Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(13):1842-1855. doi:10.7150/jca.15876 This issue Cite
Review
Long Non-coding RNAs in Urologic Malignancies: Functional Roles and Clinical Translation
1. Center for Systems Biology, Soochow University, Suzhou 215006, China
2. School of Chemistry, Biological Engineering, Suzhou University of Science and Technology, Suzhou 215011, China
3. Suzhou Dushuhu Hospital, Clinic Center, Soochow University, Suzhou 215123, China
4. Department of Urology, The Second Affiliated Hospital of Soochow University, Suzhou, 215004, China
5. Ningbo Urologic and Nephrotic Hospital, Ningbo 315000, China.
Received 2016-4-18; Accepted 2016-6-29; Published 2016-8-15
Abstract
Early diagnosis and surveillance for metastasis and recurrences are critical issues of urologic cancer. Deregulation of long non-coding RNAs (lncRNAs) has been implicated in urologic malignancies and represents potential markers or therapeutic targets. However, the utility of lncRNA as biomarkers appears to be overstated due to heterogeneous or irreproducible results from different studies. Thus, a critical and cautious review on the biomarker potential of lncRNAs is needed. This review provides an update on new findings of lncRNA-based markers for urologic cancer. The diverse mechanisms and associated examples of lncRNAs involved during the carcinogenesis of prostate cancer, bladder cancer and renal cancer were discussed in a more balanced and critical manner, as were the suitability of lncRNAs as diagnostic or prognostics markers.
Keywords: lncRNA, long non-coding RNA, urologic cancer, translation, marker.
Introduction
The global efforts in transcriptome mapping have suggested the pervasive transcriptional potential of human genome and identified numerous non-coding transcripts. Based on the length, the non-coding RNAs can be divided into small non-coding RNAs (<200 bp) and long non-coding RNAs (lncRNAs) (> 200bp) [1, 2].
Reviews on functions of lncRNAs in cancer have been published, describing repeatedly current knowledge on prominent cancer lncRNA candidates. Specific reviews on lncRNAs with regard to urological cancers have also been published [3-6].
LncRNAs may exhibit tissue- or disease-specific expression which can provide potential biomarkers. However, the utility of lncRNA as biomarkers appears to be overstated due to heterogeneous results from different studies. In many cases, initial reports could indeed not be replicated. The story of prostate cancer gene 3 (PCA3) is a good illustration of how long and tedious the path to a clinically successful biomarker really is. Thus, a review on the biomarker potential of lncRNAs should be critical and cautious.
This review provides an update on findings of lncRNAs in urological cancer by assessing the literature until March, 2016 from PubMed and discusses the suitability of lncRNAs as diagnostic or prognostics markers in a balanced and critical manner.
Mechanisms of Lncrna Function
LncRNAs function through a variety of mechanisms (Figure 1), e.g. as (i) recruiters regulating gene expression in cis and in trans; (ii) scaffolds that assemble chromatin remodeling machinery at the site of regulation; (iii) decoys that titrate away and prevent the action of effector molecules; (iv) mediators of alternative splicing; (v) precursors for small ncRNAs. Understanding this functional versatility is critical for the clinical exploitation of the lncRNAs.
Locus-Specific Recruiters
Many lncRNAs are highly enriched in the chromatin fraction where they regulate transcription regulation through the recruitment of chromatin remodeling complexes. Such lncRNAs can alter epigenetic states either in cis or in trans.
Antisense RNA-mediated Cis-Regulation
Many lncRNAs overlap protein-coding genes but are transcribed in the antisense orientation. Antisense RNAs are well suited for cis-regulation of their sense counterparts. The locally accumulated antisense RNAs are assumed to recruit chromatin remodeling proteins through either RNA-RNA or DNA-RNA recognition rules and trigger heterochromatin formation for gene silencing. A growing list of the validated sense-antisense pairs has been implicated in urologic cancer.
One typical example is the action of antisense ncRNA in the INK4 locus (ANRIL). ANRIL mediates silencing of a cluster of tumor suppressor genes, including its sense partner p15/CDKN2B through SUZ12 (a PRC2 component) and the p16/CDKN2A and p14/ARF locus via CBX7 (a PRC1 subunit).
RASSF1 antisense RNA 1 (RASSF1-AS1) is a tumor suppressor involved in cell cycle and apoptosis in prostate cancer [7]. RASSF1-AS1 forms an RNA-DNA hybrid at the RASSF1A promoter and recruits PRC2 which specifically represses RASSF1A transcription.
C Terminal Binding Protein 1 Antisense (CTBP1-AS) is a key antisense ncRNA upregulated in prostate cancer and negatively correlated with its sense gene CTBP1 [8]. CTBP1-AS recruits transcriptional repressor PSF and histone deacetylases to CTBP1 promoters and represses CTBP1 expression in cis. CTBP1-AS also inhibits tumor suppressors in trans via the PSF-dependent mechanism thus facilitating prostate cancer initiation and progression.
Another example of the RNA guiding mechanism could be seen by the action of the ncRNACCND1 expressed at the 5'-regulatory region of the cyclin D1 (CCND1) gene [9]. ncRNACCND1 is expressed upon DNA damage and recruits the RNA binding protein TLS that represses CBP/p300 activities on CCND1 in cis, causing gene-specific inhibition.
Mechanisms of lncRNA function.
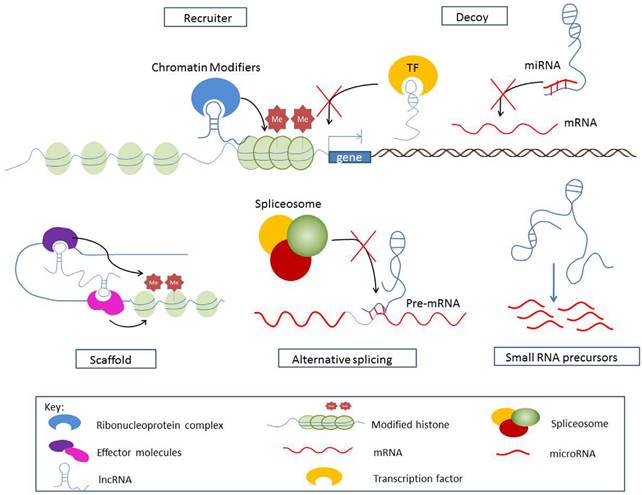
Other antisense lncRNAs were found to be inversely related to their antisense partners in urological cancer, e.g. TRPM2-AS, CBR3-AS1, 5' and 3'aHIF-1α, ZEB2NAT and MDC1-AS. But the precise mechanisms underlying the regulation of sense transcript have yet to be deciphered. TRPM2-AS [10] is consistently upregulated in prostate cancer and inversely related to its antisense transcript TRPM2, dictated by methylation degree of the CpG island. TRPM2 silencing leads to decreased susceptibility to cell apoptosis and necrosis. CBR3 antisense RNA 1 (CBR3-AS1) is significantly upregulated in primary prostate tumors and PCa cells [24]. Silencing of CBR3-AS1 downregulates AR, inhibits cell growth, and increases apoptosis in both androgen-dependent and independent LNCaP cells. Cui et al. [11] suggested a reciprocal regulation of CBR3-AS1 and AR in a positive feedback loop in prostate tumorigenesis. While others characterized 5'aHIF-1α and 3'aHIF-1α, two antisense transcripts at the 5' and 3' end of the human hypoxia-inducible factor-1α (HIF-1α) gene [12] respectively in kidney tumor specimen. 5'aHIF-1α and 3'aHIF-1α are involved in different regulation mechanisms and are activated under different environmental stress and histological cell types. 3'aHIF-1α binds to the 3'UTR of HIF-1α and down-regulates HIF-1α translation. Moreover, 3'aHIF-1α is overexpressed specifically in non-papillary kidney cancer and could discriminate between papillary and nonpapillary renal cell carcinoma [13]. 5'aHIF-1α is involved in membrane transport and mediates the HIF-1α pathway. Recently, Zhuang et al. [14] reported a novel antisense lncRNA ZEB2NAT of ZEB2 that enhances the invasion of UBC cells through the TGFβ1-ZEB2NAT-ZEB2 axis. Xue et al. [15] identified antisense lncRNA MDC1-AS that upregulates the expression of tumor-suppressing MDC1 in bladder cancer.
