Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(8):973-983. doi:10.7150/jca.15118 This issue Cite
Research Paper
ELMO1 Directly Interacts with Gβγ Subunit to Transduce GPCR Signaling to Rac1 Activation in Chemotaxis
Chemotaxis Signaling Section, Laboratory of Immunogenetics, National Institute of Allergy and Infectious Diseases, National Institutes of Health; Rockville, MD 20852, USA.
* These authors contributed equally to this work.
Received 2016-1-28; Accepted 2016-3-14; Published 2016-5-12
Abstract
Diverse chemokines bind to G protein-coupled receptors (GPCRs) to activate the small GTPase Rac to regulate F-actin dynamics during chemotaxis. ELMO and Dock proteins form complexes that function as guanine nucleotide exchange factors (GEFs) for Rac activation. However, the linkage between GPCR activation and the ELMO/Dock-mediated Rac activation is not fully understood. In the present study, we show that chemoattractants induce dynamic membrane translocation of ELMO1 in mammalian cells. ELMO1 plays an important role in GPCR-mediated chemotaxis. We also reveal that ELMO1 and Dock1 form a stable complex. Importantly, activation of chemokine GPCR promotes the interaction between ELMO1 and Gβγ. The ELMO1-Gβγ interaction is through the N-terminus of ELMO1 protein and is important for the membrane translocation of ELMO1. ELMO1 is required for Rac1 activation upon chemoattractant stimulation. Our results suggest that chemokine GPCR-mediated interaction between Gβγ and ELMO1/Dock1 complex might serve as an evolutionarily conserved mechanism for Rac activation to regulate actin cytoskeleton for chemotaxis of human cells.
Keywords: chemotaxis, G protein-coupled receptors (GPCRs), heterotrimeric G protein, small GTPase Rho, guanine nucleotide exchange factors (GEFs), ELMO/Dock complex, Rac activation.
Introduction
Chemotaxis is directional cell migration mediated by a chemokine gradient. It plays essential roles in many physiological processes, such as recruitment of immune cells to inflammation sites, wound healing, neuron patterning, and metastasis of cancer cells (1-5). Cells use G-protein-coupled receptors (GPCRs) to sense and migrate toward a chemotactic gradient (3,6,7). Binding of a chemokine to its receptor induces the dissociation of heterotrimeric G-proteins into Gαi and Gβγ subunits, which in turn activate their downstream signaling components, such as Rac and the Arp2/3 complex. This leads to the growth of the dendritic actin-cytoskeleton in the leading front of a cell, resulting in cell migration (8-10). Consequently, activation of GPCRs promotes actin polymerization through the small GTPase Rac activation, which plays critical roles in chemotaxis (6,11-14). However, the linkage between the GPCRs/GαGβγ signaling machinery and the Rac-controlled actin polymerization has not been fully understood.
ELMO/Dock complexes are evolutionarily conserved and function as guanine nucleotide exchange factors (GEFs) to regulate Rac activation. The activation of Rac promotes the growth of actin filaments that drive cell migration (12,15-18). The activity of Rac GTPases is regulated by two large classes of proteins: GEFs and GTPase-activating proteins (GAPs). GEFs activate small GTPases by promoting nucleotide exchange from GDP to GTP; and GAPs bind to GTP-bound Rac proteins to enhance their intrinsic GTPase activity by which convert Rac proteins from the active GTP-bound sate to the inactive GDP-bound state (12,17-19). Activation of cell surface receptors stimulates the GEF activity of ELMO/Dock complexes to activate Rac proteins for cell migration (18,20). Active GTP-bound Rac proteins stimulate Arp2/3 complexes to promote actin polymerization for cell migration. ELMO proteins are evolutionarily conserved and are found in organisms ranging from Dictyostelium discoideum and Caenorhabditis elegans, to mouse and human (15,16,21,22). Cell surface receptors, such as tyrosine kinase receptors (TKRs) or GPCRs, regulate activities of ELMO/Dock complexes in response to extracellular stimuli (12,16,20,22). It has been shown that activation of TKRs induces the interaction between RhoG, a small GTPase, and ELMO (20). The ELMO/Dock complex activates Rac during phagocytosis of apoptotic cells in C. elegans or during neurite outgrowth stimulated by nerve growth factor (16,20). Recently, we found that activation of GPCRs triggers an association between free Gβγ and ElmoE, an ELMO isoform in D. discoideum, promoting the ElmoE/Dock complex to activate RacB which induces actin polymerization for cell migration (22). Many components of the GPCR-mediated signaling networks that control cell migration are highly conserved; so we attempted to test the hypothesis that a GPCR-controlled pathway, consisting of Gβγ, ELMO/Dock and Rac, also regulates the actin cytoskeleton in human cells. The goal was to reveal an evolutionarily conserved molecular mechanism underlying GPCR-mediated association between Gβγ and the ELMO/Dock complex in human cells.
