Impact Factor
ISSN: 1837-9664
J Cancer 2017; 8(11):2088-2096. doi:10.7150/jca.19373 This issue Cite
Research Paper
Characterization of the Role of the Malate Dehydrogenases to Lung Tumor Cell Survival
1. Department of Physiology and Pharmacology, Karolinska Institutet, Nanna Svartz väg 2, SE-171 77, Stockholm, Sweden.
2. Experimental Biomolecular Physics, Department of Applied Physics, Royal Institute of Technology (KTH), AlbaNova University Center, SE-106 91 Stockholm, Sweden.
Received 2017-1-27; Accepted 2017-5-2; Published 2017-7-5
Abstract
Cellular compartmentalization of biochemical processes in eukaryotic cells is critical for many functions including shuttling of reducing equivalents across membranes. Although coordination of metabolic flux between different organelles is vital for cell physiology, its impact on tumor cell survival is not well understood. By using an integrative approach, we have dissected the role of the key metabolic enzymes Malate dehydrogenases (MDH1 and MDH2) to the survival of Non-small Cell Lung Carcinomas. Here, we report that while both the MDH1 (cytosolic) and the MDH2 (mitochondrial) enzymes display elevated levels in patients compared to normal counterparts, only high expression of MDH1 is associated with poor prognosis. We further show that the MDH1 enzymatic activity is significantly higher in NSCLC cells than that of MDH2. Accordingly, genetic depletion of MDH1 leads to significantly higher toxicity than depletion of MDH2. These findings provide molecular insights into the metabolic characteristics of the malate isoenzymes and mark MDH1 as a potential therapeutic target in these tumors.
Keywords: Malate dehydrogenase, Cancer Metabolism, Lung Cancer
Introduction
Eukaryotic cells compartmentalize biochemical processes to distinct organelles that depend on metabolic cycles to shuttle reducing equivalents across biological membranes. Most of the compartmentalized reactions are redox (reduction/oxidation) reactions that require nucleotide-based cofactors to support various cellular processes including the production of ATP, biosynthesis and signal transduction [1, 2]. To obtain an efficient ATP production in mitochondria, the cytosolic NADH needs to be transferred into the mitochondrial matrix to access the electron transport chain [3]. One such important system is the malate dehydrogenases (MDH) that catalyzes the interconversion of oxaloacetate and malate utilizing NAD+/NADH coenzyme system. Two main forms of malate dehydrogenases have been identified in several organisms. While the mitochondrial MDH2 participates in the tricarboxylic acid (TCA) cycle, it cooperates with its cytosolic isoenzyme MDH1 to contribute to the malate/aspartate shuttle [4]. This metabolic cycle transfers NADH into the mitochondria, allowing cytosolic reducing equivalents to be delivered to the electron transport chain [5, 6]. The malate dehydrogenases are catalyzing an energetically unfavorable (non-spontaneous) reaction as the ΔG0 is larger than 0 [7-9]. MDH1 and MDH2 are encoded by distinct genes and differ in their specificity for the NAD+/NADH. Both MDH isoenzymes exist in physiological conditions as dimers with identical subunits [8, 10].
Early investigations diverged in their conclusion indicating that the malate dehydrogenases display either elevated activity in breast and lung tumors [11-13], or lower activity in cancerous tissues [14-16], thus the contribution of MDH1 and MDH2 to cancer cell survival has not been elucidated. Moreover, it has been reported that mutations in the MDH gene have been observed in pathological conditions. A germline mutation in the MDH2 was recently reported in paraganglioma to cause a 6-12 fold decrease in its expression level [17]. MDH1 has also been shown to be highly expressed in muscle metastases from pancreatic adenocarcinomas [18]. While mutations in the MDH1 have been reported in lung cancer, it is not known if these mutations affect its enzymatic activity [19].
In this study, we have analyzed multiple cohorts of primary non-small cell lung adenocarcinomas (>300 patients), which revealed an upregulation of MDH1 and MDH2 as validated on both the level of gene and protein expression in tumor biopsies. However, we found that only MDH1 expression and not MDH2, is associated with poor prognosis of NSCLC patients. Our enzymatic, genetic and metabolomics studies uncover that MDH1 displays significantly higher activity compared to MDH2. Accordingly, we show that genetic depletion of MDH1 is selectively toxic to the cells compared to MDH2 depletion. Thus, our study reveals the contribution of MDH1 and MDH2 to cancer cell survival and mark MDH1 as a potential therapeutic target in Non-small Cell Lung Carcinomas.
Materials and Methods
Cell Culture
All tumor cell lines (NCI-H1838, NCI-H1563, NCI-H1437, NCI-H1792, NCI-H838, NCI-H2087, NCI-H522, and A549) included in the study were grown in RPMI-1640 medium (Sigma Aldrich, St Louis, MO, USA) supplemented with 10% (v/v) heat‐inactivated FBS, 100 U/ml penicillin, 100 U/ml streptomycin and 2% (w/v) glutamine. The normal lung cells (Wi-38, MRC and IMR-90) were grown in DMEM supplemented with 10% (v/v) heat‐inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 U/ml streptomycin, 2% (w/v) glutamine and 1% non-essential amino acids. The cells were grown in a humidified 5% CO2 atmosphere at 37°C to allow exponential cell growth.
