3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(14):2720-2728. doi:10.7150/jca.19130 This issue Cite
Research Paper
Down-regulation of RPS9 Inhibits Osteosarcoma Cell Growth through Inactivation of MAPK Signaling Pathway
Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, Shanghai, 200233, China
* Authors share co-first authorship.
Received 2017-1-10; Accepted 2017-4-17; Published 2017-8-22
Abstract
Objectives: Osteosarcoma is the most common malignant bone tumor in adolescents; however, the mechanisms involved in the pathogenesis and progression of osteosarcoma remain to be elucidated. Researchers have provided valuable insights into the tumorigenesis of Ribosomal protein S9 (RPS9) in some cancers. The purpose of this study was to elucidate the expression, functions, and mechanisms of RPS9 in human osteosarcoma. Methods: The expression of RPS9 in osteosarcoma tissues and cell lines was evaluated by qRT-PCR and western blotting. Knockdown of RPS9 induced by RNA interference (RNAi) method in three osteosarcoma cell lines (MNNG/HOS, MG63, and U2OS) was employed to analyze the effects of RPS9 on cell proliferation and cell cycle distribution. The host signaling pathways affected by RPS9 were detected using the intracellular signaling antibody array kit PathScan®. Results: The expression of RPS9 was found to be up-regulated in human osteosarcoma tissues and cell lines. Its expression was positively correlated with Enneking stage and the tumor recurrence. Down-regulation of RPS9 inhibited osteosarcoma cell proliferation, colony-forming ability, and cell cycle G1 phase in vitro. In addition, our data demonstrated that knockdown of RPS9 repressed the protein levels of phospho-SAPK/JNK and phospho-p38. Conclusion: RPS9 is up-regulated and has a pro-tumor effect in osteosarcoma through the activation of MAPK signaling pathway and thus can be used as a potential target for gene therapy.
Keywords: RPS9, MAPK signaling pathway, proliferation, osteosarcoma.
Introduction
Osteosarcoma, the most common primary bone malignancy, occurs most frequently in children and adolescents [1]. Although the development of combination treatment, including limb salvage surgery and neoadjuvant chemotherapy, has significantly increased the survival rates in patients with osteosarcoma from 20 to 75% [2-4], overall clinical outcomes remain unsatisfactory, especially for patients with metastasis or recurrent osteosarcoma. In recent years, accumulating evidence has validated numerous molecular alterations and tumor-related signaling pathways involved in the pathogenesis and progression of osteosarcoma [5, 6]. However, the highly complex molecular mechanism of osteosarcoma has not been elucidated. Thus, there is an urgent need to search for novel markers in order to develop new therapeutic strategies and predict clinical outcomes in osteosarcoma.
RPS9, a novel NPM1 interacting protein, is one of the first proteins that directly bind to the 18S rRNA [7-9]. RPS9 can be detected in the nucleoli and cytoplasm of mammalian cells [7] and it has been implicated in translation elongation, including mRNA unwinding and decoding accuracy. RPS9 plays an essential role in the early stages of ribosome biogenesis; knockdown of RPS9 results in the accumulation of the 45S and 30S pre-rRNAs, causing decreased global protein synthesis [10, 11]. Several studies have reported the role of RPS9 in tumorigenesis; however, the function and underlying mechanism of RPS9 in osteosarcoma remains unclear.
Mitogen-activated protein kinase (MAPK) is an insulin-mitogen activated protein (Ser/Thr) kinase [12]. Aberrant activation of MAPK can transmit extracellular signals to regulate processes such as cell growth, differentiation, migration, and apoptosis [13]. Loubeau et al. reported that NPM1 silencing reduced prostate cancer cell growth through the inhibition of MAPK signaling [14]. microRNA-7 has been known to constrain the activation of MAPK signaling pathway, thereby suppressing the proliferation and migration of NSCLC cells [15]. Similarly, extensive literatures have reported the significant role of MAPK signaling pathway in osteosarcoma [16, 17]. However, whether MAPK signaling pathway is involved in the RPS9-induced effects of osteosarcoma remains to be elucidated.
In this study, we investigated the expression of RPS9 in human osteosarcoma cell lines (MNNG/HOS, MG63, and U2OS) and in an osteoblast cell line (hFOB 1.19). RPS9 was found to be highly expressed in osteosarcoma cell lines. Similarly, high expression was found in osteosarcoma tissues compared to that in adjacent non-tumor tissues. The functions of RPS9 in osteosarcoma cells were further understood. Our results demonstrated that knockdown of RPS9 hindered osteosarcoma cell growth by inhibiting the G1 cell cycle. The underlying mechanism of RPS9 was also investigated. Reduced RPS9 levels down-regulated MAPK signaling in osteosarcoma cells. Notably, we found that RPS9 expression was positively correlated with the Enneking stage and the recurrence of osteosarcoma. To the best of our knowledge, this is the first report stating that over-expression of RPS9 might be a prerequisite for the development and progression of human osteosarcoma.