In this study, we found that chemoattractants induce dynamic membrane translocation of ELMO1 in multiple human cell lines. We also found that ELMO1 is essential for GPCR-mediated migration of cancer cells. More importantly, ELMO1 directly interacts with Gβ upon chemoattractant stimulation. The interaction between ELMO1 and Gβ is through the N-terminus of ELMO1, and is important for the membrane translocation of ELMO1. Finally, we showed that ELMO1 is required for full Rac activation upon chemoattractant stimulation. These results indicate that ELMO1 directly transduces the signals from GPCR to activate Rac, thereby regulating F-actin dynamics during migration of human cancer cells.
Experimental Procedures
Plasmids and Antibodies
YFP-tagged ORF clone of ELMO1 was purchased from GeneCopoeia (Rockville, MD). YFP-tagged truncated mutants of ELMO1 were generated by a PCR-based approach and cloned into the Af1II/NotI sites of pReceiver-M15 (GeneCopoeia). The constructs were sequenced to confirm the appropriate mutations. Goat polyclonal anti-ELMO1 and goat polyclonal anti-ELMO2 antibodies were obtained from Abcam (Cambridge, MA). Mouse monoclonal Anti-DDK antibody came from Origene (Rockville, MD). Rabbit polyclonal anti-Gβ (M-14) antibodies and goat polyclonal anti-CXCR4 antibodies were obtained from Santa Cruz biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-Dock180 (C4C12) antibodies were from Cell Signaling Technology (Beverly, MA). Mouse monoclonal anti-RhoG (1F3 B3 E5) was purchased from Millipore (Billerica, MA). Mouse monoclonal anti-Rac1 was from Cytoskeleton (Denver, CO). Mouse monoclonal anti-GFP was purchased from Roche Applied Science (Penzberg, Germany). Mouse monoclonal anti-β-actin antibody was from Sigma Aldrich (St. Louis, MO). Horseradish peroxidase-conjugated anti-mouse, anti-rabbit, and anti-goat secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Recombinant Human/Rhesus Macaque/Feline CXCL12/SDF-1α and fibronectin were purchased from R&D systems (Minneapolis, MN)
Cell Culture and Transfections
HeLa cells were maintained in Roswell Park Memorial Institute 1640 (RPMI 1640) medium from Invitrogen (Grand Island, NY) containing 10% fetal bovine serum (Invitrogen) and 1 mM HEPES (Invitrogen) at 37ºC with 5% CO2. HeLa cells were transiently transfected by Xtreme-HP transfection reagent (Roche) according to the manufacturer's instructions. HL60 cells were differentiated as previously described (23). For imaging experiments, differentiated HL60 cells were seeded on the eight-well chambered cover slides from Nalge Nunc International (Penfield, NY) coated with 0.2% gelatin. The cells were starved for 4 hours and a final concentration of 100 nM fMLP was added to the chamber.
Immunoprecipitation
Cells (2.5 × 106) grown in 100 mm dishes were transfected with 10 µg of YFP or YFP-tagged full length (FL) or truncated mutants (M1-M4) of ELMO1 one day before the experiment. The cells were then placed in serum starvation conditions for 6 hours at 24 hour post-transfection and stimulated with SDF1α (100 ng/ml) for 20 min. The collected cells were incubated with 400 µl of lysis buffer containing 150 mM NaCl, 0.1% NP40, 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 X protease inhibitor cocktail (Roche), 10 mM NaF, 1 mM Na3VO4, and 1 mM PMSF. The mixture was centrifuged at 10,000 g at 4ºC for 20 min. The supernatants were incubated with the magnetic beads conjugated with anti-GFP antibodies and washed three times with washing buffer according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany).
Generation of ELMO1 Knockdown Cells
HeLa cells were infected with lentivirus expressing either non-specific control (CTL) or elmo1 specific shRNA. The mature sense is TGGACCAGTTCAAGAG CAA (Thermo Scientific, Barrington, IL). The positive clones were selected by puromycin (2.5 µg/ml) in the growth medium.
Chemotaxis Assays
Wound-healing Assay
HeLa cells (2 × 104) were seeded on 96 well plates coated with fibronectin (10 µg/ml). Scratching wounds were made with a Cellplayer Wound Maker from Essen Bioscience Inc. (Ann Habor, MI) 16 hours after plating. The wounds were washed twice with PBS and allowed to heal in the presence or absence of 100 ng/ml pertussis toxin (PT). Images of the wounds were taken every 2 hours for a duration of 30 hours. The relative wound density was determined by IncuCyte Software (Essen Bioscience Inc.) and the data was exported to GraphPad Prism 5 for statistical analysis.
Transwell Assay
Cell migration was assessed as described previously with minor modifications (24). Briefly, the bottom wells of the 48 well Micro Chemotaxis Chamber system (30 µl well plate, 5 µm filter pore size) from Neuro Probe (Gaithersburg, MD) were filled with 30 μl of the solutions containing 0.5% BSA, 1X PBS, and the indicated concentration of SDF-1α. The microplate was then covered with the ChemoTX filter. Cell suspension (25 μl) containing 5 × 104 Hela cells in PBS/0.5% BSA was placed onto the filter over each well, and incubated for 2 hours at 37°C with 5% CO2. After incubation, cells that transmigrated through the filter were stained on the lower filter surface with Hematoxylin Eosin Stain Kit from Fisher Scientific (Pittsburg, PA). The average number of cells on the lower filter surface was determined by counting the number of cells in three random non-overlapping high power (×400) fields by light microscopy.