Immunohistochemical Analysis of Primary NSCLCs
Staining's for MDH1 and MDH2 were performed using the Autostainer 480 (Thermo Fisher Scientific, Waltham, MA, USA). Tissue slides were incubated with Ultra V block (TA-125-UB, Thermo Fisher Scientific, Waltham, MA, USA) for 5 min, and thereafter incubated with Anti-MDH1 and MDH2, respectively (The Human Protein Atlas ID: HPA027296 (MDH1) and HPA019716 (MDH2) diluted 1:1300 for 30 min. The slides were then incubated with labeled horseradish peroxidase-polymer for 30 min, followed by 3,3′-Diaminobenzidine (DAB) solution for 5 min. Slides were counterstained in Mayers hematoxylin for 5 min using the Autostainer XL (Leica), and then rinsed in lithium carbonate water for 1 min. The slides were dehydrated in graded ethanol and coverlipped (PERTEX, Histolab) using an automated glass coverslipper (CV5030, Leica) and scanned using the automated scanning system Aperio XT (Aperio Technologies, Leica, Wetzlar, Germany). Tissue Microarrays were purchased from Biomax and contained 33 normal adjacent cores and 32 tumor cores.
Metabolomics
The isotopomer analysis was performed as previously described [30]. Tumor cell lines (NCI-H1838, NCI-H1563, NCI-H1437 and NCI-H1792) were seed in 60-mm culture dishes and grown until 70-80% confluency at 37ºC in RPMI medium (US Biologicals, Salem, MA, USA) containing 10% dialyzed FBS supplemented with 4mM glutamine. When tracing glucose carbons, 10 mM U-13C-glucose (Cambridge Isotope Laboratories, Andover, MA, USA) was supplemented to the media for 8 hr. After the indicated time point, the medium was rapidly removed and cells were washed three times in cold 1xPBS, and polar metabolites was extracted in 0.8 mL of Chloroform/Methanol/Water (1:3:1) at 4ºC. The plates were place on a rocker for 1 hr at 4ºC. 5 µL of media samples were mixed with Chloroform/Methanol/Water (1:3:1) and vortexed for 5 min at 4ºC. The lysates were cleared of cellular debris by centrifugation for 3 min at 13 000x g at 4ºC. All the cell lines were analyzed in biological replicates where n=3. The samples were subjected to Hydrophilic interaction liquid chromatography (HILIC) carried out on a Dionex UltiMate 3000 RSLC system (Thermo Fisher Scientific, Hemel Hempstead, UK) using a ZIC-pHILIC column (150 mm × 4.6 mm, 5 μm column, Merck Sequant). The column was maintained at 30°C and the samples were eluted with a linear gradient (Solution A: 20 mM ammonium carbonate in water, and solution B acetonitrile, over 26 min at a flow rate of 0.3 ml/min. 80% B (0 min), 20% B (15 min), 5% B (15 min), 5% B (17 min), 80% B (17 min) and 80 % B (24 min). A Thermo Orbitrap Exactive (Thermo Fisher Scientific, Waltham, MA, USA) was operated in polarity switching mode (m/z range 70-1400,) with spray voltage at 4.5 kV and -3.5 kV the capillary temperature at 275ºC. The sheath gas flow was set to 40 units, the auxiliary gas flow to 5 units and the sweep flow was set 1 unit. The MS data acquisition was performed in a range of 70-1400 m/z with the resolution set at 50 000. Mass calibration was performed for each polarity immediately prior to each analysis. The calibration mass range was extended to cover small metabolites by inclusion of low-mass contaminants with the standard Thermo Calmix masses (< m/z 1400), C2H6NO2 for positive ion electrospray ionization (PIESI) mode (m/z 76.0393) and C3H503 for negative ion electrospray ionization (NIESI) mode (m/z 89.0244). Enhancement of calibration stability, lock-mass correction was additionally applied to each analytical run. (Thermo Scientific. raw LC-MS files were converted to mzXML format using proteowizard. The resultant mzXML files were processed using XCMS to pick peaks with mzMatch to filter, match, and annotate the peaks [31,32,33].
RNAi
100,000 cells per well were transfected in 1.8 ml of growth medium in 6-well plates. Interferin (Polyplus transfection, New York, NY, USA) was mixed with 20 nM siRNA targeting MDH1 (ON-TARGETplus Human MDH1 siRNA, J-009264-09 and J00964-12) or MDH2 (ON-TARGETplus Human MDH2 siRNA, J-008439-11 and J-008439-12), in a final volume of 200 µl and added to the cells. Non-targeting control (ON-TARGETplus non-targeting siRNA D-001810-10) from Dharmacon (Lafayette, CO, USA) was used. The knockdown was analyzed by immunoblotting 24 hr post transfection.
