are likely to have a tumor suppressor function through the modulation of CXCL14 expression. In conclusion, CXCL14 might play a pivotal role in the pathobiology of oral cancer, probably by regulating DNA methylation and leukocyte migration. The level of CXCL14 expression may be a valuable adjuvant parameter to predict the prognosis of patients with oral carcinoma and may be a potential therapeutic target.
Keywords: Chemokine, CXCL14, EGFR, DNA methylation, Irradiation, LCE
Introduction
Tumors develop in multiple steps [1-3], and tumor progression depends on the balance of expression between tumor progression-promoting and -suppressing genes, being in favor of the former at each step [4-6]. Tumor cells produce cytokines and chemokines either to weaken the immune system and/or to disguise themselves to be protected from immune cell-mediated attacks. Chemokine receptors and their functions differ between tumor cells and normal cells, which suggests that the dysregulation of chemokines may be involved in the development of malignancy [7,8]. The recent explosion of research in the field of chemokine function as mediators of tumor progression has led to the concept that these small, immunomodulatory proteins also play key roles in carcinogenesis.
CXCL14 is an ELR (Glu-Leu-Arg)-negative chemokine that is abundantly expressed in most normal tissues [9-12]. However, many epithelial cancer cell lines and primary carcinomas do not express CXCL14, suggesting that it may have a tumor suppressor function [10,12-14]. CXCL14 expression is suppressed by epidermal growth factor (EGF) and can be restored by treatment with an EGF receptor (EGFR) tyrosine kinase inhibitor in head and neck squamous cell carcinoma (HNSCC) cells [15]. EGFR signaling plays a crucial role in the aggressive features of human HNSCC [16].
Cancer is caused by the accumulation of genetic and epigenetic alterations [17,18]. Abnormal gene expression in cancer cells may be caused by epigenetic modifications, including DNA methylation, histone modifications and changes in chromatin structure [19-22]. Aberrant DNA methylation has been established as one of the major mechanisms by which tumor suppressor genes are silenced in cancer [19]. Unlike genetic alterations, epigenetic changes are potentially reversible, making them attractive and promising targets for therapeutic intervention. CXCL14 expression was demonstrated to be silenced by DNA hypermethylation in many malignant tumors, including lung cancer [23] and acute myeloid leukemia [24].
Chemokines are a superfamily of small chemotactic cytokines that direct the migration of leukocytes [25]. Late Cornified Envelope (LCE) clusters contain multiple well-conserved genes encoding stratum corneum proteins [26,27] and are located on chromosome 1q21 in a region called the epidermal differentiation complex [28]. That region is enriched for genes that are expressed during epidermal differentiation, including loricrin, involucrin, filagrin, small proline-rich protein genes and LCE genes [26]. Up-regulation of CXCL14 was accompanied by the differentiation of epithelial cells [29]. LCE genes were reported to be significantly up-regulated in response to the UV irradiation of skin cells [26]. The gene encoding CXCL14 was identified as a UV-responsive gene and we hypothesize that this gene may be turned on following radiation via a mechanism involving the hypomethylation of cytosine. Thus, CXCL14 has been suggested as a potential tumor suppressor as well as being involved in other physiological and pathological processes. However, the expression of CXCL14 protein and its clinical significance, the physiological functions of LCE genes, especially their involvement in oral cancer, are still largely unknown. In the current investigation, we show that the CXCL14 promoter is highly methylated in oral cancer cells and its expression is down-regulated in EGFR-positive cells. Restoration of CXCL14 expression in irradiation-induced Ca9-22 cells increases the expression of LCE genes in mice. We demonstrate in this study that LCE genes are a novel target of CXCL14, can interact with EGFR and might modulate DNA methylation following irradiation. This mechanism may be important for the interplay of two important cancer-related genes, CXCL14 and EGFR, and our findings indicate a possible role of CXCL14 protein in human carcinogenesis.
Materials and Methods
Surgical specimens
Formalin-fixed, paraffin-embedded samples originating from 130 cases of primary oral squamous cell carcinoma (OSCC) (73 men and 57 women; age range: 38-82 years, mean: 63.7 years) were used. The primary sites of OSCC were 63 tongue (48.4%), 35 gingiva (27.0%), 20 buccal mucosa (15.4%) and 12 oral floor (9.2%). We also analyzed 5 frozen samples of non-tumor oral mucosa (3 men and 2 women; age range, 29-46 years; mean: 36.8 years) and 5 samples each of primary (3 men and 2 women; age range, 58-68 years; mean: 64.2 years) and of metastatic OSCC (3 men and 2 women; age range, 55-72 years; mean: 65.6 years). The study plan was performed according to the ethical standards presented in the Declaration of Helsinki, and was approved by the Medical Ethics Committee of Nara Medical University, Kashihara, and the Tokyo Dental College, Ichikawa, Japan. All specimens were obtained from randomly selected patients attending Hospitals without preoperative therapy. Tumor staging was performed according to the Union for International Cancer Control TNM classification system (seventh edition), and the histologic grade of OSCC was classified according to World Health Organization criteria. Medical records and prognostic follow-up data were obtained from the patient database maintained by the hospital. The follow-up period was 31 - 97 months (mean: 53 months).
Cell culture
The human OSCC lines HSC2, HSC-3, HSC-4, Ca9-22 and Ho-1-U-1 were obtained from the Japanese Cancer Research Resources Bank. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Wako Pure Chemical Industries, Osaka, Japan) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Biowest, Riverside, MO, USA) in 5% CO2 in air at 37˚C. In an experiment, Ca9-22 cells were irradiated using an X-ray source at a dose rate of 10 Gy.
Genomic DNA extraction and Bisulfite conversion
Genomic DNA isolation was performed using a DNeasy Blood & Tissue Kit (Cat No.: 69504; Qiagen, Hilden, Germany) following the manufacturer's protocol. The amount of DNA extracted was measured using a Nanodrop (ND-1000, Thermo Fisher Scientific, Waltham, MA, USA). One µg of each genomic DNA was converted using an EpiTect Fast Bisulfite Conversion Kit (Cat No.: 59824, Qiagen, Hilden, Germany) as described in the manufacturer's instructions. Modified DNA was suspended in elution buffer and was immediately used or stored at -80˚C.