Micropipette Chemotaxis Assay
Cells were seeded in a 1 well chamber (Nalge Nunc International) coated with 0.2% gelatin and starved for 4 hours before the experiments. Cells were then imaged with a Zeiss Laser Scanning Microscope 710 with a 40×, 1.3 NA Plan-Neofluar objective lense. To visualize the gradient, chemoattractants were mixed with fluorescent dye Alex 633 from Thermo Scientific (Waltham, MA) and released from a microinjector (Eppendorf, Hamburg, Germany) connected to a Femtotips (Eppendorf).
Rac1 Activation Assay
Experimental procedures followed the manufacturer's instructions (Cytoskeleton, Denver, CO). Briefly, beads were incubated with the cell lysates prepared by the indicated conditions for 2 hours at 4˚C and then washed three times with ice cold lysis buffer. The bound Rac-GTP with beads was analyzed by SDS-PAGE, followed by western blot detection for Rac1 protein.
Microscopy
Alexa Fluor 594 was added to the stimulant solution as an indicator. Images were taken with a 40x oil immersion lense (1.3 NA Plan-Neofluar objective lense) on a LSM 710 Laser Scanning Confocal Microscope (Carl Zeiss, Göttingen, Germany) and analyzed with ZEN 2010B SP1 Software (Zeiss, Thornwood, NY) and Adobe Photoshop (San Jose, CA).
Calcium Response Assay
The cells were placed in starvation conditions for 18 hours with DMEM medium before incubation with a final concentration of 5 µg/ml Fluo4 AM (Invitrogen, Inc.) for 30 min with or without pertussis toxin (PT) at indicated concentration. The cells were then washed with DMEM medium with or without PT three times at 10 min intervals. A final concentration of 100 ng/mL SDF1α mixed with fluorescent dye Alex 633 Images were added to the cells to induce a calcium response. Images were taken and analyzed as detailed in the Microscopy section above.
Results
Chemokine GPCR signaling controls the cellular localization of ELMO1
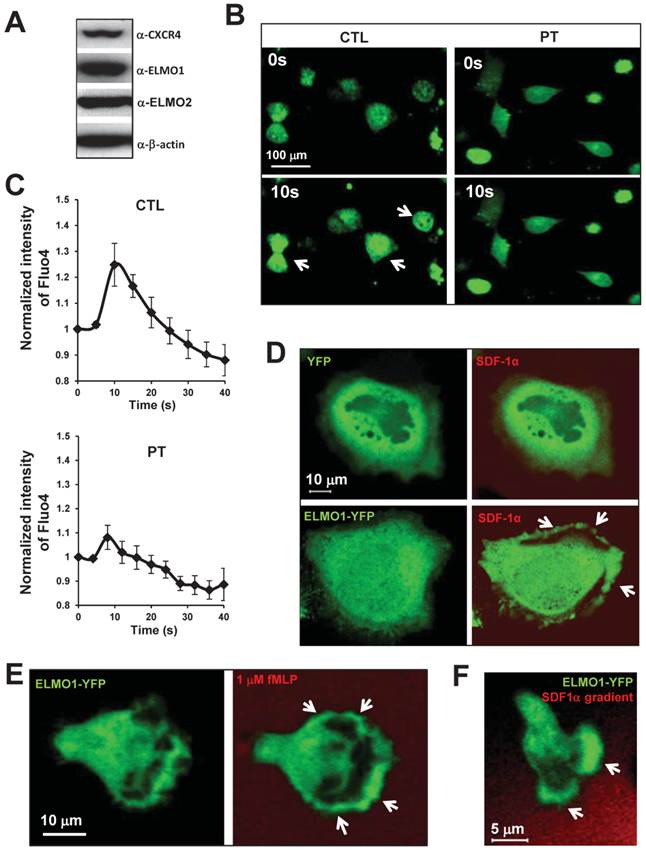
To determine whether ELMO proteins are regulated by chemokine GPCR signaling, we examined the cellular localization of ELMO1 in response to chemoattractant stimulation in various human cell lines. HeLa cells highly expressed ELMO1/2 proteins and also chemokine receptor CXCR4 (Figure 1A). Upon SDF1α (a ligand of CXCR4) stimulation, HeLa cells displayed a robust calcium response (Figure 1B-1C). The treatment of pertussis toxin (PT), a drug that blocks Gαi signaling, significantly inhibited the calcium response. Our results, as expected, indicated that SDF1α/CXCR4 controlled Gαi-dependent signaling events, such as the calcium response in the HeLa cells. We then examined the cellular localization of ELMO1-YFP in HeLa cells upon SDF1α stimulation. Before SDF1α stimulation, ELMO1-YFP was mostly distributed in the cytosol. SDF1α stimulation induced the membrane translocation of ELMO1-YFP, but not the YFP alone (Figure 1D). In human neutrophil-like HL60 cells, fMLP triggered rapid and robust membrane translocations of ELMO1-YFP (Figure 1E). When HL60 cells chemotaxed in an SDF1α gradient, ELMO1-YFP was also actively recruited to the leading edge of cells (Figure 1F). These results indicate that chemokine GPCR signaling controls the cellular localization of ELMO1 in human cells.