Western Blot
Immunoblotting was performed as described before [34]. Protein lysates prepared using Complete Lysis-M buffer (Roche Diagnostics, Risch-Rotkreutz, Switzerland) supplemented with protease inhibitors (Roche Diagnostics, Risch-Rotkreutz, Switzerland) were mixed with Laemmeli's loading buffer, boiled and separated on 15% SDS-PAGE at 40 mA followed by transfer to nitrocellulose membranes for 90 min at 120 V. Membranes were blocked for 1h with 5% non‐fat milk in TBS at room temperature and subsequently probed with the following antibodies: anti-MDH1 (ab76616, Abcam), anti-MDH2 (clone # 11908, Cell Signaling, Danvers, MA, USA) and anti-Actin (Sigma Aldrich, St Louis, MO, USA). The primary antibodies were diluted in TBS containing 1% BSA, and 0.1% NaN3. Horseradish peroxidase‐conjugated secondary antibodies (Pierce) were diluted in 2.5% blocking buffer. Clarity™ Western ECL (BioRad, Hercules, CA, USA) was used for revealing the blots.
Malate Dehydrogenase Activity Assay
For the mitochondrial MDH2 activity assay, cells were permeabilized using 0.005% digitonin and supernatant containing the cytosolic fraction and MDH1, and the pellet with crude mitochondria were lysed using a douncer on ice in 100μl using ice cold MDH assay buffer. The following reaction mix was used per reaction: 36 μl MDH Assay Buffer 2 μl MDH Enzyme Mix, 10 μl MDH Developer, 2 μl MDH Substrate. 2 μl MDH was used as positive control and the background control mix contained the whole reaction mix without substrate. Absorbance at 450 nm was measured using a Biomax machine (Promega), from 10 min up to 30 min. The activity was normalized to protein concentration.
NAD+/NADH assay
The experiment was performed according to manufacturer's description (NAD+/NADH Assay Kit, Abcam, ab176723). 7 x106 cells were scraped in ice cold PBS and centrifuged for 5 min at room temperature. 5x106 cells were subsequently resuspended in 100 μl lysis buffer and incubated for 15 min at room temperature. 25 μl of NADH extraction buffer or NAD+ extraction buffer was added to each sample and then incubated for 15 min. 75 μl NADH reaction mixture was added into each well and the reaction was incubated for 30 min while protected from light. Fluorescence was measured for up to 120 min on a Biomax microplate reader (Promega) at Ex/Em 540/590.
TRAST spectroscopy
TRAST measurements were performed on a home-built, epi-illuminated, microscope, the basic configuration and operation principles of which has been described elsewhere [20]. Cytosolic fractions were isolated from cells are described previously [21], in brief cells were permeabilized using 0.005% digitonin in PBS, incubated on ice for 5min and centrifuged. Supernatants containing the cytosolic fraction were used for the TRAST experiments. In short, a continuous-wave, solid state laser (Cobolt Zouk, 20 mW, 355 nm) was used for excitation, modulated in time by an acousto-optic modulator (MQ110-A1-UV, AA Opto-Electronics, Orsay). The laser beam was focused into the sample by a water immersion objective (40x, NA 1.2, Zeiss, C-Apochromat), resulting in a beam radius of 1.3 μm (1/e2), and an average excitation irradiance of 140 kW / cm2 in the focal plane. Fluorescence from the sample was collected through the same objective and separated from the excitation light by a dichroic beamsplitter (Di02-R405, Semrock) and a band-pass emission filter (BrightLine FF01-403/95, Semrock) in front of the EM-CCD camera (Andor, Luca). Control of the excitation pulses as well as synchronisation with the detection was handled by a digital I/O card (PCI-6602, National Instruments).
Data analysis: As previously described [20], the redox kinetics of NADH were determined from so-called TRAST curves, showing how the detected time-averaged NADH fluorescence varied with the excitation pulse-train characteristics (pulse duration)]. In the cellular lysates, 355 nm irradiation resulted in fluorescence from both NADH and flavin compounds in the samples. Most of the flavin fluorescence could be spectrally removed by the emission filter (down to a few percent of the NADH fluorescence signal), but an additional linear un-mixing step was applied to further isolate the NADH signal. In this step, the large difference in triplet state kinetics (gray part in the model, Figure 3C) between NADH and FAD was exploited, which made it possible to separate the two signals based on their μs-timescale behavior. The effects of photo-oxidation are observed on a longer timescale (approx. 100 μs to 1 ms) and are not affected by this analysis. The resulting TRAST curve from NADH was then analyzed as described in [20].
Results
Elevated expression levels of MDH1 and MDH2 in primary NSCLCs
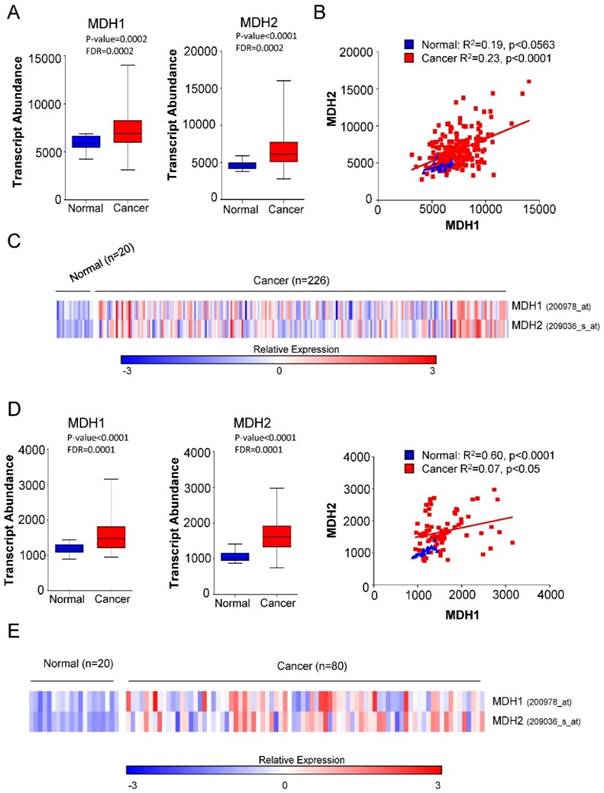
We first investigated the expression of MDH1 and MDH2 in patients with Non-small Cell Lung Carcinoma (NSCLC) and normal healthy cases. Microarray gene expression data in two independent cohorts (GSE31210 and GSE33532) comprising 40 healthy controls and 306 tumor samples from Gene Expression Omnibus (GEO) database were analyzed. Our analysis revealed that both MDH1 and MDH2 were significantly elevated in NSCLC cancer patients (Figure 1A-1E).