Methylation specific PCR (MSP)
MSP was carried out using 10 ng modified DNA in a 25 μl volume using a QuantiTect SYBR Green PCR Kit (Qiagen, Hilden, Germany). Methyl Primer Express® 1.0 Software (Thermo Fisher Scientific, Waltham, MA, USA) was used to design the PCR primers targeting bisulfite-converted genomic DNA. Two pairs of primers specific for CXCL14 were as follows: [for methylated DNA F 5'-TTCGGAAAAGTTGGTTTGTC-3' and R 5'-ACGTCCCGCTCTACCTTTA-3' and for unmethylated DNA F 5'-GGTTTGGAAAAGTTGGTTTGTT-3' and R 5'-ACATCCCACTCTACCTTTAAA-3' yielded a 110 bp PCR product]; [for methylated DNA F 5'-TTTTTAAAGGTAGAGCGGGAC-3' and R 5'-CGAAAAAAAACCCGCTATC-3' and for unmethylated DNA F 5'-GGTTTTTTAAAGGTAGAGTGGGAT-3' and R 5'-CAAAAAAAAACCCACTATC
ACA-3' yielded a 164 bp PCR product]. The amplification conditions were as follows: one cycle of 95°C for 15 min, followed by 55 cycles of 94°C for 15 s, 30 s at 57˚C and 30 s at 72°C. EpiTect Control DNAs (Cat No.: 59568, Qiagen, Hilden, Germany) (completely methylated or completely unmethylated bisulfite converted DNAs) were used for methylation analysis. The PCR products were analyzed by high-resolution capillary electrophoresis on a QIAxcel system (Qiagen, Hilden, Germany) using a QIAxcel DNA High Resolution kit. The high-resolution gel cartridge allows the separation of DNA fragments with a resolution of 3-5 bp and DNA fragment sizes were measured by the inclusion of the QX DNA Size Marker pUC18/HaeIII.
Bisulfite sequencing PCR (BSP)
Bisulfite-treated DNA was amplified using primers flanking the targeted regions, including the MSP-amplified region and the transcriptional start site. Three pairs of sequencing primers were as follows: F 5'-GTGTAAAACGACGGCCAGTTTTTTAGGTTTTTGAGGAT-3', R 5'-GCAGGAAACAGCTATGACCACCTTTAAAAAACCCAAA-3' (146 bp); F 5'-GTGTAAAACGACGGCCAGTTGGGTTTTTTAAAGGTAGA-3', R 5'- GCAGGAAACAGCTATGACCATAACTCACTAAAATTTCTCAAT-3' (236 bp); F 5'-GTGTAAAACGACGGCCAGTTATTGAGAAATTTTAGTGAGTTAT-3', R 5'- GCAGGAAACAGCTATGACCACTCTACTCRACTTTCTCTACC-3' (238 bp). PCR cycle conditions were as follows: 95°C × 10 min; 5 cycles (92°C × 30 s, 49°C × 10 s, 72°C × 2 min); 35 cycles × (95°C × 30 s, 50°C × 10 s, 72°C × 2 min) and 72°C × 60 min for 1 cycle. Amplified PCR products were then incubated with ExoSAP at 37°C for 30 min and then at 80°C for 15 min to deactivate the enzymes. The PCR products were then purified, cloned into the pGEM-T Easy vector system (Promega KK, Tokyo, Japan) and the individual clones were sequenced using an ABI 3730 DNA sequencer (Thermo Fisher Scientific, Waltham, MA, USA).
RNA extraction and quantitative RT-PCR (QRT-PCR) analysis
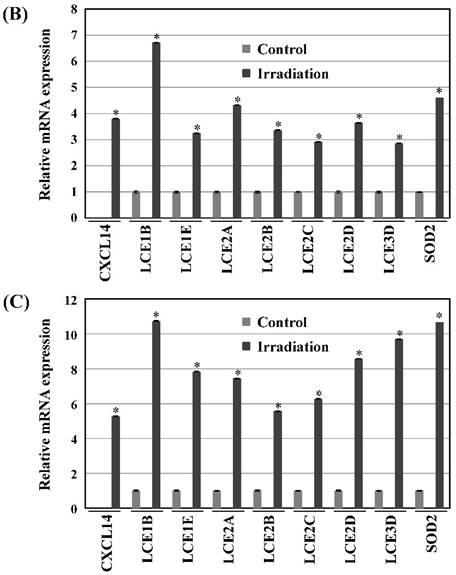
RNA extraction and QRT-PCR analysis was performed according to our previous report [30]. In brief, total RNA was extracted from the tissues and cells using an RNeasy Mini Kit (Qiagen, Tokyo, Japan), and was reverse-transcribed using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems, Foster City, CA, USA). For RT-PCR, the reaction mixture (20 μL) contained 1 μL of each diluted cDNA sample and 10 pmol of each pair of oligonucleotide primers. The relative expression levels of target mRNAs, compared to the level of β-actin RNA, were analyzed by real time PCR with the corresponding TaqMan MGB probes (Hs01557413_m1for CXCL14, Hs00866755_s1 for LCE1B, Hs02380094_g1 for LCE1E, Hs00820278_sH for LCE2A, Hs04194422_s1 for LCE2B, Hs02390636_s1 for LCE2C, Hs02390641_s1 for LCE2D, Hs00754375_s1 for LCE3D, Hs00167309_m1 for superoxide dismutase 2 (SOD2) and Hs99999903_m1 for β-actin) using QuantStudio 6 Real Time PCR System (Applied Biosystems). The thermal cycling conditions were according to the TaqMan Fast Universal PCR protocol.
Treatment with 5-aza-2′deoxycytidine (5-aza-dc) and/or trichostatin A (TSA)
Oral carcinoma Ca9-22 cells were seeded at a density of 1X10 6⁄mL in 10-cm dishes. After overnight culture, cells were treated with 10 μM of the DNA demethylating agent 5-aza-dc (Sigma-Aldrich, St. Louis, MO, USA) for 96 h. Where indicated, the cells were further treated with 100 nM of the histone deacetylase inhibitor TSA (Sigma-Aldrich, St. Louis, MO, USA) for an additional 24 h. After treatment, the cells were harvested for RNA extraction.
Reactive Oxygen Species (ROS) detection
Irradiated Ca9-22 and CXCL14 over-expressing cells were assayed for intracellular ROS using an Image-iT™ LIVE green ROS detection kit (Molecular Probes, Inc, Eugene, OR). They were then counterstained with Hoechst 33342 to label nuclei and were imaged using a confocal laser scanning microscope (BZ-8000, Keyence Corporation, Osaka, Japan). Transient transfection of expression plasmid for human CXCL14 (OriGene Technologies, Inc. Rockville, MD, USA) was performed using the Lipofectamine 2000 system (Invitrogen, Carlsbad, CA, USA).