Materials and Methods
Cell lines and cell culture
Three osteosarcoma cell lines, namely MNNG/HOS, MG63 and U2OS, and one human osteoblast cell line hFOB 1.19 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). They were cultured in either DMEM (MNNG/HOS and MG63) or RPMI-1640 (U2OS) containing 10% fetal bovine serum (Biowest, South America), 100 U/ml penicillin (Sigma-Aldrich, St Louis, MO, USA), and 100 mg/ml streptomycin (Sigma-Aldrich, St Louis, MO, USA). The cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. The hFOB 1.19 cell line was cultured according to established ATCC protocols.
Human osteosarcoma samples
From 2013 to 2014, a total of 50 patients with osteosarcoma were treated at the Shanghai Jiao Tong University Affiliated Sixth People's Hospital, who received primary surgical treatment and preoperative and postoperative neoadjuvant therapy. Each osteosarcoma sample, together with the corresponding non-tumor tissue sample, was obtained during surgery. The samples were immediately frozen in liquid nitrogen after resection and stored at -80 °C in a refrigerator. Ethics approval was obtained from the local hospital ethics committees, and written informed consent was obtained from each patient prior to sample collection.
RNA extraction and qRT-PCR analysis
Total RNA of human tissue samples and cultured cells was purified using Trizol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using a PrimeScript RT Reagent Kit (Takara, Shiga, Japan). qRT-PCR was performed using SYBR Green Premix Ex Taq (Takara, Shiga, Japan) on an ABI 7500 PCR system (Applied Biosystems). All reactions were performed in 10-μL reaction volumes in triplicate. The following primer sequences were used: RPS9: Forward: 5'-GAAGCGGAGCCAACATGC-3', Reverse: 5'-ATACTCGCCGATCAGCTTCAG-3'; β-actin: Forward: 5'-TTGTTACAGGAAGTCCCTTGCC-3', Reverse: 5'-ATGCTATCACCTCCCCTGTGTG-3'.
Protein extraction and western blotting analysis
Lysates were extracted from cultured cells using T-PER Protein Extraction Reagent (Thermo Fisher Scientific) containing PhosSTOP (Roche, Basel, Switzerland) and Complete Mini (Roche, Basel, Switzerland). Equal amounts of proteins were electrophoresed and transferred onto Nitrocellulose membranes or PVDF membranes (Millipore, Billerica, MA, USA). After blocking in 5% non-fat milk, the membranes were incubated with the following primary antibodies: RPS9 (Proteintech, 1:250), SAPK/JNK (total) (Cell Signaling Technology, 1:1000), SAPK/JNK (Thr183/Tyr185) (Cell Signaling Technology, 1:1000), p38 (total) (BBI Life Sciences, 1:500), p38 (Thr180/Tyr182) (SAB Signalway Antibody, 1:500), or β-actin (Sigma-Aldrich, 1:20,000). The secondary antibody used in the study was anti-rabbit IgG (1:5000, Sigma-Aldrich). Visualization was performed using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific).
siRNA transfection
Human RPS9 siRNA and siRNA nonspecific control were synthesized by Ribobio (Guangzhou, China) and were transfected into cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. The sequence of siRNA targeting RPS9 was: GTCTCGACCAAGAGCTGAA. For proliferation and cell cycle assays, as well as for RNA extraction and western blotting, cells were used 48 h after transfection.
Cell proliferation assay and colony formation assay
Cell proliferation assay
48 h after transfection, cells were seeded into a 96-well plate (3000 cells/well). A 10-μL aliquot of CCK-8 (Dojindo, Kumamoto, Japan) was added to the wells in triplicate, incubated for 2 h, and the absorbance at 450 nm was measured. Each measurement was performed in triplicate, and all experiments were repeated twice.
Colony formation assay
48 h after siRNA transfection, MNNG/HOS, MG63, or U2OS cells (1 × 103 cells/well) were cultured in 6-well plates for 10 days, followed by 100% methanol fixation for 30 min and 0.1% crystal violet staining for 30 min. The cell colonies were counted; all assays were independently performed in triplicate.
Cell cycle analysis
Forty-eight hours after transfection, cells were collected and fixed with 70% ethanol. Subsequently, the cells were stained with 50 μg/mL propidium iodide (PI) (Kaiji, China) containing RNaseI (Kaiji, China), followed by analysis on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). The results were analyzed using the ModFit software (BD Biosciences). Assays were performed three times independently.
Intracellular signaling array
Forty-eight hours after transfection, the cells were harvested and lysed on ice with 0.1 ml of cell lysis buffer containing a cocktail of protease inhibitors for 5 min. The lysates were centrifuged at 10,000 g at 4 °C for 10 min. Intracellular signaling molecules were detected using a PathScan® intracellular signaling array kit (Cell Signaling Technology, #7744) according to the manufacturer's procedure. The fluorescent images of the slide were captured with Odyssey® Infrared Imaging System (LI-COR) and intensities of the spots were quantified using Image Studio analysis software.