To further investigate whether the higher mRNA expression levels of malate dehydrogenases in NSCLC also translates into an elevated enzymatic activity, the total malate dehydrogenase activity, including MDH1 and MDH2, was measured in a panel including 3 normal lung (IMR-90, MRC and Wi-38) and 8 NSCLC (NCI-H1838, NCI-H1563, NCI-H1437, NCI-H1792, NCI-H838, NCI-H2087, NCI-H522, and A549) cell lines. The results revealed that the activity in tumor cells was significantly higher compared to normal lung cell lines (Figure 2A), strongly suggesting that beyond elevated expression levels, the malate dehydrogenases might also have enhanced enzyme activities during tumorigenesis of NSCLCs. These data provided a rationale to further explore the role of malate dehydrogenases to the survival of Non-small Cell Lung Carcinomas. We analyzed if the expression of MDH1 and MDH2 had any clinical relevance and was associated with poor prognosis in NSCLCs. Although the expression levels of MDH1 and MDH2 were both elevated, our Kaplan-Meier analyses revealed that only MDH1 expression was associated with poor prognosis in NSCLCs (Figure 2B and Supplemental Figure S1A). These data suggest that while malate dehydrogenases are generally elevated in NSCLCs [17, 18], they show diversity in the prognostic value in patients.
Genetic Depletion of MDH1 displays toxicity in Non-small Cell Lung Carcinomas
To study the protein expression of MDH1 and MDH2 in primary NSCLC cases versus matched normal cases, a tissue microarray analysis was performed. Both MDH1 and MDH2 were significantly elevated on the protein level in the tumor tissues compared to corresponding normal cases (Figure 2C). Western blot analysis of expression of MDH1 and MDH2 in the cell line panel further confirmed similar observations as in primary material (Figure 2D). Combined, these observations show that the differential expression of MDHs in tumor versus normal translates to the cell lines.
To probe the functional relevance of the observed elevated expression of the malate dehydrogenases to NSCLC tumor cell survival, we undertook a genetic approach to deplete MDH1 or MDH2 in the lung cancer cell line panel by two independent siRNAs targeting the dehydrogenases, respectively. Knockdown of MDH1 significantly induced cell death and showed selective toxicity to the NSCLC compared to the normal cell lines (Figure 2E). Contrastingly, while, MDH2 depletion also caused selective toxicity to tumor cells compared to normal lung cell lines, its depletion had only a moderate effect (Figure 2E), and was not as pronounced as compared to MDH1 suppression in NSCLC.
Differential Enzymatic Activity of MDH1 and MDH2 in tumor cells
To evaluate the activity of the malate dehydrogenases in intact cells, a targeted metabolomics analysis was performed based on uniformly 13C-labeled glucose in 4 cell lines (NCI-H1838, NCI-H1563, NCI-H1792 and NCI-H1437) (Figure 3A). MDHs interconvert malate and oxaloacetate, which is metabolized to aspartate by glutamic oxaloacetic transaminase (Figure 3A). However, as oxaloacetate is rapidly degraded upon extraction of polar metabolites, we analyzed the fraction of 13C transferred from malate to aspartate (a surrogate metabolite for oxaloacetate). The isotopomer analysis showed that glucose is metabolized into the reductive branch of the TCA cycle and 13C incorporation was detected in Succinate, Fumarate, Malate and Aspartate (Figure 3A). Our findings are in line with recent findings showing that aspartate is required for cell proliferation [22, 23]. However, although rapidly proliferating cells have distinct metabolic requirements compared to slowly proliferating cells, the molecular network of metabolic enzymes are essentially the same. This means that proliferating cells must re-wire their metabolism in a way that supports the biosynthetic needs to generate a new cell.
Elevated expression of MDH1 and MDH2 in primary NSCLCs. (A) Transcript abundance (probe intensity) of the indicated genes in primary NSCLCs from the GSE31210 dataset. (B) Linear regression analysis was performed using GraphPad Prism to determine the relationship between the gene expression levels of MDH1 and MDH2 in the GSE31210 dataset. (C) Heat map representation of the relative mRNA levels of the MDH1 and MDH2 from the GSE33532 cohort. (D) Transcript abundance (probe intensity) of the indicated genes in primary NSCLCs from the GSE33532 dataset. (E) Heat map representation of the relative mRNA levels of the MDH1 and MDH2 genes from GSE33532 dataset. Differential expression was determined using Mann-Whitney U test combined with the false discovery rate (FDR) correction. Microarray gene expression data of two cohorts (GSE31210 and GSE33532) from Gene Expression Omnibus (GEO) database were analyzed. Differential expression was determined using Mann-Whitney U test combined with the false discovery rate (FDR) correction.