Cis-silencers in Genomic Imprinting
Several cis-regulatory lncRNAs are also functionally involved in genomic imprinting, which is an epigenetic event whereby one parental allele is preferentially silenced to maintain the allele-specific expression. Disturbed imprinting in XIST, CDKN1C/KCNQ, H19/IGF2 and DLK1/MEG3 loci is commonly observed in urologic cancers.
X-inactive specific transcript (XIST) is a 19 kb long lncRNA expressed exclusively from the inactive X chromosome. It mediates X chromosome inactivation in female mammalian cells to balance the X-chromosome genes dosage between the male and female. XIST recruits PRC2 to the polycomb group transcription factor YY1, which can load the XIST-PRC2 complex onto the Xic and initiate H3K27me3 throughout the Xi. XIST promoter is hypomethylated in prostate cancer tissues and cell lines [16, 17].
An additional example is the KCNQ1 opposite strand transcript 1 (KCNQ1OT1) in the KCNQ1 imprinted gene cluster. KCNQ1OT1 is paternally expressed and bidirectionally silences cis-linked imprinted genes of the centromeric domain. KCNQ1OT1 binds both PRC2 and G9a to promote two different repressive histone marks H3K27me3 and H3K9me3, thereby mediating the silencing of the flanking genes. On the maternal chromosome where ICR2 promoter is methylated, KCNQ1OT1 is silenced and the imprinted genes are transcribed. Altered expression and functional implication of KCNQ1OT have been investigated in bladder cancer [18], prostate cancer [19], Beckwith-Wiedemann syndrome (BWS) [20, 21], Wilms' tumors [22] as a consequence of disturbed imprinting.
Defective imprinting in H19/IGF2 locus is also a common event in bladder cancer and Wilms tumors. H19/IGF2 harbors conversely imprinted IGF2 and H19 expressed from the paternal and the maternal alleles respectively. In bladder cancer, the H19 promoter is paternally hypomethylated [23], causing biallelic expression of H19 and silencing of IGF2. The induced H19 promotes cell growth by regulating ID2 expression [24] and also contributes to metastasis by associating with enhancer of zeste homolog 2 (EZH2) and inhibiting E-cadherin transcription [25]. Unlike the situation in bladder cancer, Wilms tumor is linked to a gain in methylation on the maternal allele [26, 27]. Biallelic methylation of the H19 promoter silences H19 and causes biallelic expression of IGF2, thereby stimulating cell growth. In BWS, the epigenetic H19 silencing increases the risk of developing childhood tumors [21].
DLK1/MEG3 locus is structurally and epigenetically similar to the IGF2/H19 domain. The locus contains the paternally imprinted non-coding MEG3 and the maternally imprinted DLK1. DLK1 and MEG3 are prevalently downregulated in prostate cancer [19], bladder cancer [28, 29] and primary renal cell carcinoma (RCC) tissues [30] caused by the epigenetic silencing across the 14q32 imprinted gene cluster.
lncRNA Action in Trans
Despite prominent examples of lncRNA action in cis, lncRNAs also affect a large number of genes in trans.
The HOX transcript antisense RNA (HOTAIR) guides both histone modification complexes PRC2 and LSD1 to their target genes genome-wide and mediates epigenetic silencing. HOTAIR expression is elevated in castration resistant prostate cancer [31] and renal carcinoma cell lines [32].
linc-UBC1 also functions as recruiter in trans. linc-UBC1 is overexpressed in primary bladder cancer. It recruits PRC2 complex and increases H3K27Me3 in the promoter of CCND2 and HOXA5 and depressed their expression. Knockdown of linc-UBC1 inhibits proliferation and compromises invasiveness, cell motility and metastasis.
Molecular Scaffolds
Emerging evidence indicates that lncRNAs serve as molecular scaffolds that mediate chromosomal looping between distal enhancer and promoter.
Yang et al. [33] suggested that PCa non-coding RNA 1 (PRNCR1) and prostate cancer 2 gene expression marker 1 (PCGEM1) mediate chromosomal looping to regulate androgen receptor (AR) dependent transcription. PRNCR1 interacts with the AR and recruits DOT1-like histone H3K79 methyltransferase (DOT1L), which in turn methylates AR. The activated AR binds to PCGEM1, which recruits pygopus family PHD finger 2 (PYGO2). PYGO2 recognizes H3K4me3 at gene promoters. Subsequently, a complete loop from a distal upstream enhancer to the promoter is established leading to the activation of the androgen-responsive genes.
However, the role of PRNCR1 and PCGEM1 in AR activation was challenged by later researches. Prensner et al. [34] observed no evidence for their interaction with the AR. Parolia et al. [35] showed that PCGEM1 is uniformly distributed in PCa cell nucleus and cytoplasm, and remains unaltered upon AR activation. Hung et al. [36] found that PCGEM1 promotes proliferation not via AR activation, but through c-Myc activation. Therefore, the role of PCGEM1 and PRNCR1 in castration resistant prostate cancer (CRPC) remains uncertain.
A recent study [37] indicated two additional lncRNAs MALAT1 and Taurine upregulated gene 1 (TUG1) serving as sub-nuclear molecular scaffolds that mediate growth control gene relocation between nuclear structures. TUG1 is upregulated in bladder urothelial carcinoma. It selectively guides the methylated Pc2 on growth control gene promoters to the co-repressor complexes in PcG bodies for transcription repression. MALAT1 interacts with unmethylated Pc2 in the activation environment of the ICGs for transcription activation.
Target Decoy or Mimicry
Another means by which lncRNAs regulate transcription is to act as decoys. In this archetype, lncRNAs sequester biomolecules away from natural target and prevent them from fulfilling functions. This kind of lncRNAs always antagonize transcription regulators, so knockdown of these lncRNAs may lead to gain-of-function of its targets [38].
Decoy at Transcriptional Level
One particular type of lncRNA decoy involves proteins. lncRNA can bind to transcription-factors that would otherwise bind to the DNA promoters and thereby inhibit gene transcription.
The second chromosome locus associated with prostate-1 (SCHLAP1) is a protein-interacting lncRNA implicated in tumor invasion and metastasis and upregulated in aggressive PCa [39]. Mechanistically, SCHLAP1 co-immunoprecipitates with SNF5, a core unit of the SWI/SNF complex and impairs the SWI/SNF-mediated epigenetic regulation. SWI/SNF complex is a well-documented chromatin regulator with tumor suppressor function. This complex remodels the chromatin and physically moves nucleosomes at gene promoters to control transcription.
In contrast to SCHLAP1 which is pro-survival, Growth-Arrest Specific 5 (GAS5) is a tumor suppressor with a pro-apoptotic role in prostate cancer. GAS5 has a stem-loop structure in its sequence that mimics the glucocorticoid responsive gene. GAS5 binds to glucocorticoid receptors and prevents its genomic binding. This results in the suppression of GR responsive genes such as anti-apoptotic cIAP2 [40] and ultimately induces growth arrest [41]. GAS5 can also inhibit transcriptional stimulation of progesterone receptor and androgen receptor in a similar fashion. Since androgen receptor is critical in prostate cancer cells survival, down regulation by GAS5 may inhibit pro-survival signaling via the androgen receptor pathway.
Decoy at Translational Level
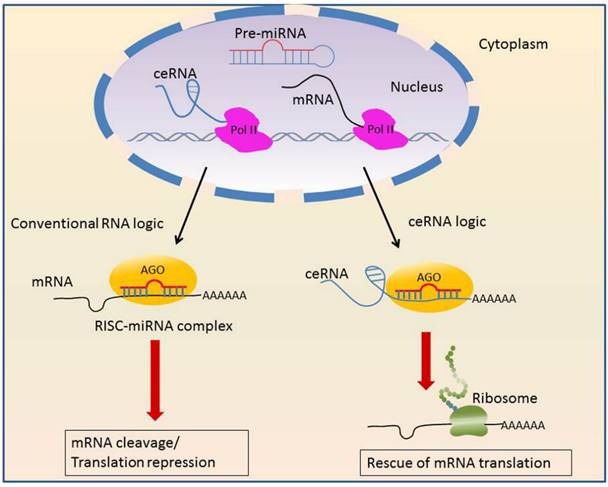
LncRNA decoys not only proteins, but also microRNAs. In this context, lncRNAs act as competing endogenous RNAs (ceRNAs) that sponge microRNAs to interfere with their function (Figure 2).