Chemokine GPCR signaling controls the cellular localization of ELMO1. A. HeLa cells highly express ELMO1/2 proteins and chemokine receptor CXCR4. B. Montage shows calcium response of HeLa cells with or without pertussis toxin (PT) treatment upon the stimulation of 100 ng/ml SDF1α. The cells were treated with calcium indicator 1 μM Fluo4 (Green, Molecular Probe) 30 min before the experiments. Arrows point to the cells display strong calcium response. The quantitation of calcium response in each group of cells is presented in C. The fluorescent intensity of cells at time 0 is normalized to 1. Mean ± SD is shown, n=7 or n=10 in CTL or PT-treated cells, respectively. D. Montage shows membrane targeting of YFP tagged ELMO1 (Green) upon chemoattractant SDF1α (Red) stimulation. HeLa cells were transiently transfected with plasmids encoded YFP only or YFP tagged with ELMO1. 100 ng/ml SDF1α mixed with fluorescent dye Alexa 633 (Red) was uniformly applied to the cells. E. Robust membrane translocation of ELMO1-YFP in HL-60 cells upon a uniformly applied chemoattractant fMLP stimulation. 1 μM fMLP mixed with Alexa 633 (Red) was applied to HL60. F. Leading edge localization of ELMO1 in chemotaxing HL-60 cells in a SDF1α gradient. In D-F, arrows point on the sites of cells with ELMO1-YFP.
ELMO1 plays a role in chemokine GPCR-mediated cell migration
To reveal the potential role of ELMO1 in chemokine GPCR-mediated cell migration, we established an ELMO1 knockdown cell line (elmo1kd), in which the expression of ELMO1 was stably knocked down using an elmo1 specific lentiviral shRNA (Figure 2A). ELMO1 shRNA specifically knocked down the expression of ELMO1, but not that of ELMO2 or CXCR4 (Figure S1A). We also established a control cell line (CTL), which was stably infected by lenti-virus particles encoding non-specific shRNAs (Figure 2A). SDF1α stimulation triggered robust calcium responses in both CTL and elmo1kd cells (Figure S1B-S1C), indicating that some of the CXCR4-mediated signaling were not affected by knocking down ELMO1 expression.
ELMO1 plays a role in GPCR mediated cell migration. A. The establishment of ELMO1 stably knocked down cell lines (elmo1kd) by elmo1 specific Lentiviral shRNA technology. Control (CTL) or elmo1 (elmo1 kd) specific targeting lentivirus was used to transfect HeLa cells. B. Montage shows scratched wounds at 0 hour (0 H) and healed wounds after 17 hours (17 H) in CTL or elmo1kd cells in the presence or absence of 100 nM pertussis toxin (PT) treatment. Its quantitative measurement at hour 17 is presented in C. Statistical analysis are from two independent experiments with 10 samples in each group, p<0.01. D. Montage shows the chemotaxed CTL or elmo1kd cells by transwell chemotaxis assay with indicated concentration of SDF1α gradient. Quantitative measurement of cell counts per view is presented in E. Statistical analysis is from three independent experiments with 6 samples in each group, p<0.01.
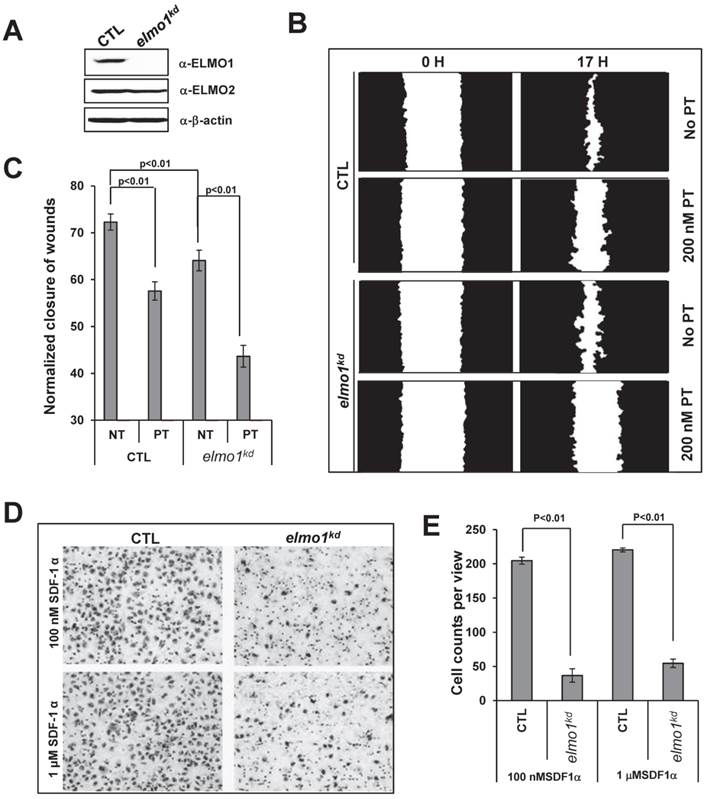
We then examined the role of ELMO1 in cell migration. Using a wound healing assay, we found that elmo1kd cells showed slower wound closure when compared to CTL cells (Figure 2B-2C). Interestingly, we found that PT treatment significantly decreased the wound closure of CTL cells, indicating that wound healing process involves GPCR/Gi-mediated cell migration. We also used a Transwell assay to further examine the role of ELMO1 in SDF1α-mediated cell migration (Figure 2D-2E). CTL cells migrated across the membrane toward SDF1α, whereas elmo1kd cells were significantly impaired in SDF1α-mediated migration. Our results suggest that ELMO1 plays a role in CXCR4/Gi-mediated cell migration.