The malate dehydrogenases are utilizing the NAD/NADH as co-factors for their enzymatic activities, thus the level or the ratio of NAD+ or its reduced form might be at distinct levels in tumor cells compared to normal cells. Hence, the level of NAD/NADH ratio was measured in the cell line panel. The results show that no statistically significant variation was observed among the tumor cell lines and also not compared to normal cells (Figure 3B). To further provide an independent correlate to these findings, we interrogated the redox environment regulating the balance between NADH and NAD+, based on the intrinsic fluorescence of NADH, using (Transient State) TRAST spectroscopy [24-26]. In the TRAST measurements, the relative populations and switching kinetics between NADH and NAD+ was monitored via its fluorescence, generated upon laser pulse-train excitation, in cellular lysates from the different cell lines (see methods and materials below). In the lysates, no differences in the relative fractions of NADH and NAD+ could be observed, also not in the exchange rates between the two redox forms (Figure 3C and 3D). Therefore, no major differences in their redox environments influencing the NADH/NAD+ balance and switching kinetics are to be expected. In contrast, a clear difference between the lysate samples and a sample with NADH in aqueous solution could be observed, with the effective photo-oxidation rate of NADH (koff) a factor of two lower in the lysate samples. With reference to the kinetic model of Figure 3C, the observed difference can be attributed to a higher, redox-buffered, reduction rate (kred) of photo-oxidized NADH (•NADH+) back to NADH in the lysates than for NADH in aqueous solution.
MDH1 has functional and prognostic value in NSCLCs. (A) Total cellular malate dehydrogenase activity measured in 11 cell lines. The blue bar is cumulative data from the 3 normal cells and the red bar is cumulative from 8 tumor cell lines. Activity was normalized per mg protein. (B) Kaplan-Meier Analysis of MDH1 expression and overall survival in 226 NSCLC patients. (C) Tissue-Microarray analysis (TMA) of the MDH1 and MDH2 protein expression level in 33 NSCLCs and 33 normal adjacent lung tissues. Representative images are shown. (D) Expression level of the malate dehydrogenases in a cell line panel consisting of NSCLCs and normal lung cells. (E) Effect of genetic depletion on cell viability in the cell line panel, by two independent siRNAs, targeting MDH1 and MDH2, respectively. Cell death is presented as fold over control siRNA. Error bars, ±SEM. *p<0.01, **p<0.001, ***p<0.0001; two-tailed student's t-test. ns= non-significant.
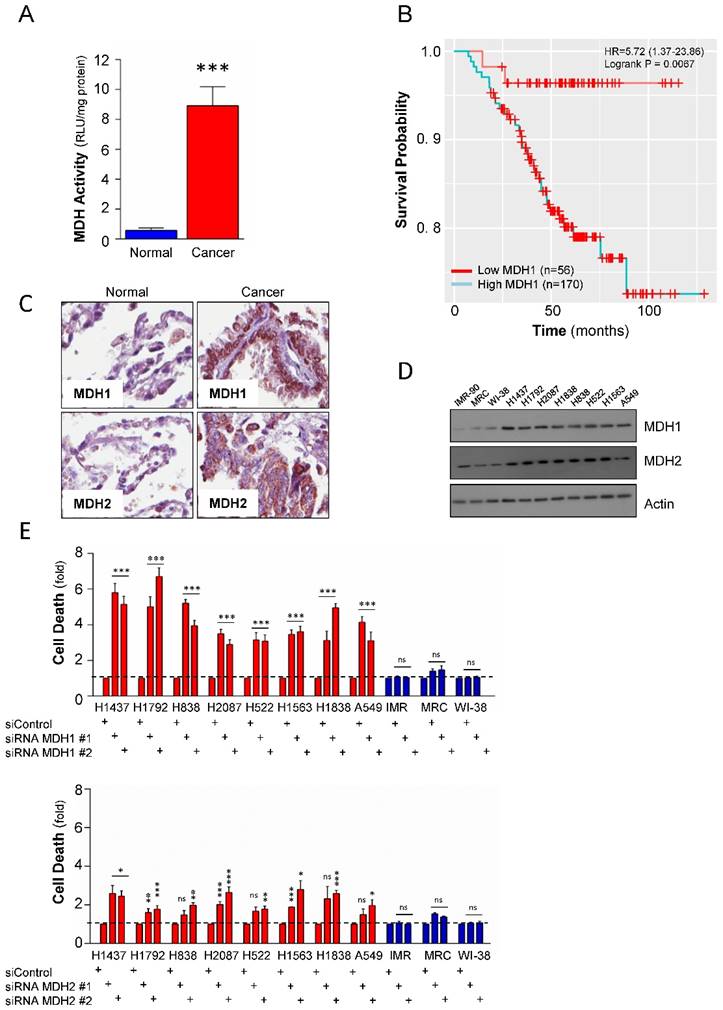