A prominent example is the PTENP1, pseudogene of the tumor suppressor PTEN [42, 43]. PTEN and PTENP1 share multiple miRNA target sites, therefore PTENP1 competes for the same set of regulatory miRNAs that normally targeted PTEN to modulate PTEN expression [44]. PTEN is commonly lost in advanced PCa [45] and clear cell renal cell carcinoma (ccRCC) [44]. The restoration of PTEN inhibits tumor cell proliferation [46]. The prostate cancer associated transcript 1 (PCAT1) is another lncRNA as miRNA antagonizer. PCAT1 is the top-ranked, upregulated lncRNA in prostate cancer tissues. PCAT1 disrupts miR-34a mediated down-regulation of cMyc [34] and promotes proliferation. Further, PCAT1 downregulates the tumor suppressor gene BRCA2 [47], similar to microRNA mediated mRNA degradation. MALAT1 also serve as a ceRNA through interfering with miR-200s and increases ZEB2 expression in ccRCC [48]. Urothelial carcinoma-associated 1 (UCA1) acts as a nature decoy that blocks miR-16 to inhibit ROS formation, thus promoting tumorigenesis in bladder cancer [49].
Mediators of Alternative Splicing
Recently lncRNAs have been shown to act at a post-transcriptional level by regulating alternative splicing of pre-mRNAs.
MALAT1 is enriched in nuclear speckles, a sub-nuclear domain that is enriched with various pre-mRNA splicing factors including Serine-Arginine (SR) protein. MALAT1 harbors a large number of SR protein binding sites. It modulates the phosphorylation of Serine-Arginine (SR) splicing factors and mediates their distribution [50] in the nuclear speckles, thereby controlling the localization of splicing factors at active transcription sites. Besides SR, MALAT1 also interacts with other pre-mRNA splicing factors, e.g. PRP6 and TDP-43. MALAT1 loss-of-function alters alternative splicing pattern of pre-mRNAs such as tissue factors and endoglin which are important in cancer biology.
Precursors of Small NcRNAs
LncRNAs can function as primary precursors for other small ncRNAs deregulated in cancer. It is estimated that at least 4% of lncRNAs host small ncRNAs. For example, H19 hosts miR-675 in first exon. miR-675 is a tumor suppressor involved in the regulation of developmental genes [51]. It inhibits cell proliferation in response to stress or oncogenic signals [52].
GAS5 encodes 10 different small nucleolar RNAs (snoRNAs) in its introns [53]. Interestingly, only a subset GAS5-encoded snoRNAs are upregulated in progressing PCa [54], suggesting that RNA products derived from the same precursor transcript could function through separate mechanisms at post-transcriptional levels.
Clinical Utility of Lncrnas in Urological Cancer Management
Many lncRNAs are expressed in a tissue or cancer specific manner. They are often stable in biological fluids and could be easily detected using qRT-PCR amplification. The specificity and stability of lncRNAs have prompted efforts to evaluate their potential as biomarkers for diagnosis, prognosis, therapy response and therapeutic targets. A list of putative lncRNA biomarkers is summarized in Table 1.
Mechanisms of competing endogenous RNAs.
lncRNAs involved in urologic cancer.
| Name | Cancer | Location | Source | Expression | Function | Clinical utility |
|---|---|---|---|---|---|---|
| 3'aHIF-1α | Kidney | 14q23.2 | Tissue | ↑ | Downregulates HIF-1α translation by binding to its 3' UTR. | Marker for nonpapillary RCC |
| H19 | Bladder | 11p15.5 | Tissue/ Cell line | ↑ | Paternally hypomethylated; Silences IGF2 and NKD1, activates Wnt/β-catenin, promotes EMT | Prognosis |
| H19 | Kidney | 11p15.5 | Tissue | ↓ | Maternally methylated; Causes biallelic expression of IGF2; Precursor of miR-675 | Risk prediction |
| HOTAIR | Prostate/ Bladder | 12q13.13 | Tissue/ Cell line | ↑ | Guides PRC2 and LSD1/CoREST/REST complexes to HOXD locus for epigenetic silencing | Prognosis |
| Linc00963 | Prostate | 9q34.11 | Cell line | ↑ | Inhibits EGFR expression; Attenuates motility and invasion; Enhances apoptosis | Prognosis |
| lincRNA-p21 | Prostate | Urine/ Exosome | ↓ | suppresses expression of the genes transcriptionally regulated by p53 by binding to the hnRNP-K complex | Diagnosis | |
| linc-UBC1 | Bladder | 1q32.1 | Tissue | ↑ | Recruits PRC2 complex and regulates histone methylation of target genes | Prognosis |
| MALAT1 | Bladder/Kidney/Prostate | 11q13.1 | Tissue | ↑ | Interacts with unmethylated Pc2; Activates growth control gene; Influences alternative splicing | Diagnosis and Prognosis |
| MD-miniRNA | Prostate | 11q13.1 | Plasma | ↑ | Diagnosis | |
| PCA3 | Prostate | 9q21-22 | Tissue/ Urine | ↑ | Modulates AR signaling | Diagnosis |
| PCAN-R1 | Prostate | 1q32.1 | Tissue | ↑ | Inhibits cell growth and soft-agar colony formation | Prognosis |
| PCAN-R2 | Prostate | 9q22.32 | Tissue | ↑ | Inhibits cell proliferation and soft-agar colony formation | Prognosis |
| PCAT1 | Prostate | 8q24.21 | Tissue/ Cell | ↑ | Downregulates BRCA2; Disrupts miR-34a mediated down-regulation of cMyc | Prognosis |
| PCAT18 | Prostate | 18q11.2 | Tissue | ↑ | Inhibits apoptosis and migration | Prognosis |
| PCAT29 | Prostate | 15q23 | Tissue/ Cell line | ↓ | Suppresses growth and metastasis | Prognosis |
| PCGEM1 | Prostate | 2q32 | Tissue | ↑ | Promotes cell proliferation; Delays p21 and p53 expression; Inhibits apoptosis | Risk prediction/ Prognosis |
| PRNCR1 | Prostate | 8q24 | Tissue | ↑ | Increases viability in the androgen-dependent LNCaP cells | Risk prediction |
| RASSF1-AS1 | Prostate | 3p21.3 | Tissue/ Cell line | ↑ | Represses RASSF1A transcription via H3K27Me3 | Prognosis |
| RCCRT1 | Kidney | Tissue | ↑ | Associates with clinicopathologic parameters | Prognosis | |
| SCHLAP1 | Prostate | 2q31.1 | Tissue | ↑ | Impairs SWI/SNF-mediated epigenetic regulation | Prognosis |
| SPRY4-IT1 | Kidney | 5q31.3 | Tissue | ↑ | Enhances proliferation, migration, and invasion | Prognosis |
| TRPM2-AS | Prostate | 21q22.3 | Tissue | ↑ | Silences TRPM2 expression | Prognosis Therapeutic target |
| UCA1 | Bladder | 19p13.12 | Urine | ↑ | Regulates cell cycle distribution via CREB through PI3-K dependent pathway | Diagnosis |
| XIST | Prostate | Xq13.2 | Serum | - | Hypomethylated in PCa; Recruits PRC2 onto the Xic and initiates H3K27me3 | Diagnosis |
Risk Prediction
Recently several lncRNAs transcribed from cancer risk loci have been identified. 8q24 gene desert region, for example, harbors several lncRNA genes, e.g. PCAT1 and PRNCR1. 8q24 is a key prostate cancer predisposition locus associated with PCa susceptibility. GWAS have discovered within the lncRNAs multiple prostate cancer-associated SNPs for risk prediction. rs378854 in 8q24 region is associated with expression of oncogenic lncRNA PVT1 in PCa [55]. A further lncRNA at the 8q24 region is PRNCR1 [56] transcribed between rs1456315 and rs7463708. It is most significantly related to PCa susceptibility in the Japanese. SNPs in the H19 gene are also associated with the risk of bladder cancer [57]. rs2839698TC and rs2107425CT in H19 significantly reduce risk of non-muscle-invasive bladder cancer. rs16834898 and rs6434568 in PCGEM1 [58] indicate a lower PCa risk with Chinese population. However, how these variants affect cancer susceptibility remains to be elucidated.