In vivo experiments
Approximately 5x106 Ca9-22 cells were injected subcutaneously into both sides of the dorso-lateral region of 10 female athymic nude mice (BALB/cAJcl-nu/nu, Clea Japan, Tokyo). Treatment was initiated when the average tumor volume reached 100 mm3. Mice were exposed to X-rays using a linear accelerator radiation therapy system (HL-1500; Hitachi Medical Corp., Tokyo, Japan). For radiation fractionation experiments, exposure to 2 Gy was done daily for 3 weeks with mice restrained in a thermoplastic shell. The total radiation dose was 30 Gy. The tumor tissues at day 18 were used for experiments. Mice were housed in our approved animal holding facility and were treated according to the guidelines of the Kanagawa Dental University on Animal Care.
Immunohistochemistry and double immunofluorescence staining
Immunohistochemistry was performed according to a modified protocol of our previous report [31]. Briefly, formalin-fixed, paraffin-embedded tissue sections were immunostained for CXCL14 and EGFR using a CSA II System (Dako, Carpinteria, CA, USA), in accordance with the manufacturer's instructions. Sections were initially immersed in Target Retrieval Solution (DAKO, Carpinteria, CA, USA) at 95°C for 12 min, and then were cooled for 30 min. Endogenous peroxidase activity was blocked with REAL Peroxidase-Blocking Solution (S2023, DAKO, Carpinteria, CA, USA) for 30 min. Antibodies against CXCL14 (1:100; Proteintech Group, Inc, Rosemont, IL, USA) and EGFR (1:100; Abcam, Cambridge, MA, USA) were used as primary antibodies and were incubated overnight at 4°C. The secondary antibodies conjugated to peroxidase (Nichirei Biosciences, Tokyo, Japan) were incubated at room temperature for 30 min. After rinsing with PBS, all specimens were color developed with a 3,3′-diaminobenzidine tetrahydrochloride (DAB) chromogen kit (Dako, Carpinteria, CA, USA), counterstained with hematoxylin, and examined by light microscopy. The immunostaining of all specimens was performed simultaneously to ensure the same antibody reaction and DAB exposure conditions. In each specimen, the percent of positive cytoplasmic stained cells was calculated in 4 different fields using a 10x objective. For CXCL14, we classified the cases into 3 categories according to their signal intensity as follows: High: ≥25% total cells showed weak/moderate staining, Low: ≤25% total cells stained positive, Negative: immunoreactivity was completely absent. For double immunofluorescence staining, after incubation in protein block solution (DAKO, Carpinteria, CA, USA) for 1 h, the first antibody CXCL14 (1:150, Abcam) was prepared with Zenon Alexa Fluor 594 (Molecular Probes, Eugene, OR, USA) according to the manufacturer's instructions, applied to the samples and incubated in a humidified dark chamber for 2 h. The sections were then washed in PBS for 25 min and were subsequently incubated with the secondary antibody to EGFR (1:150, Abcam) prepared with Zenon Alexa Fluor 488 for 2 h. Following 3 additional washes in TBS-T, ProLong® Gold Antifade Reagent was used for nuclear counterstaining. Red fluorescence represented CXCL14 positive expression, while EGFR positive expression was recognized by green fluorescence. Cells lacking fluorescence signals were defined as CXCL14 and/or EGFR negative.
Statistical analysis
Statistical analysis was performed using Fisher's exact test, and the Chi square test. Disease-free survival was analyzed using the Kaplan-Meier method, and the differences between groups were calculated using the log-rank test. All statistical analyses were conducted using R software.
Results
Expression of CXCL14 in human oral cancer cell lines and tissues
Expression of CXCL14 in human OSCC cell lines and tissues. (A) CXCL14 mRNA expression in OSCC cell lines. Nearly confluent cells were cultured overnight with or without serum. Total RNA was extracted from each cell line and CXCL14 mRNA expression was analyzed using QRT-PCR. Differential expression of CXCL14 was found in HSC-2, HSC-3, HSC-4 and Ho-1-U-1 cells and a complete loss of CXCL14 expression was detected only in Ca9-22 cells with or without serum, inferring an aberrant gene silencing of CXCL14 in OSCC cells. (B) Gene expression analysis in frozen samples of non-tumor oral mucosa, primary and metastatic cancer tissue samples. QRT-PCR was performed using RNA isolated from each tissue sample. A significant decrease in CXCL14 mRNA levels was observed in primary and metastatic cancers compared with non-tumor tissues.
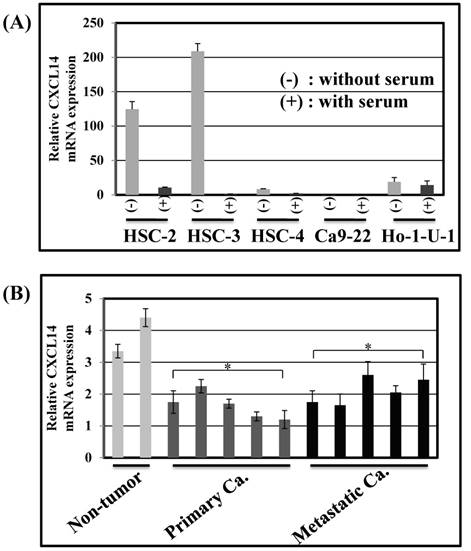
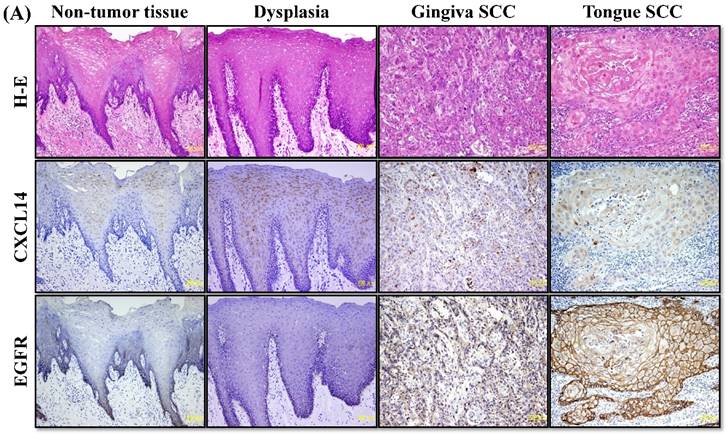
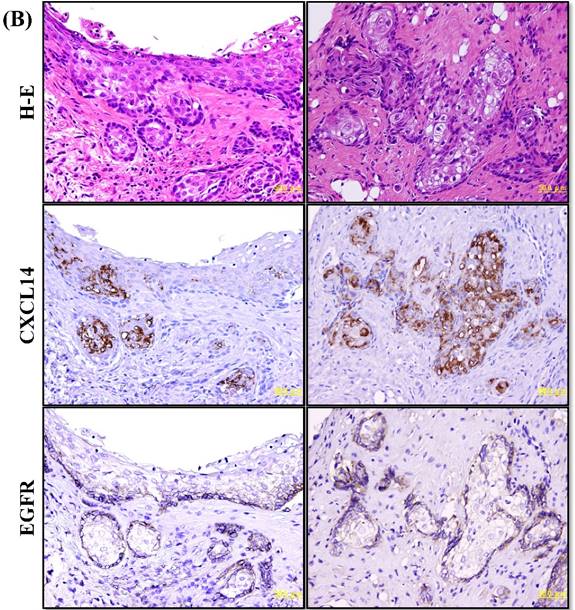