Differential Enzymatic Activity in cells is not driven by distinct co-factor levels. (A) 13C isotopomer analysis of uniformly labeled glucose. Schematic overview of the metabolic pathway. 13C-enrichment of glucose-derived 13C- Succinate 13C-Fumarate, 13C-Malate, and 13C-Aspartate. (B) Analysis of the NAD+ and NADH ratio in the cell line panel. (C) Model for the redox kinetics of NADH [28, 29] used to analyze the TRAST data. I: Following excitation of ground state NADH to an excited singlet state (NADH*) with rate k01, fluorescence can be generated upon return to the ground state (k10), intersystem crossing (kisc) can take place to a more long-lived triplet state (TNADH), or an electron can be ejected (kox) forming a radical cation •NADH+. •NADH+ can either recombine with an electron and return to the NADH ground state (kred) or undergo deprotonation followed by a second oxidation (kox2) to form NAD+. TNADH relaxes back to the ground state with the rate kt. The oxidized form, NAD+, is not excitable at 355 nm and. recovery of NAD+ to NADH in the detection volume (kon) is therefore likely to take place mainly by diffusion rather than by a photo-physical transition. II: As a further simplification, the model can be reduced to interconversion between the fluorescent NADH and the non-fluorescent NAD+ with the effective transition rates koff and kon. (D) TRAST curves showing average fluorescence intensity as a function of excitation pulse width. Solution measurements using cytosols from tumor cell lines (red circle), normal lung cells (blue square) and a reference of NADH in PBS (green triangle). Inset: The effective photo-oxidation rate, koff, in each sample, extracted from the TRAST curves, as previously described [20, 25], using the model in Figure 3C.
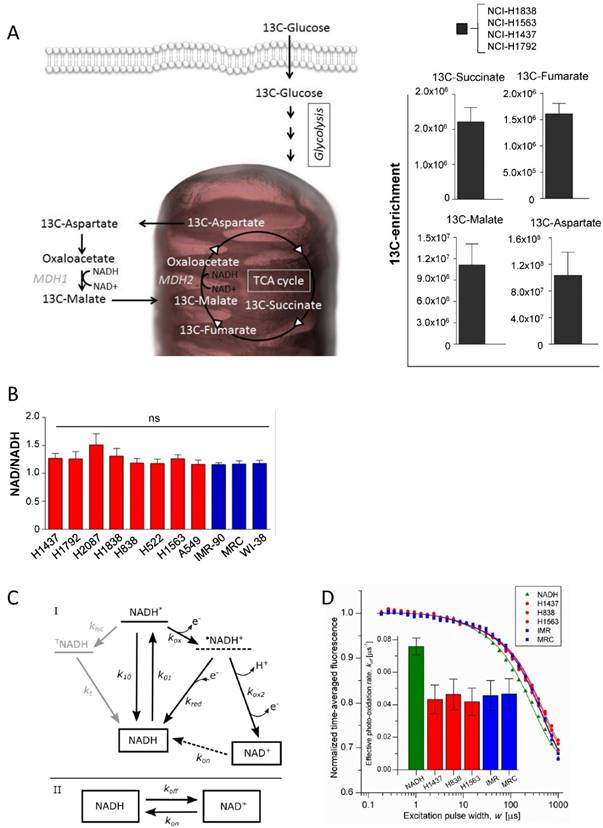
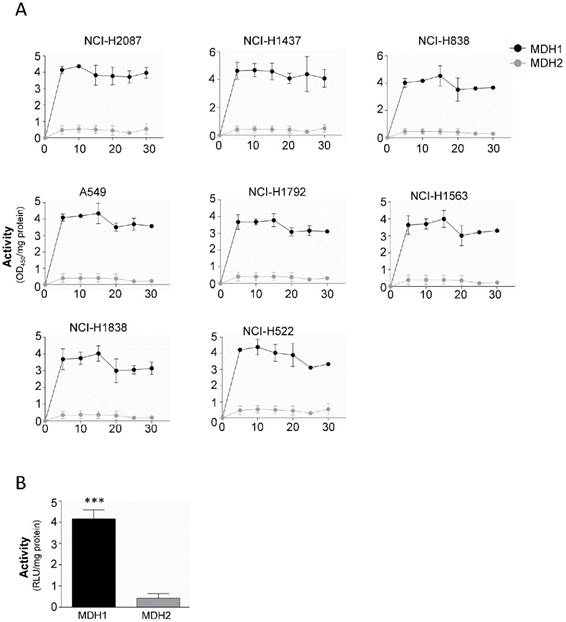
Our results indicate that both MDH1 and MDH2 are elevated in malignant tissue compared to their normal counterparts, although MDH1 is the enzyme that shows functional and prognostic value. To probe the mechanism behind this observation, we isolated mitochondria from the cytosol, to separate the enzymes, where MDH1 ends up in the cytosolic fraction and MDH2 in the mitochondrial fraction. Remarkably, analysis of the respective enzyme's kinetic properties revealed that the MDH1 exerts approximately 4-fold higher enzymatic activity compared to MDH2 (Figure 4A and 4B).
Altogether, these results show that although both enzymes are elevated in malignant tissue both on the RNA and protein level, the cytosolic MDH1 isoform accounts for a significantly higher proportion of the total cellular activity and have therefore prognostic relevance for NSCLCs.