Markers in Screening and Diagnosis
Early diagnosis of primary and recurrent cancers followed by timely treatment will significantly decrease mortality. The most common screening test for prostate cancer early detection to date is serum PSA screening. However, the use of PSA lacks specificity and fails to discriminate aggressive tumors from indolent cancers, leading to unnecessary biopsies and overtreatment. Thus there is a continued need for more sensitive and specific biomarkers for prostate cancer early detection that supplement PSA.
Numerous efforts have been applied to transcriptome-wide study of urological cancer and found numerous urological cancer associated lncRNAs. The most promising among these is the PCA3. PCA3 is specifically overexpressed in malignant prostate tissue and absent in indolent prostate conditions or cancers of non-prostate origin. PCA3 has demonstrated a superior specificity and accuracy to serum PSA. Moreover, PCA3 is detectable in post-DRE urine samples in urine and urine sediments, therefore enabling noninvasive diagnose. For this reason, PCA3 has been translated into the clinical setting. A PCA3 assay PROGENSA® has been approved by the FDA as a diagnostic method to aid in the decision of repeat prostate biopsies [59].
Despite the improved specificity, the lower sensitivity of PCA3 renders it insufficient as a standalone biomarker. However, the inclusion of PCA3 can improve the diagnostic accuracy of other clinical variables. The combination of urinary PCA3 and TMPRSS2:ERG is currently being addressed in two independent multi-center trials [60, 61]. A multiplex marker panel consisting of PCA3, PSA, TMPRSS2:ERG, sarcosine and annexin A3 also showed improved diagnostic accuracy for PCa [62]. Combining PCA3 scores with the Prostate Health Index (Phi) improves specificity in prostate cancer diagnosis than single marker alone [63].
In addition to PCA3, several other urine biomarkers were under investigation for the detection of PCa. Wang et al. [64] observed that urinary MALAT1 score outperforms the conventional %free PSA in PSA grey zone cohorts (4-10 ng/mL). MALAT1 could detect more cancer cases and save avoidable and repeated biopsies. The exosomal lincRNA-p21 in urine is diagnostically superior to PCA3 in distinguishing PCa from benign prostatic hyperplasia (BPH) [65]. lincRNA-p21 is a tumor suppressor stimulated by the p53 protein. Once transcribed, lincRNA-p21 binds to the hnRNP-K complex and suppresses expression of the genes regulated by p53. LncRNA FR0348383 [66] is upregulated in PCa compared with matched benign tissues. FR0348383 transcript in post-DRE urine has diagnostic value for patient cohorts especially in the PSA grey zone. The application of FR0348383 score could avoid 52% unnecessary biopsies without missing high grade PCa.
Besides urine, diagnostic value of lncRNAs in circulating blood was also evaluated. For example, a MALAT1 derived fragment, MD-miniRNA is differentially expressed in the serum and plasma of PCA patients [67, 68]. MD-miniRNA can distinguish positive prostate biopsies from negative at AUC of 0.841. Compared with PSA, MD-miniRNA has better specificity but lower sensitivity for PCa detection. However, the sensitivity of MD-miniRNA increases in higher grade tumor samples, which suggest that MD-miniRNA may selectively detect aggressive PCa patients. Combined MD-miniRNA and PSA could improve diagnostic sensitivity and specificity. The 5' region of the XIST gene is commonly less-methylated in PCa irrespective of XIST expression [16]. Identification of a XIST hypomethylated fragment in serum may have tumor diagnostic potential for PCa [17].
UCA1 is a well-documented oncogene in bladder cancer which promotes cancer cell growth, invasion and drug resistance [69-71]. Since its first description in 2006 [72], many studies have confirmed UCA1 to be specific and sensitive marker for bladder cancer diagnostic or follow-up [69-75]. UCA1 is especially sensitive in diagnosis of superficial high grade (G2-G3) TCC cases [74, 75].
Functionally, UCA1 may exert pro-tumor activity by activating PI3-K signaling pathway [70] and Akt signaling pathway [70, 76]. More recent studies [77] put forward that UCA1 promotes cell growth by downregulating the cell cycle inhibitor p21 via BRG1. UCA1 is upregulated in bladder cancer cells under hypoxic conditions induced by transcription factor Ets-2 [76], CCAAT/enhancer binding protein α (C/EBPα) [78] and hypoxia-inducible factor-1α (HIF-1α) [79]. The correlation between these molecules under hypoxic conditions remains unclear. UCA1 is also involved in glucose metabolism in bladder cancer [80].
The variant transcript of UCA1, UCA1a (CUDR) also plays a role in bladder cancer progression. Upregulated UCA1 and UCA1a have been shown to enhance resistance to cisplatin in bladder cancer cells [71, 81], suggesting their role in chemoresistance [71].
However, many claims above are based on single reports with small cohorts for which replication studies are mostly lacking. According to a recent report [82], the diagnostic value of UCA1 in bladder cancer is lower than previously reported and its role in the follow-up of recurring tumors is limited. Thus UCA1 is currently not recommended as standard diagnostic tools in routine urology. UCA1 can be used only to complement cystoscopy and cytology in the early diagnosis when a primary bladder cancer is suspected.
In addition to UCA1, the differential expression pattern of several other lncRNAs has been reported in bladder cancer. PCAT-1 [83] and H19 [24, 84] were up-regulated in bladder cancer compared with normal controls. On the other hand, MEG3 expression is decreased in bladder cancer [28]. SncmtRNA and the ASncmtRNAs in urine can detect tumor cells with different grades, and were suggested as a non-invasive diagnostic tool [85].
For renal cancer, there is still no reliable biomarker available for daily practice despite numerous efforts. The study of lncRNAs is limited to a few samples that were analyzed by microarrays. Five recent studies investigated the lncRNA expression profile of RCC tissues using microarray technologies. However, the samples sizes are small (n=6 [86]; n = 4 [87]; n = 6 [88]; n = 11 [89]; n=15 [90]) and lack validation cohort, thereby limiting the value of the studies. Nevertheless, it is obvious that unique lncRNA expression profiles exist in RCC. In accordance with prostate and bladder cancer, MALAT1 is also overexpressed in RCC than benign renal tissue or cell lines [48, 91]. MALAT1 silencing inhibits RCC cell proliferation and promotes apoptosis. Hirata et al. [92] found that MALAT1 is transcriptionally activated by c-Fos downstream of the VHL pathway and interacts with Polycomb protein EZH2 to induce epithelial-to-mesenchymal transition (EMT).
Marker for Prognosis
LncRNAs are also strong predictors of tumor recurrence and patient outcome. H19 has been advocated as a potential prognostic biomarker for early recurrence in bladder cancer. Hochberg et al [93] first reported altered expression of H19 in bladder cancer. H19 upregulation is related to an increased risk of biochemical recurrence and shorter metastasis-free survival [94]. linc-UBC1 overexpression is a negative prognostic marker for lymph node metastasis in bladder cancer [95].
Upregulation of HOTAIR may be closely associated with recurrent and poor prognosis of bladder cancer. HOTAIR upregulation associates with increased risk of recurrence in stage Ta/T1 bladder cancer [96] and affects invasiveness and differentiation state of urothelial cancer cells in a cell type specific manner. Upregulation of HOTAIR is also correlated with histological grades and overall survival in bladder transitional cell carcinoma (TCC) and inhibits chemosensitivity to doxorubicin [97]. Elevated SPRY4-IT1 is strongly correlated with aggressive clinicopatholigcal features and poor prognosis in bladder cancer and provides an unfavorable prognostic factor [98]. Upregulation of MALAT1 in bladder cancer corresponds to the tumor grade and metastatic stage [81, 99-101]. Experiment showed that MALAT1 induces EMT to promote bladder cancer cell migration by associating with suz12 [102] and activating Wnt signaling [100].
In PCa, high SChLAP1 expression is correlated with biochemical recurrence, clinical progression and mortality. It significantly predicts metastatic progression in high-risk PCa patients who had undergone radical prostatectomy and lethal mCRPC [39, 103]. Recently an RNA in situ hybridization (ISH) assay for SChLAP1 was developed for the detection of aggressive prostate cancers [104].