ELMO1/Dock1 complex interacts with Gβ upon activation of a chemokine GPCR
Previous studies have shown that ELMO1 and Dock1 form a complex that interacts with RhoG (20). Our previous work has shown that activation of a chemoattractant GPCR promotes an interaction between the β subunit of heterotrimeric G-protein and an ELMO/Dock complex in the eukaryotic model organism, Dictyostelium (22). To reveal potential molecular mechanisms of ELMO1 function in mammalian chemokine GPCR signaling, we sought to examine the potential interactions between ELMO1, Dock1, RhoG, and G-protein subunits in response to chemokine stimulation by immunoprecipitation (Figure 3A-3B).
ELMO1/Dock1 complex interacts with Gβ upon GPCR activation. A. ELMO1/Dock1 complex interacts with Gβ subunit upon chemoattractant stimulation determined by immunoprecipitation using anti-GFP antibody conjugated magnetic beads as bait. Western-blot detection of Dock1, RhoG, and Gβ is also shown. B. Quantitative measurement of the interaction of ELMO1 and its interacting partners in panel A. Mean ± SD from two independent experiment is shown. C. Montage shows a decreased membrane translocation of YFP-ELMO1 (Green) in PT treated HeLa cells in response to SDF1α stimulation. Fluorescent dye Alexa 633 (Red) containing 100 ng/ml SDF1α was uniformly applied to HeLa cells.
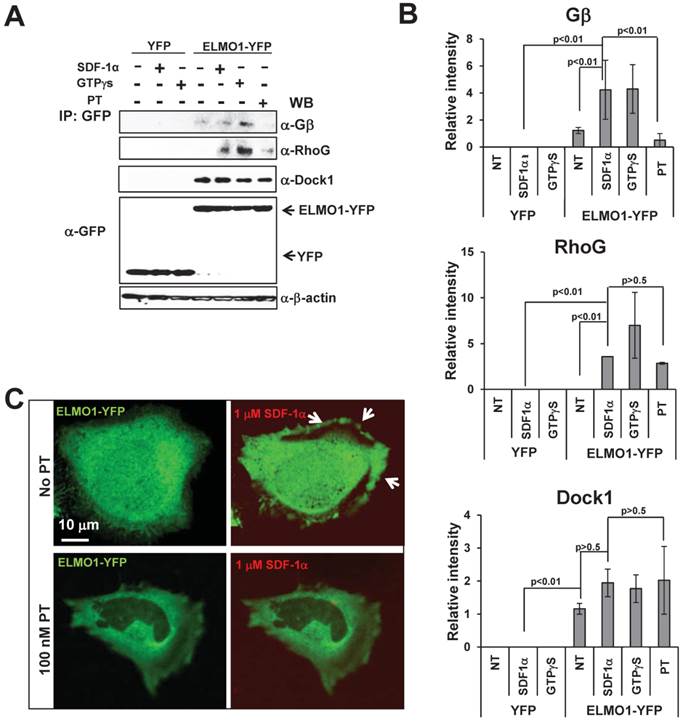
Lysates of HeLa cells expressing ELMO1-YFP or YFP alone at indicated conditions were incubated with anti-GFP beads to identify ELMO1-YFP interacting molecules. Using western blot analyses with antibodies against pan-Gβ, RhoG, or Dock1 proteins, we found that ELMO1 and Dock1 formed a stable complex independent of either chemokine GPCR signaling (- or +SDF1α) or Gi signaling (+GTPγS or +PT). Interestingly, activation of CXCR4 receptor (+SDF1α) or Gi proteins (+GTPγS) promoted association between the ELMO1/Dock1 complex with Gβ as well as RhoG. When Gi signaling was blocked (+PT), the interactions between the ELMO1/Dock1 complex with Gβ and RhoG was abolished (Figure 3A-3B). Furthermore, treatment of PT also reduced SDF1α-induced membrane translocation of ELMO1 (Figure 3C). Our data suggested that SDF1α/CXCR4 signaling activates heterotrimeric Gi proteins to promote the association of Gβγ and RhoG with the ELMO1/Dock1 complex. Additionally, it also induces the membrane translocation of the ELMO1/Dock1 complex.