MDH1 has significantly higher activity than MDH2 in tumor cells. (A) Enzyme kinetic analysis of MDH1 and MDH2 activities measured in 8 tumor cell lines for 30 min. (B) Cumulative from the tumor cell line panel (black bar) and cumulative data from the 3 normal cells (gray bar). Activity was normalized per mg protein. Error bars, ±SEM. ***p<0.0001; two-tailed student's t-test. n=4-6.
Discussion
The malate dehydrogenase isozymes are critical enzymes for central metabolic processes. However, their respective role to tumor cell survival has not been elucidated. Early investigations indicated that the total cellular malate dehydrogenase activities can be either elevated or decreased in cancerous tissues [11-16]. In this study we examined the relative contribution of the MDH1 and MDH2 to tumor cell survival. Cancer cells have distinct metabolic requirements compared to normal healthy cells [27].
Accordingly, our finding that MDH1 and MDH2 expression levels are elevated in primary tumors compared to matched normal controls indicated that during the transformation process, cancer cells have engaged a dependence on these enzymes. Surprisingly, when we interrogated the clinical relevance of the MDHs, MDH1 appears to be the dominant form as higher expression was associated with poor prognosis. This suggests that the cytosolic MDH1 might have additional tumorigenic functions that MDH2 is lacking. One potential explanation to this observation might be the availability of their co-factors, NAD+ for MDH2 and NADH for MDH1. However, analysis of the levels of the cofactors argues strongly against this hypothesis as both normal lung cells as well as tumor cells display similar levels and ratio of NAD+/NADH.
Although MDH1 and MDH2 metabolic functions are thought to be connected in the malate-aspartate shuttle, our data suggests that MDH1 might have additional functions supporting tumor cell survival. In line with this, genetic depletion of the MDHs revealed that MDH1 depletion caused significantly higher toxicity to tumor cells compared to MDH2 depletion.
In addition, our metabolomics analysis revealed that the reductive branch of the TCA cycle appears more critical for the lung cancer cells as nutrient-derived carbons from 13C-glucose was detected in Fumarate, Succinate, Malate and Aspartate. One potential mechanism for the differential importance of MDH1 and MDH2 to tumor cell survival is that aspartate generated in the mitochondria by MDH2, once exported into the cytosol can contribute to protein biosynthesis. Aspartate is also required for the de novo purine and pyrimidine biosynthesis. Alternatively, as our data shows, MDH1 displays higher enzymatic activity than MDH2, may cause a decrease of oxaloacetate (the MDH1 substrate) in the cytosol favoring malate production. Conversely, MDH1-null cells have been shown to display higher Aspartate levels [22]. Since MDH2 activity is low, it may not be sufficient to produce oxaloacetate levels that hence need to be replenished by other mechanisms potentially in the cytosol. Combined, our findings show a differential contribution of the MDHs to tumor cell survival and highlights MDH1 as a potential drug target to eliminate tumor cells.
Supplementary Material
Supplementary figure S1.
Acknowledgements
This work was supported by grants from the Swedish Research Council (VR-MH), The Swedish Association for Medical Research (SSMF), The Malin and Lennart Philipson Foundation, The Ragnar Söderberg Foundation, Swedish Research Council (VR VR-NT 2012-3045)), Knut and Alice Wallenbergs Foundation (KAW 2011.0218) Jeanssons Foundation, Alex och Eva Wallströms Foundation, Åke Wibergs Foundation, O. E. och Edla Johanssons Vetenskapliga Foundation, Magnus Bergwalls Foundation, Foundation Lars Hiertas Minne, Foundation Längmänska Kulturfonden and Karolinska Institute Research grants. We thank Yiwei Ai and Aine Henley for technical assistance. We thank Per- Henrik Edqvist at the Tissue Profiling Facility at Science for Life Laboratory for excellent technical assistance with immunohistochemistry and slide scanning.
Competing Interests
The authors declare no conflicts of interest.