Immunohistochemistry (IHC) staining analysis
A standard IHC staining procedure was followed. Briefly, paraffin-embedded sections (4-μm-thick) were dewaxed in xylene, followed by heating in a microwave at 60 °C for 20 min in EDTA buffer (pH 9.0) for antigen retrieval. For each slide, endogenous peroxidase activity was blocked by a 10-min incubation in 0.3% H2O2 followed by incubation at 37 °C with a 1:100 diluted primary antibody RPS9 (Proteintech, China). Slides were rinsed three times in PBS, incubated for 30 min with EnVision staining kit (DAKO, Denmark), followed by three additional washes in PBS, and color development over 3-10 min in a moist chamber at room temperature using 3,3′-diaminobenzidine (DAB). Thereafter, the slides were counterstained in hematoxylin and dehydrated using a graded series of ethyl alcohol (70%, 90%, and 100%). For sections used as negative controls, the primary antibody was substituted by PBS. IHC signal intensities were scored as follows: 0 (no staining), 1 (staining in <1% cells), 2 (staining in 1%-10% cells), or 3 (staining in >10% of cells). The samples scored as 0 and 1 were considered negative, while the samples scored as 2 and 3 were considered positive. Assessment of IHC staining was independently performed by two expert pathologists, and any discordance was resolved through discussion and consensus.
Statistical evaluation
Data were compiled and analyzed using SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). The differences between groups were compared using a two-tailed Student's t-test. Correlations between the IHC of RPS9 and the clinicopathological parameters were determined using the Chi-Square Test. A P < 0.05 was considered significant.
Results
RPS9 is overexpressed in osteosarcoma cell lines and human osteosarcoma tissue samples
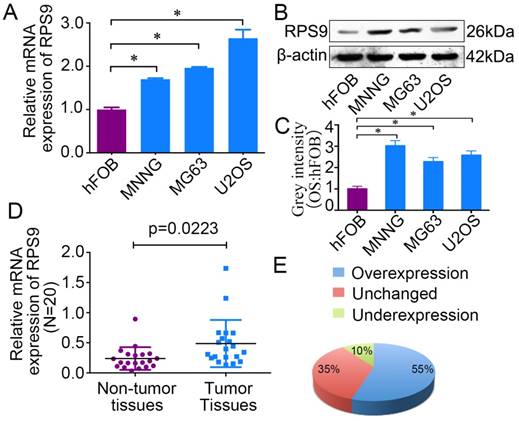
To determine the role of RPS9 in osteosarcoma, we first detected the mRNA and protein expression levels of RPS9 in osteosarcoma cell lines and in the osteoblast cell line by qRT-PCR and western blotting analysis. Compared with the osteoblast cell line, both RPS9 mRNA and protein levels were overexpressed in osteosarcoma cell lines (Figure 1A, 1B and 1C). In order to confirm the role of RPS9, RPS9 expression levels were investigated in 20 paired sets of osteosarcoma and adjacent non-tumor tissues. As shown in Figure 1E and 1F, RPS9 expression was significantly upregulated in 55% (11/20) of osteosarcoma tissues compared to that in adjacent normal tissues. These results indicate that RPS9 may have an important role in osteosarcoma tumorigenesis, which warrants further investigation.
Knockdown of RPS9 inhibits osteosarcoma cell proliferation in vitro
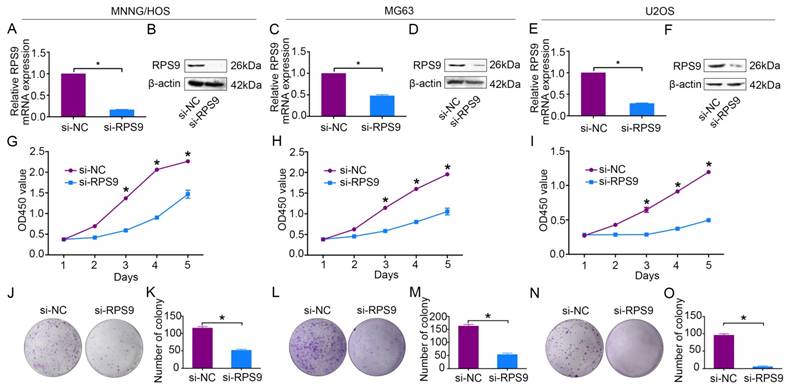
To explore the functional significance of RPS9 in osteosarcoma, RPS9-specific siRNA was used to knockdown RPS9 in osteosarcoma cells. RPS9 knockdown was validated using qRT-PCR and western blotting. RPS9 mRNA and protein expression were significantly reduced after transfection with RPS9-specific siRNA in MNNG/HOS, MG63, and U2OS cells (Figure 2A-2F). Next, cell proliferation was detected using the Cell Counting Kit-8 (CCK-8) assay [18]. A 5-day growth curve analysis showed that knockdown of RPS9 significantly inhibited the growth of osteosarcoma cells (Figure 2G-2I). The colony forming assay was carried out to determine the colony-forming capacity of osteosarcoma cells after knockdown of RPS9. It was found that both the number and the size of colonies were notably decreased in the RPS9 knockdown group compared to that in the control group (Figure 2J-2O). These results demonstrate that RPS9 plays an oncogenic role in osteosarcoma.