CXCL14 mRNA expression was detected by QRT-PCR in 5 different oral cancer cell lines at low to negative copy levels when cultured in the presence of serum (Figure 1A). The expression of CXCL14 was increased in HSC-2, HSC-3, HSC-4 and Ho-1-U-1 cells when serum-starved and a complete loss of expression was detected in Ca9-22 cells with or without serum starvation, inferring the aberrant gene silencing of CXCL14 in OSCC cells. Therefore, Ca9-22 cells were used as a positive control for methylation and irradiation assays to assess CXCL14 expression. To compare the in vivo expression profile of CXCL14 in non-tumor oral mucosa and in oral cancer, QRT-PCR was performed using RNA isolated from non-tumor oral mucosa as well as primary and metastatic cancer tissue samples. That analysis revealed a significant decrease in CXCL14 mRNA levels in primary and metastatic cancers compared with non-tumor tissues (Figure 1B). Immunohistochemistry was performed to evaluate CXCL14 and EGFR protein expression in oral cancer tissues and in their adjacent non-tumor tissues (Figure 2A). The evaluation of CXCL14 immunoreactivity also revealed differences between non-tumor, dysplasia and oral cancer tissues. CXCL14 protein was expressed predominantly in the cytoplasm in adjacent non-tumor tissues but was significantly down-regulated in OSCC tissues. In non-tumor tissues, there was weak immunoreactivity in spinous and granular cells whereas parabasal and basal cells were negative for CXCL14 or revealed only weak staining. Only the basal cells expressed EGFR in non-tumor tissues. Mild dysplasia (low-grade dysplasia; WHO 2017 classification) showed irregular elongation of the epithelium with acanthosis and stratification of the basal layer. In mild dysplasia, the expression of CXCL14 was found to extend from the basal to the superficial layers of the squamous epithelium in oral tissues. Spinous cells as well as granular cells revealed strong CXCL14 immunoreactivity with weak staining in parabasal and basal cells. EGFR expression was localized in basal and parabasal cells in dysplasia tissues. Tumor cells were stained to a variable extent in different areas, with the majority of cells displaying low CXCL14 immunoreactivity. The tumor cells infiltrated the interstitium with proliferation and invasion in gingival tumor. Negative or weak positive CXCL14 expression was observed in gingival tumor cells whereas EGFR expressed homogenous cytoplasmic staining. In contrast, CXCL14 immunoreactivity was present in the cytoplasm of the tongue tumor cells with occasional single cell keratinization and EGFR was homogenously expressed in the cell membrane of the tumor cells (Figure 2A). Representative examples of CXCL14 and EGFR immunohistochemistry in CXCL14 high-expressing tissues are shown in Figure 2B.
Immunohistochemistry to evaluate CXCL14 and EGFR protein expression in oral cancers and their adjacent non-tumor tissues. (A) Evaluation of CXCL14 immunoreactivity revealed differences between non-tumor, dysplasia and oral cancer tissues. CXCL14 protein was expressed predominantly in the cytoplasm in adjacent non-tumor tissues but was significantly down-regulated in OSCC tissues. In contrast, oral tumors exhibited strong EGFR staining and weak or negative staining in non-tumor and in dysplastic tissues. Original magnification; x10. H-E; hematoxylin and eosin. (B) Representative examples of CXCL14 and EGFR immunohistochemistry in CXCL14 high-expressing tissues. Immunohistochemical analysis was performed as described in the Materials and methods. Original magnification; x20. H-E; hematoxylin and eosin.
Association between CXCL14 expression and patient and tumor characteristics and clinical outcome
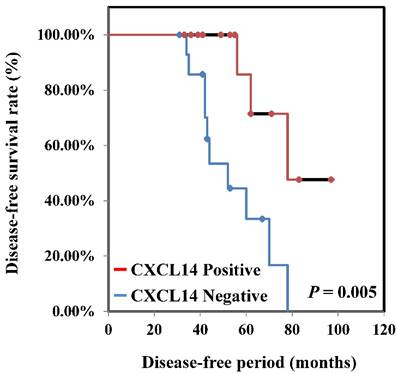
To determine if the expression of CXCL14 correlates with clinical outcome, we performed immunohistochemical analysis of oral tumors with clinical follow-up data in the Nara Medical University data set. Among the 130 patients included in that analysis, 36 had their CXCL14 expression level analyzed. Relationships between CXCL14 expression and clinicopathological parameters are summarized in Table 1. Immunoreactivity to CXCL14 was strongly associated with tumor progression (P < 0.001). Patients with stage Tis/ T1 and T2 tumors (33/83; 39.8%) were more likely to be positive for CXCL14 than were patients with T3 and T4 stage (3/47; 6.4%), whereas metastasis-negative tumors were more likely to be positive for CXCL14 (33/95; 34.7%) than were metastasis-positive tumors (3/35; 8.6%). A significant association was also observed between CXCL14 expression and clinical stage (P = 0.0013). Among 130 patients, 29 (38.7%) with CXCL14 positivity were at stages I and II, whereas only 7 of 55 patients (12.7%) expressing CXCL14 were at stages III and IV. No significant correlation was identified between the expression level of CXCL14 and other clinicopathological factors. Kaplan-Meier survival analysis indicated that patients with negative CXCL14 expression levels had worse disease-free survival than did those with positive CXCL14 expression levels (P = 0.005) (Figure 3).
Disease-free survival curve in OSCCs. Disease-free survival curve of OSCC patients was calculated by the Kaplan-Meier method. The disease-free survival among all CXCL14-negative patients was significantly worse than among CXCL14-positive patients (P = 0.005).