The N-terminal region of ELMO1 is required for the ELMO1/Gβ interaction
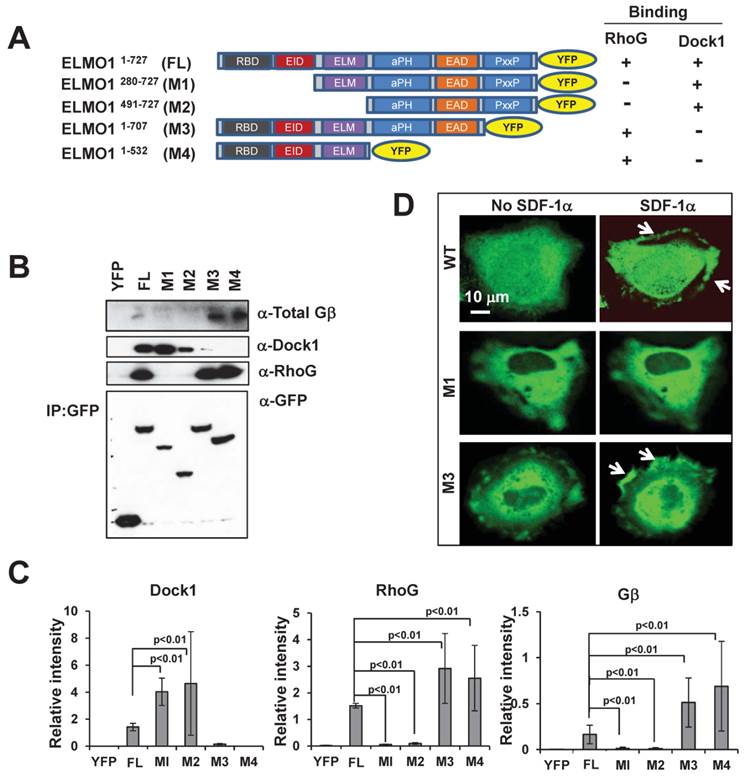
To understand the molecular mechanism of the ELMO1-Gβ interaction, we determined the regions of ELMO1 that interact with Gβ, RhoG and Dock1. Using a bioinformatics approach, we identified six domains in ELMO1: RBD, EID, ELM, aPH, EAD and PxxP (Figure 4A). We generated four YFP-tagged full length (FL) or truncated mutants (M1-M4) of ELMO1 and examined their ability to interact with RhoG, Dock1, and Gβ. In co-immunoprecipitation assay using lysates from cells expressing YFP-tagged FL, M1, M2, M3, and M4 (Figure 4A-4C), we found that two mutants, M1, which lacks the RBD and EID domains, and M2, which lacks the RBD, EID, and ELM domains, did not interact with RhoG, but could associate with Dock1. Conversely, mutant M3 that lacks PxxP and mutant M4 that lacks PH, EAD and PxxP, were able to interact with RhoG but failed to interact with Dock1. More importantly, we found that M1 and M2 also failed to associate with Gβ, while M3 and M4 still maintained the ability to bind Gβ (Figure 4B-4C). These results indicate that the 280 amino acids at the N-terminal of ELMO1 are required for the ELMO1-Gβ association. To understand the role of ELMO1-Gβ interaction in ELMO1 membrane targeting, we further examined the membrane translocation of ELMO1 mutants upon SDF1α stimulation. We found that M1, which was not able to interact with Gβ, displayed a decreased level of membrane translocation, while M3, which interacted with Gβ and RhoG but not Dock1, was still able to translocate to the membrane (Figure 4D). These results suggest that the N-terminal region (1-280 a.a) of ELMO1 is required for the ELMO1-Gβγ and ELMO1-RhoG interactions and is essential for the membrane targeting of ELMO1Complex upon the activation of CXCR4.
ELMO1 is required for chemokine-induced Rac activation and co-localizes with actin filaments
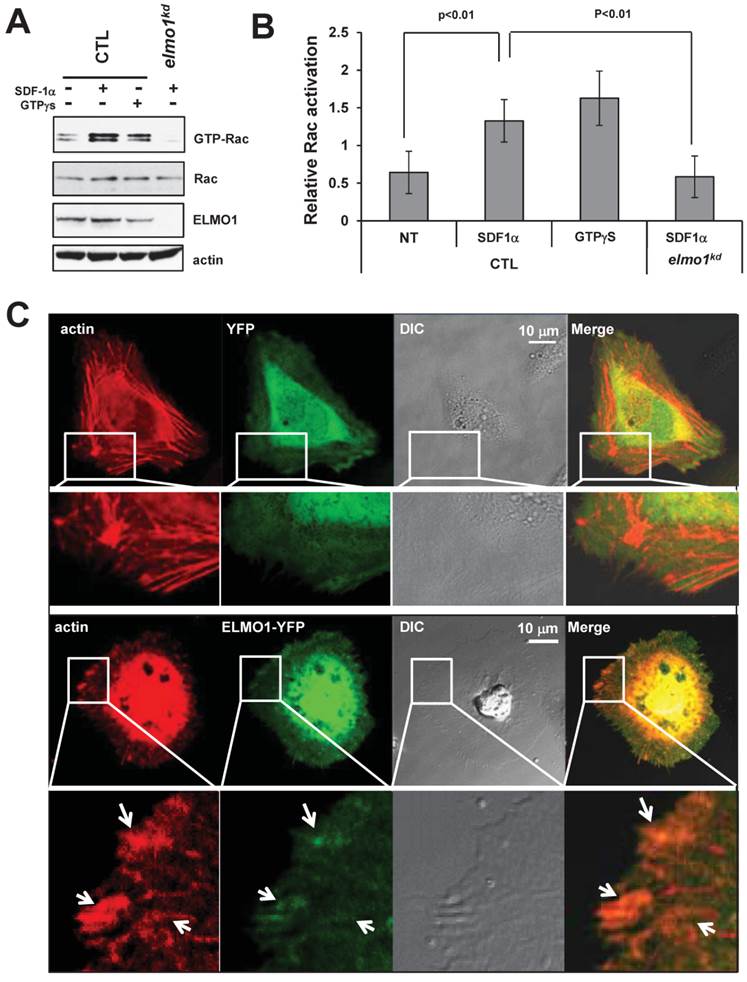
It has been shown that ELMO1 and Dock1 form a complex to function as a GEF to trigger Rac1 activation (20). To determine the role of ELMO1 in the activation of Rac in response to SDF-1α stimulation, we compared the level of Rac-GTP formed in CTL and elmo1kd cells (Figure 5A-5B). We found that treatment with either SDF1α or GTPγS induced a robust Rac1 activation in CTL cells. However, SDF1α failed to induce a clear Rac1 activation in elmo1kd cells (Figure 5A-5B), indicating that ELMO1 plays an important role in CXCR4-mediated Rac1 activation.