PCGEM1 and PRNCR1 [33] were previously identified to be highly overexpressed in aggressive PCa. While PCGEM1 is confirmed by later studies to be associated with prostate cancer, PRNCR1 does not exhibit convincing upregulated expression [34, 35]. Prensner et al. [34] contradicted the prognostic value of PRNCR1 in aggressive PCa by analyzing a large cohort of high-risk prostate cancer patients. They claimed that PRNCR1 does not demonstrate a convincing association with poor patient outcomes.
Several studies have reported a significant correlation between PCA3 score and prognostic parameters. However, the prognostic value of PCA3 is controversial. Other independent studies [105, 106] found no significant correlation between the PCA3 score and clinicopathologic variables.
HOTAIR is overexpressed in CRPC cell lines [31], tissues [107], and after androgen deprivation therapies. Further study demonstrates that the HOTAIR directly binds to AR to protect it from ubiquitin-mediated degradation, and thus drives androgen-independent AR activation and promotes CRPC. GAS5 transcript level is significantly lower in metastatic prostate cell lines compared with normal prostates. Down-regulation of GAS5 has been found to mediate the progression of CRPC. PCAT1 is specifically upregulated in high-grade PCa, independent of chromosome 8q24 amplification, and has been implicated as a prognostic biomarker for prostate cancer metastasis and poor overall survival [108]. The upregulation of TRPM2-AS associates with poor clinical outcome and has been characterized as a high-risk PCa marker [109]. PCAT18 is highly up-regulated in metastatic PCa, and is able to discriminate between localized PCa and metastatic CRPC [110]. PCAT5 was also reported as a CRPC-specific biomarker [111]. Linc00963 is differentially expressed in the androgen-independent and metastatic PCa cell line and is responsible for the androgen-independence transition [112]. Overexpression of MALAT1 in PCa tissues is related to high Gleason score, late tumor stage and CRPC. In vitro MALAT1 silencing inhibits metastasis of CRPC cancer cells and leads to cell cycle arrest. In vivo knock-out of MALAT1 inhibits growth of tumor xenografts [113, 114]. MALAT1 associates with the N-terminal of EZH2 through 3′ end and enhances EZH2-mediated migration and invasion in CRPC cell lines. Knockdown of MALAT1 impairs EZH2 recruitment to its target loci and represses oncogenetic activity of EZH2 [115].
Du et al. [116] reanalyzed array-based PCa expression profiles and identified 2 metastases relevant PCAN-R1 and PCAN-R2 as potential drivers of cancer progression. Recently, Malik [117] identified low PCAT29 expression correlated with poor prognostic outcomes.
For renal cancer, several independent indicators of clinical outcome have been identified in patients with ccRCC. SPRY4-IT1 is significantly correlated with advanced clinical stage, lymphnode metastasis and poorer prognosis in ccRCC [118]. ccRCC patients with higher H19 level have more advanced clinical stage and worse overall survival [119]. Decreased expression of NBAT-1 [120] is associated with the progression and poor prognosis of ccRCC. Upregulated RCCRT1 is correlated with tumor stage, histological grade and lymph node metastasis [121]. Recently, Malouf et al. [122] established a molecular subclass of ccRCC through lncRNA-based clustering and identified cluster C2 to associate with aggressiveness and poor overall survival.
LncRNA-Based Therapy
Anti-cancer strategies based upon lncRNA biology are rapidly emerging. The therapeutic concepts could be either to suppress the oncogene expression or to activate tumor-suppressive lncRNAs.
LncRNA Targeting Strategy
Direct targeting of oncogenic lncRNAs is an attractive strategy for treatment of cancer. A straightforward method is to interfere with RNA expression via antisense technologies. RNAi-mediated gene silencing has recently gained momentum as a treatment modality. Many studies have used RNAi to knockdown spliced lncRNAs. However, RNAi activity is restricted to the cytoplasm and cannot be deployed against a nuclear localized lncRNA.
Other possible lncRNA targeting agents are catalytic nucleic acids (CNAs) such as ribozymes and DNAzymes, which can also bind and cleave lncRNA targets and repress their expression.
An alternative approach would target lncRNA interactions and silence their function. This can be achieved by the use of small molecule inhibitors that block the binding site of their protein partners. Disruption of these molecular interactions compromises the repression of the tumor suppressors and reactivates their function. For example, using small molecular inhibitors of PRC2 to block its interaction with ANRIL would be a promising therapeutic strategy.
Targeted Gene Therapy
The tumor-specific expression of lncRNA has prompted investigation into its utility in targeted anticancer therapy. One typical example is the use of H19 regulatory sequences for the treatment of bladder cancer. The BC-819 DNA plasmid, which contains diphtheria toxin controlled under the H19 promoter, has been designed to treat target cells with overexpressed H19 [123]. When BC-819 enters a cancerous cell, the Diphtheria Toxin A (DTA) chain is synthesized under the regulation of H19. This allows precise targeting of tumors without excessive harm to healthy tissue. A multicenter trial of intravesical BC-819/PEI is ongoing in patients with superficial TCC [124]. A pivotal (Phase III) bladder cancer clinical trial has commenced in 2015.
A similar vector H19-DTA-P4-DTA [125] is also under consideration for bladder cancer therapy. H19-DTA-P4-DTA expresses the DTA fragment under the control of two separate promoters from H19 and IGF2-P4. The vector exhibits increased anti-tumor activity than the single promoter vectors.
Tumor-Suppressive LncRNAs
Alternatively, one can also manipulate the expression of tumor-suppressive lncRNAs in anticancer therapy. One example is the protein decoy GAS5. Its downregulation was described in prostate cancer cell lines [41], RCC [126] and bladder cancer [127]. GAS5 expression inhibits androgen receptor signaling, promotes growth arrest and apoptosis [41]. Therefore, upregulation of GAS5 could provide a possible treatment. Based on the observation that mTOR expression is inversely correlated with GAS5 in PCa cell lines, Yacqub et al. [128] examined mTOR inhibition as a strategy to enhance GAS5 levels in several prostate cancer cell lines.
Lncrna: Current Challenges and Future Directions
The ever-growing number of lncRNA-based marker candidates stands in sharp contrast to the lack of reliable biomarkers for urological cancer. The biggest challenge is the limited validity. The candidates generated by different labs are non-overlapping or even contradictory [129, 130]. As discussed earlier, the prognostic value of PCA3 is controversial [34]. The prognostic value of PRNCR1 in aggressive PCa remains uncertain. It remains to be proven whether UCA1 is a suitable diagnostic urinary biomarker in bladder cancer. Aberrant expression of individual lncRNAs in RCC by small-scaled studies, i.e. GAS5 [122, 126], aHIF [12], MALAT1 [122] could not be confirmed.
Most current studies aim to identify single molecules as biomarkers, but one size does not fit all. These studies ignore the dynamic molecular interactions and fail to address the heterogeneity of cancer. There is an obvious need to extend the biomarker discovery from single molecules to the functional modules, pathways and the networks. Some systematic studies of lncRNA markers have proliferated recently. As an example, our colleagues [131] have constructed a mRNA-lincRNA coexpression network in PCa using RNA-Seq expression data and characterized PCa-associated lincRNA modules as putative biomarkers.
This inter-laboratory inconsistency stems largely from the intrinsic features of cancer. First, the development of cancer is systems-oriented, involving the crosstalk between multiple biological components. Also the cancer is highly heterogeneous, not only between patients, but also within the same individual [132]. Most current studies however, aim to identify single molecules as biomarkers, but one size does not fit all. These studies ignore the dynamic molecular interactions and fail to address the heterogeneity of cancer.
There is an obvious need to extend the biomarker discovery from single molecules to the functional modules, pathways and the networks. Some systematic studies of lncRNA markers have proliferated recently. As an example, our colleagues [131] have constructed a mRNA-lincRNA coexpression network in PCa using RNA-Seq expression data and characterized PCa-associated lincRNA modules as putative biomarkers.
An additional bias lies in the deficiencies in the population under analysis. The current lncRNA marker candidates are mostly based on discovery-driven approaches that frequently use small sample sizes. This type of analysis is prone to statistical overfitting of high-dimensional data, producing the “p >> n” paradigm. Thus, the potential lncRNA markers have yet to be prospectively validated in standardized, well-controlled clinical trials across large numbers of patients. Thus, the diagnostic and therapeutic benefits of many potential lncRNA markers have yet to be prospectively validated in standardized, well-controlled clinical trials across large numbers of patients.