Association between CXCL14 expression and clinicopathological parameters in OSCCs
| Parameters | Negative | CXCL14 low | CXCL14 high | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 52 | 13 | 8 | |
| Female | 42 | 8 | 7 | 0.8443 |
| Age | ||||
| <60 | 44 | 12 | 6 | |
| >60 | 50 | 9 | 9 | 0.8449 |
| Site | ||||
| Tongue | 45 | 9 | 9 | |
| Gingiva | 25 | 7 | 3 | |
| Other | 24 | 5 | 3 | 0.8591 |
| Histological differentiation | ||||
| Well | 40 | 11 | 5 | |
| Moderately | 42 | 7 | 8 | |
| Poorly | 12 | 3 | 2 | 0.8012 |
| T classification | ||||
| Tis, T1 | 27 | 15 | 8 | |
| T2 | 23 | 4 | 6 | |
| T3 | 24 | 1 | 1 | |
| T4 | 20 | 1 | 0 | <0.0001 |
| Clinical stage | ||||
| I | 26 | 14 | 7 | |
| II | 20 | 3 | 5 | |
| III | 26 | 2 | 3 | |
| IV | 22 | 2 | 0 | 0.0013 |
| Nodal metastasis | ||||
| Negative | 62 | 18 | 15 | |
| Positive | 32 | 3 | 0 | 0.0035 |
Localization of CXCL14 and EGFR in oral tumors
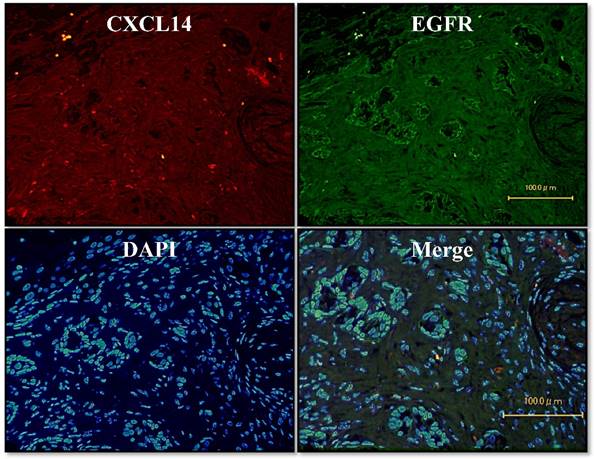
To further investigate the potential role of CXCL14, we sought to identify the relevant CXCL14-positive cells. We used an immunofluorescence assay on oral cancer tissues to survey the co-localization of EGFR with CXCL14 protein. Dual immunofluorescence labeling for CXCL14 and EGFR showed that CXCL14 positivity was largely restricted to cells that were EGFR-negative in oral cancer (Figure 4).
Hypermethylation of the CXCL14 promoter region in oral cancer cell lines and tissues
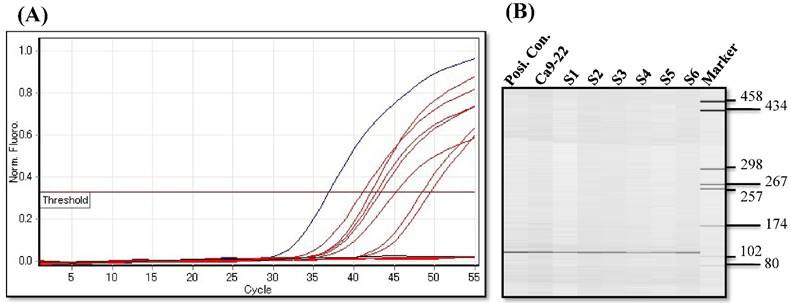
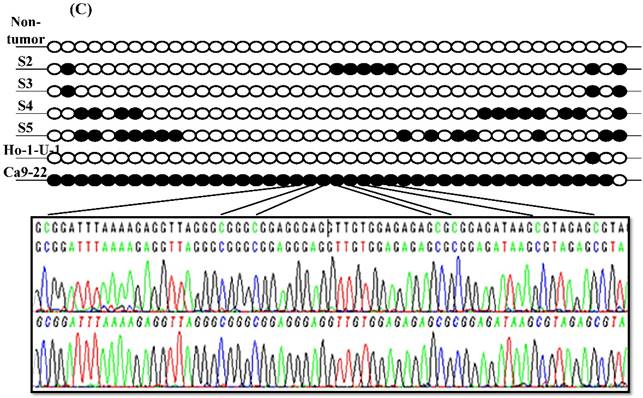
Since aberrant promoter CpG methylation is known to be related to gene silencing, we next explored the CXCL14 methylation status using MSP and BSP. Representative results of MSP for the CXCL14 promoter are shown in Figure 5A. The amplification curves of CXCL14 were sharply defined curves with a narrow peak, indicating that the established PCR method effectively amplified the target genes. Complete methylation was detected in Ca9-22 cells, and partial methylation was revealed in 74% of primary tumor tissues (Figure 5A). All CXCL14 negative tumors showed partial methylation of the CpG islands. Tumor tissues with a lower expression of CXCL14 had a predominantly higher frequency of methylated CpG sites. Ca9-22 cells and tumor tissues showed methylated PCR products (Figure 5B). To validate the MSP results and to assess the density of methylation, bisulfite sequencing of the CXCL14 promoter region was performed in non-tumor oral tissues, oral cancer tissues and cell lines. The detailed methylation status of individual CpG sites was examined using BSP (Figure 5C). The detailed BSP analyses confirmed the findings by MSP that the CXCL14 promoter was frequently methylated in Ca9-22 cells compared to non-tumor tissue and Ho-1-U-1 cell line.
Pharmacological demethylation restores CXCL14 expression in Ca9-22 cells
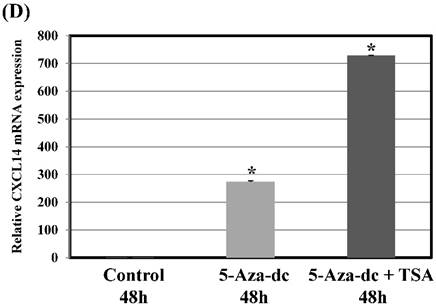
To further verify the above phenomenon, Ca9-22 oral cancer cells were treated with 5-Aza-2'-dc to recover the demethylation state of the CXCL14 gene CpG islands. Figure 5D illustrates that with 5-aza-dc treatment, CXCL14 mRNA levels were up-regulated in Ca9-22 cells compared with the control group (P = 0.019), suggesting that methylation of the CXCL14 promoter was responsible for the silencing of CXCL14 in those cells. As treatment with 5-aza-dc resulted in a lower reactivation level of CXCL14 in Ca9-22 cells, those cells were further treated with TSA. The restored expression of CXCL14 was observed more in Ca9-22 cells treated with 5-aza-dc and TSA than in those treated with 5-aza-dc only (P = 0.001) (Figure 5D).