Next, we examined the cellular localization of ELMO1 and the actin cytoskeleton in live cells. We co-transfected HeLa cells with vectors expressing mCherry-tagged actin and ELMO1-YFP or YFP alone as a control. We found that ELMO1-YFP (Green) localized with actin-filaments (Red) at the sites of the protrusions (Figure 5C), while YFP localized only in the cytosol. We also found that ELMO1 was recruited to the leading front where actin polymerization took place in chemotaxing cells in an SDF1α gradient (Figure 1F). These results indicate that ELMO1 might be commonly involved in the GPCR-mediated directional cell migration in human cells. The accumulation of ELMO1 at the leading front is consistent with the notion that ELMO1 may play a role in localizing Rac activation to promote the growth of actin polymerization to drive cell migration.
Discussion
Our previous study showed that the cAR1 GPCR regulates functions of an ELMO/Dock complex via the association between Gβγ and an ELMO protein in the model organism D. discoideum (22). Our current study revealed for the first time that a mammalian ELMO protein serves as a Gβγ effector by transducing signals from a chemokine GPCR to activate Rac leading to cell migration. The molecular mechanism underlying the association between Gβγ and ELMO1/Dock1 complex in human cells shares similarity with the discovery in D. discoideum. Specifically, we found that the activation of chemokine GPCR induces membrane translocation of ELMO1 and promotes the association between Gβγ and the ELMO1/Dock1 complex. In addition, we discovered that the ELMO1/Gβ interaction is through the N-terminal region of ELMO1 and is important for ELMO1 translocation to the plasma membrane where the ELMO1/Dock1 complex activates Rac. We showed that ELMO1 and Dock1 form a stable complex regardless of stimulations as expected (20). However, this association seems dispensable for the membrane translocation of ELMO1. We also showed that ELMO1 is required for Rac1 activation in response to chemoattractant GPCRs activation, which is consistent with previous studies (14,22).Based on these results, we propose that the Gβγ-ELMO1/Dock1 interaction might serve as an evolutionarily conserved mechanism for the GPCR-mediated rapid remodeling of actin cytoskeleton for chemotaxis from D. discoideum to human cells (Figure 6).
N-terminal of Elmo1 is required for ELMO1-Gβ interaction. A. Scheme shows YFP-tagged full length (FL) and its truncated mutants (M1-M4) of ELMO1 used for immunoprecipitation experiments. ELMO1 protein contains six major functional domains: Ras binding domain (RBD), ELMO inhibitory domain (EID), ELMO domain (ELM), atypical PH Domain (aPH), ELMO auto-regulatory domain (EAD), and proline-rich domain (PxxP). The binding characteristics of each construct with regard to RhoG and Dock1 are also indicated. B. The N-terminal of ELMO1 is required for the interaction between Gβ and ELMO1. The expression of YFP control, FL-ELMO1, or truncated mutants (M1-M4) of ELMO1 is shown in the lowest panel. Western-blot detection of ELMO1 interacting molecules of RhoG, Dock1, and Gβ are also shown. C. Quantitation of the interaction between Gβ and WT or mutants of ELMO1 shown in panel A. The density of each protein was normalized by its expression level in total cell lysates detected with an anti-GFP antibody. Mean ± SD from two independent experiments is shown. D. Montage shows the membrane translocation of WT ELMO1 and its mutants upon 100 ng/ml SDF1α stimulation. Arrows point on the sites of cells with ELMO1-YFP or its mutant.