Knockdown of RPS9 inhibits osteosarcoma cell G1 cell cycle
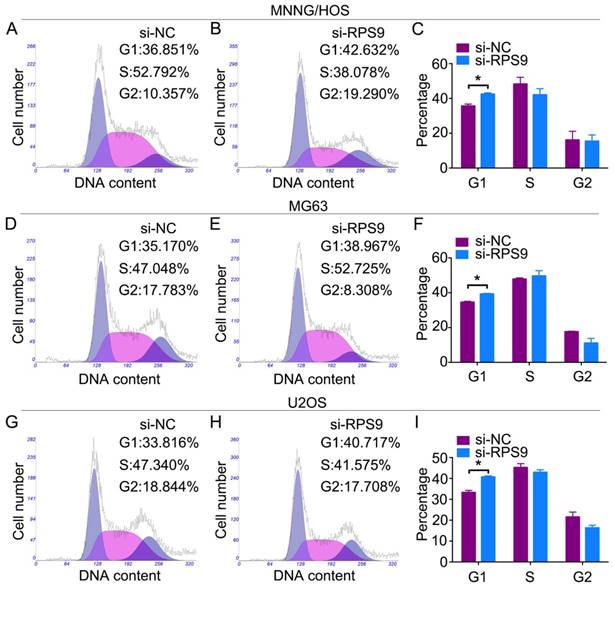
Changes in the cell cycle following RPS9 knockdown were analyzed by flow cytometry. Results of the cell cycle assay showed that RPS9-knockdown cells had a significantly higher percentage of cell population in the G1 phase of the cell cycle compared to the si-NC transfected cells (Figure 3A-3I). However, no significant difference was found in populations in the S or G2/M phases of the cell cycle after RPS9 knockdown. In conclusion, the data showed that RPS9 knockdown attenuated osteosarcoma cell proliferation by inhibiting the G1 cell cycle transition.
Knockdown of RPS9 inhibits osteosarcoma cell proliferation through MAPK signaling pathway
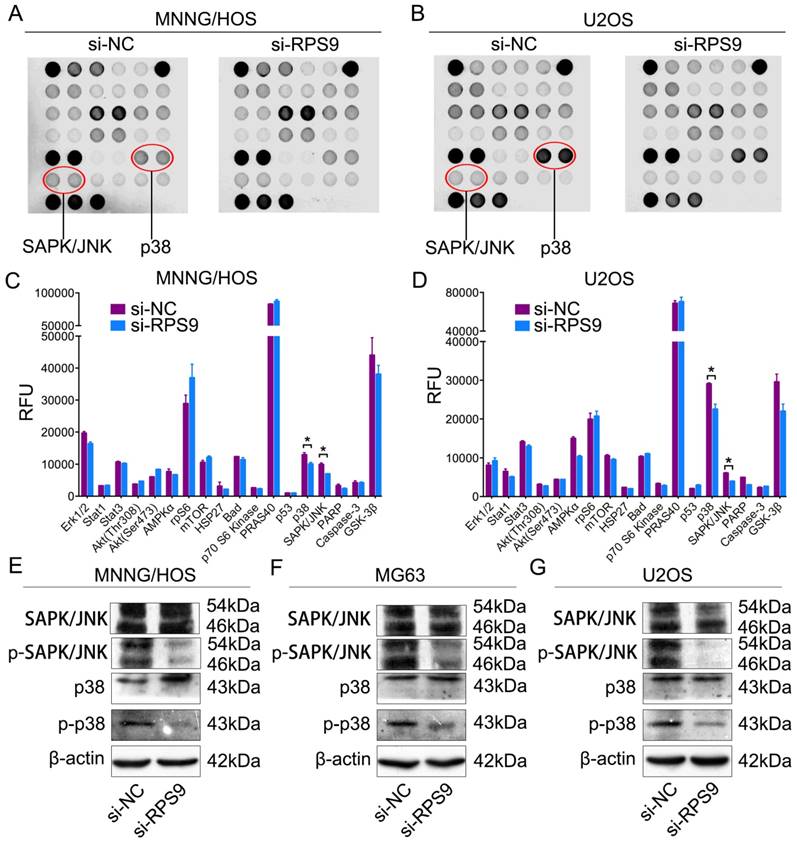
To determine the potential mechanism by which RPS9 affects the proliferation of osteosarcoma cells, intracellular signaling arrays were detected after RPS9 knockdown in MNNG/HOS cells and U2OS cells. A slight decrease in phosphorylation of some components, including p38-Thr180/Tyr182 and SAPK/JNK-Thr183/Tyr185, was detected in osteosarcoma cells (Figure 4A-4D). Subsequently, we examined whether RPS9 could influence the MAPK signaling pathway in osteosarcoma cells using western blotting analysis. The results revealed that RPS9 knockdown inhibited the phosphorylation of SAPK/JNK and p38. Thus, these results support the possibility that RPS9 plays a crucial role in osteosarcoma cell growth by influencing the MAPK signaling pathway.