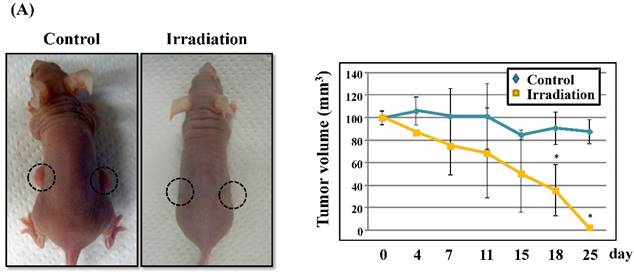
Radiation-induced expression of CXCL14 increases ROS production and suppresses in vivo tumor growth
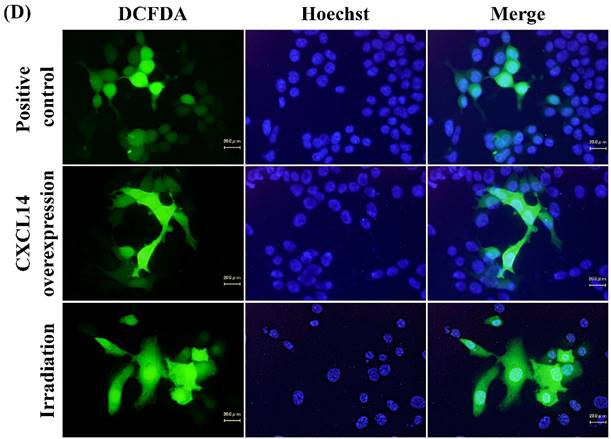
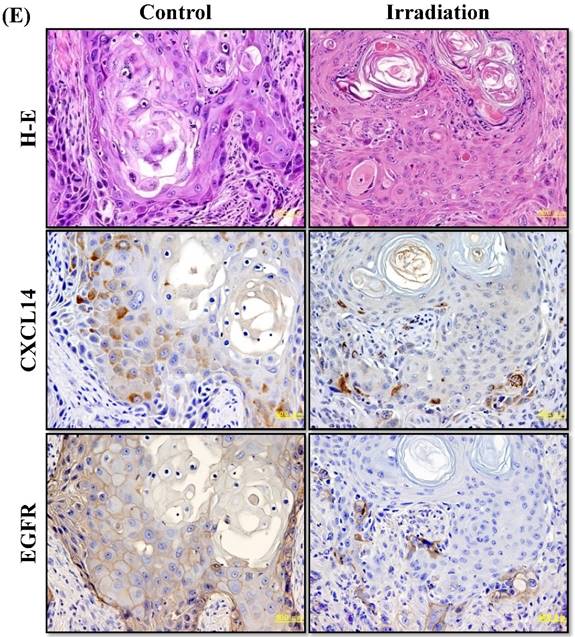
Next, we investigated whether treatment of Ca9-22 tumors with irradiation would restore CXCL14 gene expression in vivo. Irradiation exerted a significant tumor-suppressive effect against xenografted Ca9-22 cells on days 18 and 25 (P<0.001; Figure 6A). Irradiation caused a significant increase in the expression of CXCL14 mRNA levels in the Ca9-22 tumors and Ca9-22 cell line (P<0.001; Figure 6B, C). QRT-PCR analysis revealed that irradiation-treated Ca9-22 tissues and cells had markedly increased levels of LCE mRNAs and SOD2 mRNA compared to the control (P<0.001; Figure 6B, C). To determine ROS generation by CXCL14 over-expression and irradiation, we examined intracellular ROS levels in CXCL14 over-expressed and irradiated Ca9-22 cells. The results showed that CXCL14 over-expression and irradiation induced ROS generation (Figure 6D). To further confirm that correlation, we detected the expression of CXCL14 and EGFR in subcutaneous xenograft tumor tissues using immunohistochemical analysis (Figure 6E). In tumors of the radiation-treated group, the expression of CXCL14 was stronger than in tumor tissues of the control group, while a down-regulation of EGFR expression in the tumor tissue was observed (Figure 6E). These results indicate that treatment with irradiation in vitro and in vivo stimulated the expression of CXCL14 in Ca9-22 cells.
Localization of CXCL14 and EGFR in OSCC. Immunofluorescence assay of oral cancer tissues was used to characterize the co-localization of EGFR with the CXCL14 protein. CXCL14 positivity was largely restricted to cells that were EGFR-negative in OSCC. Original magnification; x20.
Hypermethylation of the CXCL14 promoter region in oral cancer cell lines and tissues. (A) Normalized graph for CXCL14 expression. Representative results of MSP for the CXCL14 promoter are shown. Complete methylation was detected in Ca9-22 cells, and partial methylation was revealed in primary tumor tissues. 5B) High-resolution capillary electrophoresis of MSP reaction products - electropherogram view: Positive control: 110 bp amplicon for CXCL14; Ca9-22: OSCC cell line; S1 - S6: Oral tumor tissues 1 - 6; Marker: QX DNA Size Marker pUC 18/Hae III. (C) Bisulfite sequencing of the CXCL14 promoter region. The detailed methylation status of individual CpG sites was examined in Ca9-22 cells by BSP. Consistent with the MSP results, dense methylation in the promoter region of CXCL14 was found in Ca9-22 cells. (D) Pharmacological demethylation restores CXCL14 expression. Ca9-22 cells were treated with 5-aza-dc to recover the demethylation state of CXCL14 gene CpG islands. Ca9-22 cells were restored to up-regulate the CXCL14 mRNA level compared with the control group (P=0.005). Ca9-22 cells were then further treated with TSA. The restored expression of CXCL14 was observed more in Ca9-22 cells treated with 5-aza-dc and TSA than in Ca9-22 cells treated with 5-aza-dc only (P<0.001).
Discussion
We highlighted specific biological processes in disease progression as a basis toward a better understanding of the molecular mechanisms that drive oral cancer development. In particular, we emphasized and further documented the loss of CXCL14 expression during cancer progression. To our knowledge, this is the first report that reveals the expression of epidermal growth factor receptor (EGFR) and CXCL14 in oral carcinomas and indicates a role for the EGFR in oral carcinomas correlating with CXCL14. EGFR is highly expressed in several types of carcinomas including HNSCC and high expression levels of EGFR reduce recurrence-free or overall survival rates in 70% of patients and act as a strong prognostic indicator [32]. The activation of EGFR often coincides with the down-regulation of CXCL14 in HNSCC [10,15]. It has to take into account that most studies on the role of CXCL14 in malignancies have been based on results of the CXCL14 mRNA level [11,33,34]. In this study, we employed immunohistochemistry and immunofluorescence to identify the homeostatic chemokine CXCL14 protein as a target of EGFR regulation in normal and cancerous epithelial cells. Normal epithelial cells expressed CXCL14 protein consistently, whereas cells with malignant potential significantly suppressed CXCL14. An analysis of correlations with clinical features indicated that the expression level of CXCL14 in oral metastatic carcinoma tissues was significantly lower than that of non-metastatic tumor tissues. The relative expression levels of CXCL14 in oral carcinoma tissue at pathological stages III and IV were lower than those of tumors at stages I and II. Additionally, the expression levels of CXCL14 in tumor tissues were significantly correlated with the prognosis of oral carcinoma. Furthermore, CXCL14 protein expression was examined in correlation with clinico-pathological parameters in oral cancer tissue samples. In addition, spinous cells as well as granular cells revealed strong CXCL14 immunoreactivity with weak staining in parabasal and basal cells in dysplasia compared to non-tumor tissues. Several studies showed that oral dysplasia has a significant high rate of transformation to cancer and enhanced CXCL14 expression may suppress this neoplastic progression. Thus, we identify CXCL14 as an important prognostic factor, whose loss is not only associated with more aggressive primary oral cancers with a worse outcome, but also with an increased risk of metastasis formation. These findings give rise to the hypothesis that the loss of CXCL14 expression may allow tumor cells to gain a selective advantage in vivo. Central to explaining the implications for the loss of CXCL14 in tumors is an understanding of the normal biological function of CXCL14. Although we proved the co-expression and colocation relationship between EGFR and CXCL14, the exact molecular biological mechanism still has not been established.