Many more important questions remain to be addressed.
In most cases lncRNA function is not fully understood. It is also not clear whether deregulated lncRNAs are drivers in the disease etiology or they are merely a consequence of the disease. Although aberrant expression of lncRNAs argues in favor of functional significance, but most publication have not described a direct association or causal role for lncRNAs in cancer, therefore its functional role in urological biology is largely unknown.
Assigning functions to lncRNAs may seem like a daunting task. For most lncRNAs, function is solely predicted by bioinformatic tools without any mechanistic understanding and experimental validation. The guilt by association analysis [133] has also been used to infer the function of an lncRNA based on their expression correlations with neighboring protein-coding genes with known functions. However, direct experimental test is still important to understand the lncRNA function. The classical experimental strategy would involve the depletion of lncRNA and assessment of the effects on gene expression and tumor-relevant phenotypes. Identification of protein interaction partners is also informative to decipher lncRNA mechanisms at the molecular level. A direct way would be the RNA affinity purification, or RIP-Seq to identify lncRNA-interacting proteins. Two methods, called chromatin isolation with RNA purification (ChIRP) [134] and capture analysis of RNA targets (CHART) [135], represent complementary approaches to study RNA-chromatin interactions. ChIRP and CHART have uncovered thousands of PCGEM1 occupancy sites. These methods will help to create an experimentally validated interactome of lncRNAs.
Unraveling the function of lncRNAs is more difficult than miRNA. The difficulties come from the relatively low sequence conservation of lncRNAs. However, the main functionality of lncRNAs may reside in its secondary structure motifs imperative for specific interaction. This is demonstrated by the double stem-loop structure of HOTAIR for PRC2 binding and the structure of MEG3 critical for its tumor suppressor functionality [84]. It will be highly informative to understand the sequence motives and functionally active RNA domains that govern lncRNA functionality.
The lncRNA-based therapeutics also has many important questions to be addressed. Targeting lncRNAs with small-interfering RNAs is not trivial due to the large transcripts size and the extensive secondary structures in lncRNAs.
In addition, the development of robust delivery system, optimization of dosage regimes and amelioration of off-target effects require intensive investigation in the future.
Conclusions
LncRNA is shedding light on the understanding of the urological malignancies. Their potential utility in cancer biology and therapy can be tremendous. We remain cautiously optimistic that when the limitations described above are overcome, lncRNAs may be used as promising markers or therapeutic targets to benefit patients.
Abbreviations
ANRIL: antisense ncRNA in the INK4 Locus; CAN: catalytic nucleic acid; CBR3: Carbonyl Reductase 3; CBR3-AS1: CBR3 antisense RNA 1; CBX7: chromobox homolog 7; CCND1: cyclin D1; ccRCC: clear cell renal cell carcinoma; CDKN1C: Cyclin-dependent kinase inhibitor 1C; CHART: capture analysis of RNA targets; ChIRP: chromatin isolation with RNA purification; cIAP2: cellular inhibitor of apoptosis 2; CRPC: castration resistant prostate cancer; CTBP1-AS: C terminal binding protein 1 antisense; DLK1: delta like homolog 1; DOT1L: DOT1-like histone H3K79 methyltrasferase; DTA: Diphtheria Toxin A; EMT: epithelial -mesenchymal transition; EZH2: enhancer of zeste homolog 2; GAS5: growth-arrest specific 5; GDF15: growth differentiation factor 15; GWAS: Genome Wide Association Studies; HIF-1α: hypoxia-inducible factor 1α; HOTAIR: HOX transcript antisense RNA; HOXC: Homeobox C; ICG: interchromatin granule; IGF2: Insulin-like growth factor 2; LOI: loss of imprinting; MALAT1: metastasis-associated lung adenocarcinoma tran-script-1; MEG3: maternally expressed 3; mTOR: mammalian target of rapamycin; NKD1: naked cuticle homolog 1; PCA3: prostate cancer gene 3; PCAT-1: prostate cancer associated transcript 1; PCAT29: prostate cancer-associated transcript 29; PcG: Polycomb body; PCGEM1: prostate cancer 2 gene expression marker 1; PRC1: polycomb repressive complex 1; PRC2: polycomb repressive complex 2; PRNCR1: PCa non-coding RNA 1; PYGO2: pygopus family PHD finger 2; RASSF1A: Ras-association domain family member 1A; RCC: Renal cell carcinoma; RIP-Seq: RNA immunoprecipitation and sequencing; SCHLAP1: second chromosome locus associated with prostate-1; SUZ12: suppressor of zeste 12 homolog; SWI/SNF: switch/sucrose non-fermenting; TCC: transitional cell carcinoma; TUG1: taurine upregulated gene 1; Xist: X-inactive specific transcript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China grants (31400712, 31470821, 91530320) and Technology R&D Program of Suzhou (SYN201409).
Conflict of Interest
The authors declare no conflict of interest.
References
1. A R, H P, C T, L P. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325-9
2. Severin AJ, Peiffer GA, Xu WW, Hyten DL, Bucciarelli B, O'Rourke JA. et al. An integrative approach to genomic introgression mapping. Plant physiology. 2010;154:3-12
3. Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. European urology. 2014;65:1140-51
4. Weiss M, Plass C, Gerhauser C. Role of lncRNAs in prostate cancer development and progression. Biological chemistry. 2014;395:1275-90
5. Bolton EM, Tuzova AV, Walsh AL, Lynch T, Perry AS. Noncoding RNAs in prostate cancer: the long and the short of it. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:35-43
6. Zhou S, Wang J, Zhang Z. An emerging understanding of long noncoding RNAs in kidney cancer. Journal of cancer research and clinical oncology. 2014;140:1989-95
7. Beckedorff FC, Ayupe AC, Crocci-Souza R, Amaral MS, Nakaya HI, Soltys DT. et al. The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation. PLoS genetics. 2013;9:e1003705
8. Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K. et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. The EMBO journal. 2013;32:1665-80
9. Wang X, Arai S, Song X, Reichart D, Du K, Pascual G. et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126-30 (3 July 2008) | doi: 10.1038/nature06992
10. Orfanelli U, Jachetti E, Chiacchiera F, Grioni M, Brambilla P, Briganti A. et al. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene. 2014:34
11. Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y. et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urologic oncology. 2013;31:1117-23
12. Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. Journal of the National Cancer Institute. 1999;91:143-51
13. Bertozzi D, Iurlaro R, Sordet O, Marinello J, Zaffaroni N, Capranico G. Characterization of novel antisense HIF-1alpha transcripts in human cancers. Cell cycle. 2011;10:3189-97
14. Zhuang J, Lu Q, Shen B, Huang X, Shen L, Zheng X. et al. TGFbeta1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Scientific reports. 2015;5:11924
15. Xue Y, Ma G, Zhang Z, Hua Q, Chu H, Tong N. et al. A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer. Oncotarget. 2015;6:484-93
16. Laner T, Schulz WA, Engers R, Muller M, Florl AR. Hypomethylation of the XIST gene promoter in prostate cancer. Oncology research. 2005;15:257-64
17. Song MA, Park JH, Jeong KS, Park DS, Kang MS, Lee S. Quantification of CpG methylation at the 5'-region of XIST by pyrosequencing from human serum. Electrophoresis. 2007;28:2379-84
18. Hoffmann MJ, Florl AR, Seifert HH, Schulz WA. Multiple mechanisms downregulate CDKN1C in human bladder cancer. International journal of cancer Journal international du cancer. 2005;114:406-13
19. Ribarska T, Goering W, Droop J, Bastian KM, Ingenwerth M, Schulz WA. Deregulation of an imprinted gene network in prostate cancer. Epigenetics: official journal of the DNA Methylation Society. 2014;9:704-17
20. Weksberg R, Nishikawa J, Caluseriu O, Fei YL, Shuman C, Wei C. et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Human molecular genetics. 2001;10:2989-3000
21. Bliek J, Maas SM, Ruijter JM, Hennekam RC, Alders M, Westerveld A. et al. Increased tumour risk for BWS patients correlates with aberrant H19 and not KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in familial cases of BWS. Human molecular genetics. 2001;10:467-76
22. Hatada I, Inazawa J, Abe T, Nakayama M, Kaneko Y, Jinno Y. et al. Genomic imprinting of human p57KIP2 and its reduced expression in Wilms' tumors. Human molecular genetics. 1996;5:783-8
23. Takai D, Gonzales FA, Tsai YC, Thayer MJ, Jones PA. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Human molecular genetics. 2001;10:2619-26
24. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. The FEBS journal. 2013;280:1709-16
25. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer letters. 2013;333:213-21