RPS9 is up-regulated in osteosarcoma tissues and cell lines. (A) (B) The mRNA and protein expression of RPS9 were measured by qRT-PCR and western blotting in osteosarcoma cell lines (MNNG/HOS, MG63, and U2OS) and in a human normal osteoblast cell line (hFOB 1.19). (C) The grey intensity was calculated in osteosarcoma cell lines and osteoblast cell line. β-actin was used as an internal control. (D) (E) The expression of RPS9 was detected in 20 pairs of osteosarcoma tissues and their adjacent non-tumor tissues. The expression of RPS9 was overexpressed in osteosarcoma tissues compared to that in non-tumor tissues. * P < 0.05.
Knockdown of RPS9 inhibits osteosarcoma cell growth in vitro. (A)-(F) Levels of mRNA and protein expression were validated after si-RPS9 transfection in MNNG/HOS cells, MG63 cells, and U2OS cells by qRT-PCR and western blotting, respectively. (G)-(I) Cell Counting Kit-8 (CCK-8) assays were performed after siRNA transfection. (J)-(O) Colony formation assays were performed for RPS9-silenced osteosarcoma cells and control cells. Data are representative of results from three independent experiments. *P < 0.05.
Knockdown of RPS9 inhibits osteosarcoma cell G1 cell cycle transition. Representative images of the cell cycle assays in MNNG/HOS cells (A) (B), MG63 cells (D) (E), and U2OS cells (G) (H) after transfection with si-NC or si-RPS9. (C) (F) (I) Diagrams showing the results of cell cycle assay in MNNG/HOS cells, MG63 cells, and U2OS cells respectively. *P < 0.05.
Knockdown of RPS9 inhibits MAPK signaling pathway in osteosarcoma cells. (A) (B) MNNG/HOS and U2OS cell extracts were prepared and analyzed using the intracellular signaling antibody array kit PathScan® (#7744). Images were captured using Odyssey® Infrared Imaging System (LI-COR). (C) (D) Diagrams show quantification of results in MNNG/HOS and U2OS cells respectively. (E) (F) (G) Western blotting analysis of MAPK signaling pathway, including SAPK/JNK and p38, in MNNG/HOS cells, MG63 cells, and U2OS cells respectively transfected with si-NC or si-RPS9.
RPS9 expression is correlated with osteosarcoma Enneking stage and tumor recurrence
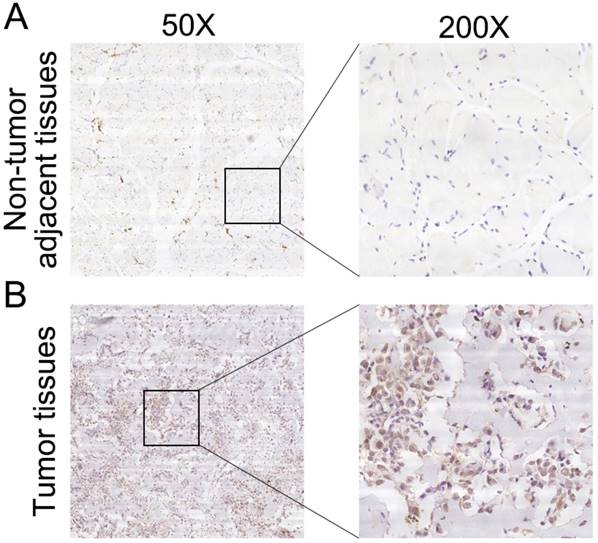
To further determine the clinicopathological significance of RPS9 in osteosarcoma, we performed an IHC analysis of RPS9 in 50 human osteosarcoma tissue samples and the corresponding non-tumor tissues. Representative IHC images of RPS9 expression in osteosarcoma tissues and non-tumor tissues are shown in Figure 5. Correlations between RPS9 expression level and clinicopathological characteristics of patients with osteosarcoma are summarized in Table 1. RPS9 expression levels were found to be higher in osteosarcoma tissue samples than in the corresponding non-tumorous tissues (P = 0.012). The expression level of RPS9 was higher in patients at a clinically advanced Enneking stage than in patients at an early stage (P = 0.011). Further analysis found that the RPS9 level was positively correlated with recurrence (P = 0.024), indicating that RPS9 is involved in the recurrence of osteosarcoma. No correlation was found between RPS9 and any of the other factors, such as gender, age, tumor location, tumor necrosis rate, cortical destruction, and metastasis. Taken together, these results indicate that RPS9 is upregulated in osteosarcoma and potentially plays an important role in the progression of osteosarcoma.