References
1. Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides-small molecules with a multitude of functions. The Biochemical journal. 2007;402:205-18
2. Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochemical and biophysical research communications. 2017;482:426-31
3. Purvis JL, Lowenstein JM. The relation between intra- and extramitochondrial pyridine nucleotides. The Journal of biological chemistry. 1961;236:2794-803
4. Minarik P, Tomaskova N, Kollarova M, Antalik M. Malate dehydrogenases-structure and function. General physiology and biophysics. 2002;21:257-65
5. Dasika SK, Vinnakota KC, Beard DA. Characterization of the kinetics of cardiac cytosolic malate dehydrogenase and comparative analysis of cytosolic and mitochondrial isoforms. Biophysical journal. 2015;108:420-30
6. Lanoue KF, Williamson JR. Interrelationships between Malate-Aspartate Shuttle and Citric Acid Cycle in Rat Heart Mitochondria. Metabolism. 1971;20:119 -+
7. Murphey WH, Kitto GB, Everse J, Kaplan N. Malate dehydrogenases. I. A survey of molecular size measured by gel filtration. Biochemistry. 1967;6:603-10
8. Roderick SL, Banaszak LJ. The conformation of mitochondrial malate dehydrogenase derived from an electron density map at 5.3-A resolution. The Journal of biological chemistry. 1983;258:11636-42
9. Kun E, Eanes RZ, Volfin P. Selective modification of mitochondrial malate dehydrogenase activity by changes in ionic strength. Nature. 1967;214:1328-30
10. Holbrook JJ, Wolfe RG. Malate dehydrogenase. X. Fluorescence microtitration studies of D-malate, hydroxymalonate, nicotinamide dinucleotide, and dihydronicotinamide-adenine dinucleotide binding by mitochondrial and supernatant porcine heart enzymes. Biochemistry. 1972;11:2499-502
11. Balinsky D, Greengard O, Cayanis E, Head JF. Enzyme activities and isozyme patterns in human lung tumors. Cancer research. 1984;44:1058-62
12. Balinsky D, Platz CE, Lewis JW. Isozyme patterns of normal, benign, and malignant human breast tissues. Cancer research. 1983;43:5895-901
13. Balinsky D, Platz CE, Lewis JW. Enzyme activities in normal, dysplastic, and cancerous human breast tissues. Journal of the National Cancer Institute. 1984;72:217-24
14. Kawakatsu K, Mori M. Histochemical evaluation of enzymatic activities in human squamous-cell cancer. Cancer research. 1963;23:539-45
15. Wenner CE. Progress in tumor enzymology. Advances in enzymology and related areas of molecular biology. 1967;29:321-90
16. Wenner CE, Spirtes MA, Weinhouse S. Metabolism of neoplastic tissue. II. A survey of enzymes of the citric acid cycle in transplanted tumors. Cancer research. 1952;12:44-9
17. Cascon A, Comino-Mendez I, Curras-Freixes M, de Cubas AA, Contreras L, Richter S. et al. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene. Journal of the National Cancer Institute. 2015:107
18. Chaika NV, Yu F, Purohit V, Mehla K, Lazenby AJ, DiMaio D. et al. Differential expression of metabolic genes in tumor and stromal components of primary and metastatic loci in pancreatic adenocarcinoma. PloS one. 2012;7:e32996
19. Li G, Yi S, Yang F, Zhou Y, Ji Q, Cai J. et al. Identification of mutant genes with high-frequency, high-risk, and high-expression in lung adenocarcinoma. Thoracic cancer. 2014;5:211-8
20. Chmyrov V, Spielmann T, Hevekerl H, Widengren J. Trans-cis isomerization of lipophilic dyes probing membrane microviscosity in biological membranes and in live cells. Analytical chemistry. 2015;87:5690-7
21. Norberg E, Gogvadze V, Ott M, Horn M, Uhlen P, Orrenius S. et al. An increase in intracellular Ca2+ is required for the activation of mitochondrial calpain to release AIF during cell death. Cell death and differentiation. 2008;15:1857-64
22. Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540-51
23. Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552-63
24. Sanden T, Persson G, Thyberg P, Blom H, Widengren J. Monitoring kinetics of highly environment sensitive states of fluorescent molecules by modulated excitation and time-averaged fluorescence intensity recording. Analytical chemistry. 2007;79:3330-41
25. Sanden T, Persson G, Widengren J. Transient state imaging for microenvironmental monitoring by laser scanning microscopy. Analytical chemistry. 2008;80:9589-96
26. Widengren J. Fluorescence-based transient state monitoring for biomolecular spectroscopy and imaging. Journal of the Royal Society, Interface / the Royal Society. 2010;7:1135-44
27. Stanley IA, Ribeiro SM, Gimenez-Cassina A, Norberg E, Danial NN. Changing appetites: the adaptive advantages of fuel choice. Trends in cell biology. 2014;24:118-27
28. Boldridge DW, Morton TH, Scott GW. Formation Kinetics and Quantum Yield of Photon-Induced Electron Ejection from Nadh in Aqueous-Solution. Chem Phys Lett. 1984;108:461-5
29. Czochralska B, Lindqvist L. Biphotonic One-Electron Oxidation of Nadh on Laser Excitation at 353 Nm. Chem Phys Lett. 1983;101:297-9
30. Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H, Norberg E. PHGDH Defines a Metabolic subtype in lung adenocarcinomas with poor prognosis. Cell Rep. 2017Jun13;19(11):2289-2303
31. Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC bioinformatics. 2008;9:504
32. Scheltema RA, Jankevics A, Jansen RC, Swertz MA, Breitling R. PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Analytical chemistry. 2011;83:2786-93
33. Chokkathukalam A, Jankevics A, Creek DJ, Achcar F, Barrett MP, Breitling R. mzMatch-ISO: an R tool for the annotation and relative quantification of isotope-labelled mass spectrometry data. Bioinformatics. 2013;29:281-3
34. Norberg E, Karlsson M, Korenovska O, Szydlowski S, Silberberg G, Uhlen P. et al. Critical role for hyperpolarization-activated cyclic nucleotide-gated channel 2 in the AIF-mediated apoptosis. The EMBO journal. 2010;29:3869-78
Author contact
![]() Corresponding author: Erik Norberg, E-mail: Erik.Norbergse; Phone: +46-736392887.
Corresponding author: Erik Norberg, E-mail: Erik.Norbergse; Phone: +46-736392887.

 Global reach, higher impact
Global reach, higher impact