26. Frevel MA, Sowerby SJ, Petersen GB, Reeve AE. Methylation sequencing analysis refines the region of H19 epimutation in Wilms tumor. The Journal of biological chemistry. 1999;274:29331-40
27. Yuan E, Li CM, Yamashiro DJ, Kandel J, Thaker H, Murty VV. et al. Genomic profiling maps loss of heterozygosity and defines the timing and stage dependence of epigenetic and genetic events in Wilms' tumors. Molecular cancer research: MCR. 2005;3:493-502
28. Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y. et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Molecular bioSystems. 2013;9:407-11
29. Greife A, Knievel J, Ribarska T, Niegisch G, Schulz WA. Concomitant downregulation of the imprinted genes DLK1 and MEG3 at 14q32.2 by epigenetic mechanisms in urothelial carcinoma. Clinical epigenetics. 2014;6:29
30. Kawakami T. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Human molecular genetics. 2006;15:821-30
31. Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S. et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PloS one. 2013;8:e70372
32. Wu Y, Liu J, Zheng Y, You L, Kuang D, Liu T. Suppressed expression of long non-coding RNA HOTAIR inhibits proliferation and tumourigenicity of renal carcinoma cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:11887-94
33. Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W. et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598-602
34. Prensner JR, Sahu A, Iyer MK, Malik R, Chandler B, Asangani IA. et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5:1434-8
35. Parolia A, Crea F, Hui X, Wang Y, Fan M, Ramnarine VR. et al. The long non-coding RNA PCGEM1 is regulated by androgen receptor activity in vivo. Molecular cancer. 2015;14:1-7
36. Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G. et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18697-702
37. Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD. et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773-88
38. Chen J, Wang R, Zhang K, Chen LB. Long non-coding RNAs in non-small cell lung cancer as biomarkers and therapeutic targets. Journal of Cellular & Molecular Medicine. 2014;18:2425-36
39. Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L. et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature genetics. 2013;45:1392-8
40. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science signaling. 2010;3:ra8
41. Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochimica et biophysica acta. 2013;1832:1613-23
42. Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM. et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Science signaling. 2010;3:ra29
43. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-8
44. Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U. et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344-57
45. Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grander D. et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nature structural & molecular biology. 2013;20:440-6
46. Gan Y, Weimin Y, Kiranmai G, Anping L, Ji W, Wei X. et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear-cell renal cell carcinoma progression. Molecular Cancer Therapeutics. 2014;13:3086-97
47. Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S. et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer research. 2014;74:1651-60
48. Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J. et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015:6
49. Li HJ, Li X, Pang H, Pan JJ, Xie XJ, Chen W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Japanese journal of clinical oncology. 2015;45:1055-63
50. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39:925-38
51. Cai X. The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna-a Publication of the Rna Society. 2007;13:313-6
52. Schmitz KJ, Helwig J, Bertram S, Sheu SY, Suttorp AC, Seggewiss J. et al. Differential expression of microRNA-675, microRNA-139-3p and microRNA-335 in benign and malignant adrenocortical tumours. Journal of clinical pathology. 2011;64:529-35
53. Smith CM. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Molecular & Cellular Biology. 1998;18:6897-909
54. Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J. et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978-91
55. Meyer KB, Maia A-T, O'Reilly M, Ghoussaini M, Prathalingam R, Porter-Gill P. et al. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS genetics. 2011;7:545-7
56. Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N. et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer science. 2011;102:245-52
57. Verhaegh GW, Verkleij L, Vermeulen SH, den Heijer M, Witjes JA, Kiemeney LA. Polymorphisms in the H19 gene and the risk of bladder cancer. European urology. 2008;54:1118-26
58. Xue Y, Wang M, Kang M, Wang Q, Wu B, Chu H. et al. Association between lncrna PCGEM1 polymorphisms and prostate cancer risk. Prostate cancer and prostatic diseases. 2013;16:139-44 S1
59. Lee GL, Dobi A, Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nature reviews Urology. 2011;8:123-4
60. Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB. et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. European urology. 2014;65:534-42
61. Lin DW, Newcomb LF, Brown EC, Brooks JD, Carroll PR, Feng Z. et al. Urinary TMPRSS2:ERG and PCA3 in an active surveillance cohort: results from a baseline analysis in the Canary Prostate Active Surveillance Study. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:2442-50
62. Cao DL, Ye DW, Zhang HL, Zhu Y, Wang YX, Yao XD. A multiplex model of combining gene-based, protein-based, and metabolite-based with positive and negative markers in urine for the early diagnosis of prostate cancer. The Prostate. 2011;71:700-10
63. Perdonà S, Bruzzese D, Ferro M, Autorino R, Marino A, Mazzarella C. et al. Prostate health index (phi) and prostate cancer antigen 3 (PCA3) significantly improve diagnostic accuracy in patients undergoing prostate biopsy. The Prostate. 2013;73:227-35
64. Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y. et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5:11091-102
65. Işın M, Uysaler E, Özgür E, Köseoğlu H, Şanlı Ö, Yücel ÖB. et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Frontiers in Genetics. 2015;6:168
66. Zhang W, Ren SC, Shi XL, Liu YW, Zhu YS, Jing TL. et al. A novel urinary long non-coding RNA transcript improves diagnostic accuracy in patients undergoing prostate biopsy. The Prostate. 2015;75:653-61
67. Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J. et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. European journal of cancer. 2013;49:2949-59
68. Dong X, Zhou CX, Shi YB, Hao L, He XZ. MD-miniRNA could be a more accurate biomarker for prostate cancer screening compared with serum prostate-specific antigen level. Tumor Biology. 2015;36:1-7
69. Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS letters. 2008;582:1919-27
70. Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496:8-16
71. Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X. et al. Long non-coding RNA UCA1a(CUDR) promotes proliferation and tumorigenesis of bladder cancer. International journal of oncology. 2012;41:276-84
72. Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ. et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:4851-8
73. Rorive S, Sandras F, Biskri L, Fossion C, Catteau X, Roumeguere T. et al. PP103 RT-PCR-based UCA1 expression detection in urine samples as non-invasive reliable method for urothelial cancer diagnosis. European Journal of Cancer Supplements. 2009;7:28-9
74. Zhang Z, Hao H, Zhang CJ, Yang XY, He Q, Lin J. [Evaluation of novel gene UCA1 as a tumor biomarker for the detection of bladder cancer]. Zhonghua Yi Xue Za Zhi. 2012;92:384-7
75. Srivastava AK, Singh PK, Rath SK, Dalela D, Goel MM, Bhatt ML. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:11435-42
76. Wu W, Zhang S, Li X, Xue M, Cao S, Chen W. Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PloS one. 2013;8:e73920
77. Wang X, Gong Y, Jin B, Wu C, Yang J, Wang L. et al. Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncology reports. 2014;32:1281-90
78. Xue M, Li X, Wu W, Zhang S, Wu S, Li Z. et al. Upregulation of long non-coding RNA urothelial carcinoma associated 1 by CCAAT/enhancer binding protein alpha contributes to bladder cancer cell growth and reduced apoptosis. Oncology reports. 2014;31:1993-2000
79. Xue M, Li X, Li Z, Chen W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biology the Journal of the International Society for Oncodevelopmental Biology & Medicine. 2014;35:6901-12
80. Li Z, Li X, Wu S, Xue M, Chen W. Long non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase 2 through the mTOR-STAT3/microRNA143 pathway. Cancer science. 2014;105:951-5
81. Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F. et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. The FEBS journal. 2014;281:1750-8
82. Milowich D, Le Mercier M, De Neve N, Sandras F, Roumeguere T, Decaestecker C. et al. Diagnostic value of the UCA1 test for bladder cancer detection: a clinical study. SpringerPlus. 2015;4:349