Discussion
Osteosarcoma is the most common type of malignant bone tumor characterized by high levels of genetic instability, with a peak incidence in the second and third decades of life [19]. The survival rate of patients has increased four-fold from the 1960s to the 1980s. However, the 5-year survival rates have remained unchanged during the last three decades, owing to the development of insensitivity to chemotherapy [20]. Uncovering the molecular mechanisms underlying the carcinogenesis and progression of osteosarcoma may help to identify effective therapies for treatment of the disease.
Correlation analyses of RPS9 protein expression in relation to clinicopathologic variables of 50 patients with osteosarcoma
| Clinicopathologic parameters | Number of cases | RPS9 expression level | χ2 | p value | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Gender | 0.002 | 0.963 | |||
| Male | 29 | 15 | 14 | ||
| Female | 21 | 11 | 10 | ||
| Age (years) | 3.120 | 0.077 | |||
| <18 | 21 | 14 | 7 | ||
| ≥18 | 29 | 12 | 17 | ||
| Location | 3.356 | 0.187 | |||
| Femur | 25 | 16 | 9 | ||
| Tibia | 17 | 6 | 11 | ||
| Elsewhere | 8 | 4 | 4 | ||
| Tumor necrosis rate (%) | 0.037 | 0.848 | |||
| <90 | 39 | 20 | 19 | ||
| ≥90 | 11 | 6 | 5 | ||
| Cortical destruction | 1.745 | 0.187 | |||
| Yes | 45 | 22 | 23 | ||
| No | 5 | 1 | 4 | ||
| Recurrence | 5.128 | 0.024* | |||
| Yes | 10 | 2 | 8 | ||
| No | 40 | 24 | 16 | ||
| Metastasis | 0.087 | 0.768 | |||
| Yes | 24 | 13 | 11 | ||
| No | 26 | 13 | 13 | ||
| Ennecking stage | 6.462 | 0.011* | |||
| Ⅱ | 39 | 24 | 15 | ||
| Ⅲ | 11 | 2 | 9 | ||
*P<0.05
Clinical significance of RPS9 in patients with osteosarcoma. (A) Representative IHC image of RPS9 in non-tumor tissues. (B) Representative IHC image of RPS9 in osteosarcoma tissues. Original magnification: 50×, 200×.
The ribosome is composed of two subunits consisting of a complex of ribosomal RNAs (rRNAs) and ribosomal proteins (RPs) [21]. Recently, ribosomal proteins have gained much attention for their role in extraribosomal processes, including DNA repair, apoptosis, proliferation, and metastasis [22, 23]. RPS3, a component of the 40S ribosomal submit, inhibits the invasion of fibrosarcoma cells by blocking the ERK pathway and MMP-9 secretion [24]. Proteomic profiling of an experimental tumor metastasis model determined 45 ribosomal proteins, of which several proteins were differentially expressed in metastatic M4A4 cells, including RPS10 and RPL39 [25]. Chen et al. reported that down-regulation of RPS6 inhibited the growth of non-small cell lung cancer by inducing G0-G1 cell cycle arrest and shortened the overall survival and relapse-free survival in p-rpS6-overexpressing patients [26].
In this study, we demonstrated that RPS9, a member of ribosomal proteins, was upregulated in osteosarcoma cell lines and osteosarcoma tissue samples in comparison with that in corresponding adjacent non-tumor tissues. Knockdown of RPS9 impaired osteosarcoma cell proliferation, as well as the colony-formation ability, and led to a G1 cell cycle arrest. These results indicated that RPS9 played a critical role in osteosarcoma cell growth by acting as an oncogene. Tian et al. reported that RPS9 showed an aberrant promoter hypermethylation and played a critical role in colorectal tumorigenesis [27]. Another study showed that RPS9 silencing impaired 18S rRNA production and induced the activation of p53 in glioma cells, which subsequently resulted in the inhibition of cell proliferation [11]. However, Lemoine et al. reported that RPS9 was down-regulated in pancreatic cancer tissues compared to in normal pancreas, thus suggesting its role as a tumor-suppressor gene in pancreatic cancer [28]. Through the analysis of putative prognostic markers (CDK4, IGFBP2, MMP2, and RPS9) in a set of 43 astrocytic gliomas, 41 glioblastomas, and 7 adjacent normal brain tissues, RPS9 was found to be a significant favorable prognostic marker in gliomas [29]. Similarly, in the present study, we found that the expression of RPS9 was positively correlated with the recurrence and Enneking stage of osteosarcoma, as evidenced by the IHC staining results.