CXCL14 exerts an anti-tumor effect on oral carcinoma in vitro and in vivo, and its mRNA and protein expression levels decline in oral carcinoma tissues compared to non-tumor tissues. Our results, along with previous studies, imply that a reduction in expression of the CXCL14 gene may contribute to the progress and development of oral cancer. CXCL14 expression is highly sensitive to serum in vivo, and serum-free conditions lead to a significant up-regulation of CXCL14. HNSCC cells treated with an EGF tyrosine kinase inhibitor increase their expression of CXCL14 protein in culture [15]. This is important in the context of the potential role of EGFR in tumorigenesis as the complete loss of this gene in oral tumors is comparatively rare and a reduced CXCL14 expression is far more common. Our current observations provide guidance and new clues toward understanding these multifactorial aspects of cancer progression.
Radiation-induced CXCL14 increases ROS production and suppresses in vivo tumor growth. (A) Irradiation exerted a significant tumor-suppressive effect against xenografted Ca9-22 cells on days 18 and 25 (P<0.001). (B) and (C) Irradiation caused a significant increase in the expression of CXCL14 mRNA levels in Ca9-22 tumors and Ca9-22 cell line (P<0.001). QRT-PCR analysis revealed that irradiation-induced Ca9-22 tissues and cells had markedly increased levels of LCE mRNAs and SOD2 mRNA compared to the control (P<0.001). (C) ROS generation by irradiation and CXCL14 over-expression. CXCL14 over-expression and irradiation induced ROS production. (D) Expression of CXCL14 and EGFR in subcutaneous xenograft tumor tissues. The expression of CXCL14 in tumor tissues was stronger in tumors of the radiation-treated group than in the control group, while a down-regulation of EGFR expression in the tumor tissue was observed. Original magnification; x20. H-E; hematoxylin and eosin.
Recent studies have provided compelling evidence that aberrant DNA methylation plays a pivotal role in cancer initiation and progression [35,36]. CXCL14 is epigenetically silenced in prostate and lung cancers [23,37]. Aberrant methylation may ultimately serve two vital roles in cancer: as biomarkers for detection and prognosis and as targets for epigenetic therapy [38-41]. CXCL14 may participate in both arenas. MSP and BSP of the promoter region of the CXCL14 gene showed dense methylation in Ca9-22 cells. The silencing of CXCL14 can be reversed by pharmacological demethylation, inferring that methylation is the predominant mechanism for the inactivation of CXCL14. As treatment with 5-Aza-dc results in the reactivation of CXCL14 to normal levels in Ca9-22 cells, we tested whether histone modification mediates CXCL14 silencing in those cells using the histone deacetylase inhibitor TSA. Further restored expression of CXCL14 was observed in Ca9-22 cells treated with 5-Aza-dc together with TSA compared to cells treated with 5-Aza-dc only, suggesting that histone modifications also play a role in the transcriptional silencing of CXCL14. MSP and BSP results showed that the promoter of CXCL14 is partly methylated in oral cancer tissues. Thus, hypermethylation of the CXCL14 promoter may represent a promising biomarker to aid in treatment decisions and reversing the promoter hypermethylation of CXCL14 could be a feasible approach to restore anti-tumor immune responses to treat oral cancers.
CXCL14 expression is lost at a high frequency in a wide range of epithelial tumors, including HNSCC [12,14,33,34,42]. The loss of CXCL14 expression in HNSCC was associated with a decreased infiltration of leukocytes, supporting the potential role of CXCL14 as a chemotactic and tumor-suppressive factor. Paralleling and complementing our findings in humans, our studies with mice show that CXCL14 is critical to the development of carcinomas, and that CXCL14 expression following irradiation at least requires and might act completely through leukocyte expression. In xenograft models, irradiation induced CXCL14 expression in Ca9-22 cancer cells, where it is constitutively silenced by DNA methylation, and activated ROS. Growing evidence suggests that ROS act as a second messenger in intracellular signaling cascades that induce and maintain the oncogenic phenotype of tumor cells. ROS have been reported to induce proliferation, survival and cellular migration [43]. CXCL14 over-expressing cells showed a clear increase of ROS with respect to their counterparts. Irradiation also increased levels of cellular ROS (Figure 6B, C, D). These results suggest that the generation of ROS acts as a mediator of the CXCL14-induced survival of oral tumor cells.
A primary finding of this work is that LCE genes respond to irradiation, thus demonstrating a similarity to the CXCL14 gene. The LCE gene cluster contains multiple conserved genes encoding stratum corneum proteins [26-28]. Our study demonstrates that most members of the LCE family are transcriptionally regulated by the tumor suppressor CXCL14 in xenograft models although the induction levels varied. CXCL14 genes were originally identified as UV-responsive genes and it is believed that their irradiation-dependent up-regulation changes the properties of the cornified envelope/stratum corneum. The CXCL14 protein acts as a molecular sensor for damage generated by irradiation through mediating leukocyte migration and ROS production in damaged keratinocytes. Taken together, cells may possess the function to express LCE family genes through CXCL14 activation to eliminate dangerous cells that have DNA damage.
Our findings endorse the concept that not only leukocytes but also epithelial keratinocytes are key regulators of cancer immunity. In conclusion, our study demonstrates that oral cancers frequently have altered CXCL14 expression and that increased CXCL14 expression following irradiation contributes to the suppression of cancer cells and the migration of leukocytes.