83. Liu L, Liu Y, Zhuang C, Xu W, Fu X, Lv Z. et al. Inducing cell growth arrest and apoptosis by silencing long non-coding RNA PCAT-1 in human bladder cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:7685-9
84. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. Journal of molecular endocrinology. 2012;48:R45-53
85. Rivas A, Burzio V, Landerer E, Borgna V, Gatica S, Avila R. et al. Determination of the differential expression of mitochondrial long non-coding RNAs as a noninvasive diagnosis of bladder cancer. BMC urology. 2012;12:37
86. Brito GC, Fachel AA, Vettore AL, Vignal GM, Gimba ER, Campos FS. et al. Identification of protein-coding and intronic noncoding RNAs down-regulated in clear cell renal carcinoma. Molecular carcinogenesis. 2008;47:757-67
87. Qin C, Han Z, Qian J, Bao M, Li P, Ju X. et al. Expression pattern of long non-coding RNAs in renal cell carcinoma revealed by microarray. PloS one. 2014;9:e99372
88. Yu G, Yao W, Wang J, Ma X, Xiao W, Li H. et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PloS one. 2012;7:e42377
89. Fachel AA, Tahira AC, Vilella-Arias SA, Maracaja-Coutinho V, Gimba ER, Vignal GM. et al. Expression analysis and in silico characterization of intronic long noncoding RNAs in renal cell carcinoma: emerging functional associations. Molecular cancer. 2013;12:140
90. Blondeau JJ, Deng M, Syring I, Schrodter S, Schmidt D, Perner S. et al. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clinical epigenetics. 2015;7:10
91. Zhang HM, Yang FQ, Chen SJ, Che J, Zheng JH. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2947-55
92. Hiroshi H, Yuji H, Varahram S, Guoren D, Koichi N, Z Laura T. et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer research. 2015:75
93. Ariel I, Lustig O, Schneider T, Pizov G, Sappir M, De-Groot N. et al. The imprinted H19 gene as a tumor marker in bladder carcinoma. Urology. 1995;45:335-8
94. Ariel I, Sughayer M, Fellig Y, Pizov G, Ayesh S, Podeh D. et al. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Molecular pathology: MP. 2000;53:320-3
95. He W, Cai Q, Sun F, Zhong G, Wang P, Liu H. et al. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochimica et biophysica acta. 2013;1832:1528-37
96. Yan TH, Lu SW, Huang YQ, Que GB, Chen JH, Chen YP. et al. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:10249-57
97. Shang C, Guo Y, Zhang H, Xue YX. Long noncoding RNA HOTAIR is a prognostic biomarker and inhibits chemosensitivity to doxorubicin in bladder transitional cell carcinoma. Cancer chemotherapy and pharmacology. 2016;77:507-13
98. Zhao XL, Zhao ZH, Xu WC, Hou JQ, Du XY. Increased expression of SPRY4-IT1 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. International journal of clinical and experimental pathology. 2015;8:1954-60
99. Han Y, Liu Y, Nie L, Gui Y, Cai Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology. 2013;81:209 e1-7
100. Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Molecular bioSystems. 2012;8:2289-94
101. Zhang J, Zhang B, Wang T, Wang H. LncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: evidence from a meta-analysis. International Journal of Clinical & Experimental Medicine. 2015:8
102. Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F. et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:1531-41
103. Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM. et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. The Lancet Oncology. 2014;15:1469-80
104. Mehra R, Shi Y, Udager AM, Prensner JR, Sahu A, Iyer MK. et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16:1121-7
105. Auprich M, Chun FK, Ward JF, Pummer K, Babaian R, Augustin H. et al. Critical assessment of preoperative urinary prostate cancer antigen 3 on the accuracy of prostate cancer staging. European urology. 2011;59:96-105
106. Hessels D, van Gils MP, van Hooij O, Jannink SA, Witjes JA, Verhaegh GW. et al. Predictive value of PCA3 in urinary sediments in determining clinico-pathological characteristics of prostate cancer. The Prostate. 2010;70:10-6
107. Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D. et al. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015;13:209-21
108. Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC. et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742-9
109. Orfanelli U, Jachetti E, Chiacchiera F, Grioni M, Brambilla P, Briganti A. et al. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene. 2015;34:2094-102
110. Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A. et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5:764-74
111. Antti Y, Kati K, Annika K, Matti A, Leena L, Mauro S. et al. Transcriptome Sequencing Reveals PCAT5 as a Novel ERG-Regulated Long Noncoding RNA in Prostate Cancer. Cancer research. 2015:75
112. Wang L, Han S, Jin G, Zhou X, Li M, Ying X. et al. Linc00963: a novel, long non-coding RNA involved in the transition of prostate cancer from androgen-dependence to androgen-independence. International journal of oncology. 2014;44:2041-9
113. Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851-8
114. Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F. et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. The Journal of urology. 2013;190:2278-87
115. Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ. et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6:41045-55
116. Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M. et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nature structural & molecular biology. 2013;20:908-13
117. Malik R, Patel L, Prensner JR, Shi Y, Iyer MK, Subramaniyan S. et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Molecular cancer research: MCR. 2014;12:1081-7
118. Zhang HM, Yang FQ, Yan Y, Che JP, Zheng JH. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. International journal of clinical and experimental pathology. 2014;7:5801-9
119. Wang L, Cai Y, Zhao X, Jia X, Zhang J, Liu J. et al. Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma. 2015;62:412-8
120. Xue S, Li QW, Che JP, Guo Y, Yang FQ, Zheng JH. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. International journal of clinical and experimental pathology. 2015;8:3765-74
121. Song S, Wu Z, Wang C, Liu B, Ye X, Chen J. et al. RCCRT1 is correlated with prognosis and promotes cell migration and invasion in renal cell carcinoma. Urology. 2014;84:730 e1-7
122. Malouf GG, Zhang J, Yuan Y, Comperat E, Roupret M, Cussenot O. et al. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Molecular oncology. 2015;9:32-43
123. Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Current opinion in molecular therapeutics. 2010;12:607-16
124. Sidi AA, Ohana P, Benjamin S, Shalev M, Ransom JH, Lamm D. et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 over expressing superficial bladder cancer refractory to bacillus Calmette-Guerin. The Journal of urology. 2008;180:2379-83
125. Amit D, Tamir S, Birman T, Gofrit ON, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of IGF2-P3 and IGF2-P4 regulatory sequences. International journal of clinical and experimental medicine. 2011;4:91-102
126. Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pacific journal of cancer prevention: APJCP. 2013;14:1077-82
127. Liu Z, Wang W, Jiang J, Bao E, Xu D, Zeng Y. et al. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PloS one. 2013;8:e73991
128. Yacqub-Usman K, Pickard MR, Williams GT. Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor action in prostate cancer cells. The Prostate. 2015;75:693-705
129. Wang Y, Chen J, Li Q, Wang H, Liu G, Jing Q. et al. Identifying novel prostate cancer associated pathways based on integrative microarray data analysis. Computational biology and chemistry. 2011;35:151-8
130. Zhang W, Zang J, Jing X, Sun Z, Yan W, Yang D. et al. Identification of candidate miRNA biomarkers from miRNA regulatory network with application to prostate cancer. Journal of translational medicine. 2014;12:66
131. Cui W, Qian Y, Zhou X, Lin Y, Jiang J, Chen J. et al. Discovery and characterization of long intergenic non-coding RNAs (lincRNA) module biomarkers in prostate cancer: an integrative analysis of RNA-Seq data. BMC genomics. 2015;16(Suppl 7):S3
132. Tang Y, Yan W, Chen J, Luo C, Kaipia A, Shen B. Identification of novel microRNA regulatory pathways associated with heterogeneous prostate cancer. BMC systems biology. 2013;7(Suppl 3):S6
133. Klomp JA, Furge KA. Genome-wide matching of genes to cellular roles using guilt-by-association models derived from single sample analysis. BMC research notes. 2012;5:370
134. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667-78
135. Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA. et al. The genomic binding sites of a noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20497-502
Author contact
![]() Corresponding author: professor Bairong Shen, email: bairong.shenedu.cn.
Corresponding author: professor Bairong Shen, email: bairong.shenedu.cn.

 Global reach, higher impact
Global reach, higher impact