It has been reported that depletion of RPS9 can impair the production of 18S ribosomal RNA and thus reinitiate the differentiation process by inducing p53 activity [11]. Another study reported that RPS9 bound to NPM1 and that the expression of a subset of RPS9 deletion mutants resulted in altered nucleolar morphology, as evidenced by changes in the localization patterns of NPM1 [7]. In the present study, in order to further investigate the underlying mechanisms of RPS9, we used the intracellular signaling antibody array kit PathScan® to identify the host signaling pathways that might be affected by knockdown of RPS9. The data revealed a slight decrease in phosphorylation of some components of MAPK pathway, including SAPK/JNK and p38, in RPS9-knockdown cells. Next, we detected changes in the MAPK pathway after RPS9 knockdown by western blotting. The results showed that knockdown of RPS9 inhibited the phosphorylation of SAPK/JNK and p38, indicating that RPS9 influences osteosarcoma progression partly through the MAPK signaling pathway. The possible underlying mechanism of RPS9 to regulate MAPK signaling pathway is as follows. It was reported that RPS9 was a novel NPM1 interacting protein [7]. Meanwhile, a previous literature reported that NPM1 silencing reduced prostate cancer cell growth through the inhibition of MAPK signaling [14]. Therefore, RPS9 may interact with NPM1 to regulate MAPK signaling pathway in osteosarcoma cells.
MAPK signaling pathway, which has been confirmed to be tightly associated with the proliferation of tumor cells [30, 31], including osteosarcoma cells [32], is an important intracellular signaling pathway. Cheng et al. reported that knockdown of S100A9 inhibited osteosarcoma cell proliferation through the inactivation of MAPK and NF-κB signaling pathways [33]. Zhang et al. reported that KiSS1 functioned as a tumor suppressor in osteosarcoma cells through inhibition of the MAPK pathway [34]. In addition, Li et al. showed that PLA2G16 enhanced metastatic osteosarcoma by activating the MAPK pathway [35].
In conclusion, we demonstrated, for the first time, that RPS9 was up-regulated in human osteosarcoma cell lines and clinical tissue samples. High expression of RPS9 was positively correlated with the Enneking stage and recurrence of osteosarcoma, which might be explained by the fact that RPS9 increased the proliferation of osteosarcoma cells. More importantly, we showed that RPS9 exerted oncogenic function through the MAPK signaling pathway in osteosarcoma cells. These data provide compelling evidence that RPS9 might serve as a potential therapeutic target for osteosarcoma.
Abbreviations
RNAi: RNA interference; RPS9: Ribosomal protein S9; MAPK: Mitogen-activated protein kinase; IHC: immunohistochemistry; DAB: diaminobenzidine; rRNAs: ribosomal RNAs; RP: ribosomal protein; CCK-8: Cell Counting Kit-8.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of Shanghai Jiao Tong University and with the principles of Declaration of Helsinki.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Liu W, Xu G, Liu H. et al. MicroRNA-490-3p regulates cell proliferation and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 2015;589:3148-53
2. Amankwah EK, Conley AP, Reed DR. Epidemiology and therapies for metastatic sarcoma. Clin Epidemiol. 2013;5:147-62
3. Isakoff MS, Bielack SS, Meltzer P. et al. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J Clin Oncol. 2015;33:3029-35
4. Chen Y, Guo Y, Yang H. et al. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget. 2015;6:23708-19
5. Zhang S, Zhao Y, Wang L. MicroRNA-198 inhibited tumorous behaviors of human osteosarcoma through directly targeting ROCK1. Biochem Biophys Res Commun. 2016;472:557-65
6. Sakamoto A, Iwamoto Y. Current status and perspectives regarding the treatment of osteosarcoma: chemotherapy. Rev Recent Clin Trials. 2008;3:228-31
7. Lindström MS. Elucidation of motifs in ribosomal protein S9 that mediate its nucleolar localization and binding to NPM1/nucleophosmin. PLoS One. 2012;7:e52476
8. Lindström MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem. 2008;283:15568-76
9. O'Donohue MF, Choesmel V, Faubladier M. et al. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol. 2010;190:853-66
10. Ferreira-Cerca S, Pöll G, Kühn H. et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell. 2007;28:446-57
11. Lindström MS, Nistér M. Silencing of ribosomal protein S9 elicits a multitude of cellular responses inhibiting the growth of cancer cells subsequent to p53 activation. PLoS One. 2010;5:e9578