Acknowledgements
The authors wish to express their gratitude to Emeritus Professor Y. Tanaka (Tokyo Dental College), Professor T. Sakurai and Dr. T. Ikoma (Kanagawa Dental University), Prof. H. Kuniyasu and Dr. T. Sasahira (Nara Medical University), Dr. C. Taguchi and Dr. F. Zhang (Nihon University School of Dentistry at Matsudo) for their help and contributions to this study. This research was partially supported by a Grant-in-Aid for Scientific Research of Ministry of Education, Culture, Sports, Science and Technology, Japan.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Vogelstein B, Fearon ER, Hamilton SR. et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-32
2. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70
3. Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210-9
4. Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46-54
5. Akhurst RJ. TGF-β antagonists: why suppress a tumor suppressor? J Clin Invest. 2002;109:1533-6
6. Benatti P, Basile V, Merico D, Fantoni LI, Tagliafico E, Imbriano C. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008;36:1415-28
7. Ruffini PA, Morandi P, Cabioglu N, Altundag K, Cristofanilli M. Manipulating the chemokine-chemokine receptor network to treat cancer. Cancer. 2007;109:2392-404
8. Mantovani A, Savino B, Locati M, Zammataro L, Allavena P, Bonecchi R. The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev. 2010;21:27-39
9. Kurth I, Willimann K, Schaerli P, Hunziker T, Clark-Lewis I, Moser B. Monocyte selectivity and tissue localization suggests a role for breast and kidney-expressed chemokine (BRAK) in macrophage development. J Exp Med. 2001;194:855-61
10. Ozawa S, Kato Y, Komori R, Maehata Y, Kubota E, Hata R. BRAK/CXCL14 expression suppresses tumor growth in vivo in human oral carcinoma cells. Biochem Biophys Res Commun. 2006;348:406-12
11. Schwarze SR, Luo J, Isaacs WB, Jarrard DF. Modulation of CXCL14 (BRAK) expression in prostate cancer. Prostate. 2005;64:67-74
12. Shurin GV, Ferris RL, Tourkova IL. et al. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. J Immunol. 2005;174:5490-8
13. McKinnon CM, Lygoe KA, Skelton L, Mitter R, Mellor H. The atypical Rho GTPase RhoBTB2 is required for expression of the chemokine CXCL14 in normal and cancerous epithelial cells. Oncogene. 2008;27:6856-65
14. Shellenberger TD, Wang M, Gujrati M. et al. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res. 2004;64:8262-70
15. Ozawa S, Kato Y, Ito S. et al. Restoration of BRAK/CXCL14 gene expression by gefitinib is associated with antitumor efficacy of the drug in head and neck squamous cell carcinoma. Cancer Sci. 2009;100:2202-9
16. Ang KK, Berkey BA, Tu X. et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350-6
17. Zhang W, Glockner SC, Guo M. et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764-72
18. Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181-206
19. Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-16
20. Merlo A, Herman JG, Mao L. et al. 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686-92
21. Zheng J, Wang J, Sun X. et al. HIC1 modulates prostate cancer progression by epigenetic modification. Clin Cancer Res. 2013;19:1400-10
22. Ramachandran I, Thavathiru E, Ramalingam S. et al. Wnt inhibitory factor 1 induces apoptosis and inhibits cervical cancer growth, invasion and angiogenesis in vivo. Oncogene. 2012;31:2725-37
23. Tessema M, Klinge DM, Yingling CM, Do K, Van Neste L, Belinsky SA. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29:5159-70
24. Sonnet M, Claus R, Becker N. et al. Early aberrant DNA methylation events in a mouse model of acute myeloid leukemia. Genome Med. 2014;6:34
25. Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. 2001;2:123-8
26. Jackson B, Tilli CM, Hardman MJ, Avilion AA, MacLeod MC, Ashcroft GS, Byrne C. Late cornified envelope family in differentiating epithelia-response to calcium and ultraviolet irradiation. J Invest Dermatol. 2005;124:1062-70
27. Marshall D, Hardman MJ, Nield KM, Byrne C. Differentially expressed late constituents of the epidermal cornified envelope. Proc Natl Acad Sci USA. 2001;98:13031-6
28. Bergboer JG, Tjabringa GS, Kamsteeg M. et al. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am J Pathol. 2011;178:1470-7
29. Ikoma T, Ozawa S, Suzuki K. et al. Calcium-calmodulin signaling induced by epithelial cell differentiation upregulates BRAK/CXCL14 expression via the binding of SP1 to the BRAK promoter region. Biochem Biophys Res Commun. 2012;420:217-22
30. Bhawal UK, Lee HJ, Arikawa K. et al. Micromolar sodium fluoride mediates anti-osteoclastogenesis in Porphyromonas gingivalis-induced alveolar bone loss. Int J Oral Sci. 2015;7:242-9
31. Sasahira T, Kurihara M, Nakashima C, Kirita T, Kuniyasu H. LEM domain containing 1 promotes oral squamous cell carcinoma invasion and endothelial transmigration. Br J Cancer. 2016;115:52-8
32. Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37:S9-15
33. Hromas R, Broxmeyer HE, Kim C. et al. Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem Biophys Res Commun. 1999;255:703-6
34. Frederick MJ, Henderson Y, Xu X. et al. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am J Pathol. 2000;156:1937-50
35. Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148-59
36. Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400-13
37. Song EY, Shurin MR, Tourkova IL, Gutkin DW, Shurin GV. Epigenetic mechanisms of promigratory chemokine CXCL14 regulation in human prostate cancer cells. Cancer Res. 2010;70:4394-401
38. Belinsky SA, Klinge DM, Stidley CA. et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089-93
39. Belinsky SA, Liechty KC, Gentry FD. et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338-44
40. Gore SD, Baylin S, Sugar E. et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361-9
41. Yang AS, Doshi KD, Choi SW. et al. DNA methylation changes after 5-aza-2'-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495-503
42. Schwarze R, DePrimo SE, Grabert LM, Fu VX, Brooks JD, Jarrard DF. Novel pathways associated with bypassing cellular senescence in human prostate epithelial cells. J Biol Chem. 2002;277:14877-83
43. Arnold RS, Shi J, Murad E. et al. Hydrogen peroxide mediates the cell growth and transformation caused by the mitogenic oxidase Nox1. Proc Natl Acad Sci USA. 2001;98:5550-5
Author contact
![]() Corresponding author: Ujjal K. Bhawal, Department of Biochemistry and Molecular Biology, Nihon University School of Dentistry at Matsudo, 2-870-1 Sakae-cho Nishi, Matsudo, Chiba 271-8587, Japan; Tel: +81-47-360-9328; Fax: +81-47-360-9329; E-mail: bhawal.ujjal.kumarac.jp
Corresponding author: Ujjal K. Bhawal, Department of Biochemistry and Molecular Biology, Nihon University School of Dentistry at Matsudo, 2-870-1 Sakae-cho Nishi, Matsudo, Chiba 271-8587, Japan; Tel: +81-47-360-9328; Fax: +81-47-360-9329; E-mail: bhawal.ujjal.kumarac.jp

 Global reach, higher impact
Global reach, higher impact