12. Avruch J. MAP kinase pathways: the first twenty years. Biochim Biophys Acta. 2007;1773:1150-60
13. Sui X, Kong N, Ye L. et al. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014;344:174-9
14. Loubeau G, Boudra R, Maquaire S. et al. NPM1 silencing reduces tumour growth and MAPK signalling in prostate cancer cells. PLoS One. 2014;9:e96293
15. Cao Q, Mao ZD, Shi YJ. et al. MicroRNA-7 inhibits cell proliferation, migration and invasion in human non-small cell lung cancer cells by targeting FAK through ERK/MAPK signaling pathway. Oncotarget. 2016
16. Chen HJ, Lin CM, Lee CY. et al. Kaempferol suppresses cell metastasis via inhibition of the ERK-p38-JNK and AP-1 signaling pathways in U-2 OS human osteosarcoma cells. Oncol Rep. 2013;30:925-32
17. Tsagaraki I, Tsilibary EC, Tzinia AK. TIMP-1 interaction with αvβ3 integrin confers resistance to human osteosarcoma cell line MG-63 against TNF-α-induced apoptosis. Cell Tissue Res. 2010;342:87-96
18. Chen Z, Wang Z, Guo W. et al. TRIM35 Interacts with pyruvate kinase isoform M2 to suppress the Warburg effect and tumorigenicity in hepatocellular carcinoma. Oncogene. 2015;34:3946-56
19. Cheng DD, Yu T, Hu T. et al. MiR-542-5p is a negative prognostic factor and promotes osteosarcoma tumorigenesis by targeting HUWE1. Oncotarget. 2015;6:42761-72
20. Yan GN, Lv YF, Guo QN. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016;370:268-74
21. Melnikov S, Ben-Shem A, Garreau de Loubresse N. et al. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol. 2012;19:560-7
22. Kim TH, Leslie P, Zhang Y. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget. 2014;5:860-71
23. Du J, Shi Y, Pan Y. et al. Regulation of multidrug resistance by ribosomal protein l6 in gastric cancer cells. Cancer Biol Ther. 2005;4:242-7
24. Kim SH, Kim J. Reduction of invasion in human fibrosarcoma cells by ribosomal protein S3 in conjunction with Nm23-H1 and ERK. Biochim Biophys Acta. 2006;1763:823-32
25. Kreunin P, Yoo C, Urquidi V. et al. Differential expression of ribosomal proteins in a human metastasis model identified by coupling 2-D liquid chromatography and mass spectrometry. Cancer Genomics Proteomics. 2007;4:329-39
26. Chen B, Zhang W, Gao J. et al. Downregulation of ribosomal protein S6 inhibits the growth of non-small cell lung cancer by inducing cell cycle arrest, rather than apoptosis. Cancer Lett. 2014;354:378-89
27. Tian X, Sun D, Zhao S. et al. Screening of potential diagnostic markers and therapeutic targets against colorectal cancer. Onco Targets Ther. 2015;8:1691-9
28. Crnogorac-Jurcevic T, Efthimiou E, Capelli P. et al. Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene. 2001;20:7437-46
29. Zhou YH, Hess KR, Liu L. et al. Modeling prognosis for patients with malignant astrocytic gliomas: quantifying the expression of multiple genetic markers and clinical variables. Neuro Oncol. 2005;7:485-94
30. Qu JL, Qu XJ, Zhao MF. et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41:875-80
31. Kamiyama M, Naguro I, Ichijo H. In vivo gene manipulation reveals the impact of stress-responsive MAPK pathways on tumor progression. Cancer Sci. 2015;106:785-96
32. Yao J, Weng Y, Yan S. et al. NOV inhibits proliferation while promoting apoptosis and migration in osteosarcoma cell lines through p38/MAPK and JNK/MAPK pathways. Oncol Rep. 2015;34:2011-21
33. Cheng S, Zhang X, Huang N. et al. Down-regulation of S100A9 inhibits osteosarcoma cell growth through inactivating MAPK and NF-κB signaling pathways. BMC Cancer. 2016;16:253
34. Zhang Y, Tang YJ, Li ZH. et al. KiSS1 inhibits growth and invasion of osteosarcoma cells through inhibition of the MAPK pathway. Eur J Histochem. 2013;57:e30
35. Li L, Liang S, Wasylishen AR. et al. PLA2G16 promotes osteosarcoma metastasis and drug resistance via the MAPK pathway. Oncotarget. 2016;7:18021-35
Author contact
![]() Corresponding authors: Cun-yi Fan, Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, No. 600, Yishan Road, Shanghai, 200233, China. E-mail: fancunyi888com Tel: +86-21-64369181 Fax: +86-21-64701361 Qing-cheng Yang, Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, No. 600, Yishan Road, Shanghai, 200233, China.E-mail: tjyqccom
Corresponding authors: Cun-yi Fan, Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, No. 600, Yishan Road, Shanghai, 200233, China. E-mail: fancunyi888com Tel: +86-21-64369181 Fax: +86-21-64701361 Qing-cheng Yang, Department of Orthopedics, Shanghai Jiao Tong University Affiliated Sixth People's Hospital, No. 600, Yishan Road, Shanghai, 200233, China.E-mail: tjyqccom

 Global reach, higher impact
Global reach, higher impact