Rac activation is impaired in elmo1kd cells upon chemokine stimulation. A. In vivo measurement of Rac activation in control (CTL) and elmo1 knocked-down (elmo1kd) cells upon SDF1α stimulation. The total cell lysates of CTL and elmo1kd cells were probed for the expression of total Rac, ELMO1, and β-actin. Active Rac was purified by pull-down assay using a specific antibody against active Rac1 as detailed in Material and Methods. B. Quantification of Rac activation in cells upon stimulation is shown. The means ± SD of three independent experiments is shown. The intensity of each GTP-Rac band is normalized by that of the total Rac. C. Montage shows the colocalization of ELMO1 and actin in the HeLa cells. YFP only or YFP-tagged ELMO1 and mCherry-tagged actin were co-expressed in the Hela cells. Arrows point at the colocalization sites of EMLO1 and actin in the cell.
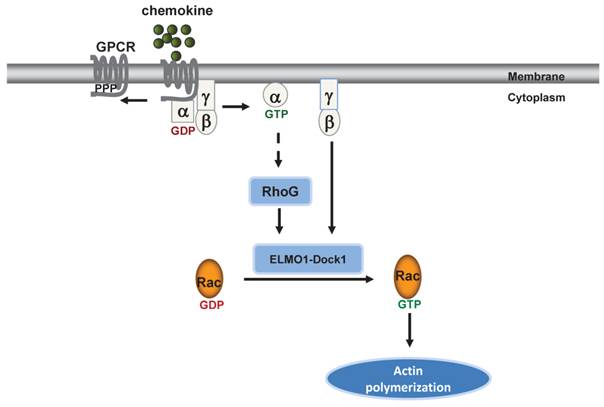
ELMO1 directly transduces GPCR signaling to Rac1 to regulate F-actin dynamics in chemotaxing cells. GPCR activation induces several signaling pathways to promote actin polymerization in human cells.
We showed that the N-terminal region of ELMO1 interacts with Gβ subunit. Previous studies have uncovered different domains of the ELMO proteins and proposed their roles for auto-inhibitory regulation of ELMO functions (25,26). It has been shown that the PH domain (residues 555-676) of ELMO1 contributes to the formation of a trimeric complex with Dock1 and the nucleotide-free Rac, and is critical for the Rac-activating function of ELMO/DOCK complex (27). ELMO1 binds Dock1 through its last 200 C-terminal residues, which include the proline-rich domain (28). In fact, new lines of evidence suggested that this PxxP domain contributes to the increased affinity and half-life of the ELMO1/Dock1 complex (29). Interestingly, we found a stronger association between Gβ and the mutant lacking the C-terminal PH, EAD and PxxP domains of ELMO1. It is possible that the interaction between Gβ and ELMO1 or between RhoG and ELMO1 is more efficient without the ELMO1-binding domain for Dock1. Since they do not share a common binding interface, Gβ and RhoG should not directly compete with Dock1 for binding ELMO1. An auto-inhibitory switch through a Ras-Binding Domain (RBD), ELMO-inhibitory Domain (EID), and ELMO-Autoregulatory Domain (EAD) has been discovered (30). The intramolecular interaction of EID and EAD folds the protein and negatively regulates the function of ELMO proteins. Meanwhile, RhoG and Dock1 compete with the EAD and RBD-EID for binding, respectively, interrupt the other components in the auto-inhibitory switch to relieve the negative regulation (30). The importance of ELMO1 conformational control has been highlighted by disruption of ELMO auto-inhibition and the consequential promotion of DOCK1 and Rac-dependent cell migration (30). We discovered that ELMO1 mutants missing the proline-rich domain (PxxP) retained the ability to bind Gβ and RhoG, and that the Gβ-binding site is encompassed in the RBD and EID regions. We propose that, upon SDF-1α stimulation, free Gβγ binds ELMO1 through its N-terminal regions, competing with Ras, RhoG, and Arf family GTPase, such as Arl4A, for membrane recruitment. Once activated, the ELMO1/Dock1 complex is recruited to the membrane by RhoG and Arl4A, leading to Rac-GTP loading and membrane protrusions.
In this study, we found that ELMO1 knockdown led to a defect in tumor cell migration and prolonged the gap closure time in a wound-healing assay. This result agrees with the previous report that ELMO1 is important for Rac-directed cell migration (14,18). We also discovered that Knocking down ELMO1 impaired SDF-1α mediated Rac-activation and directional membrane protrusions. HeLa cells are not highly mobile, but there was a clear difference between ELMO-silenced cells and control cells in both membrane ruffles and pseudopodia generated in response to the chemoattractant gradient. Therefore, ELMO1 plays an important role in GPCR-mediated directional cell migration, and this function correlates with ELMO1's direct interaction with the G protein β subunit. These findings demonstrate a novel pathway for GPCR-mediated cell signaling during tumor cell migration. We have previously found that in breast cancer cells, CXCR4 regulates the ELMO1/Dock1 complex via Gαi2 to activate Rac for metastasis of breast cancer cells (14). It appears that chemokine GPCRs activate Gαi and Gγβ subunits, each of which is able to interact with the ELMO1/Dock1 complex to activate Rac and to promote the migration of different cancer cells. Future studies are needed to reveal molecular mechanisms underlying the interactions between Gαi or Gβγ and the ELMO1/Dock1 complex in different cancer cells, and to establish the value of signaling components in these pathways as biomarkers for the diagnosis and as targets for the treatment of various cancers.
Abbreviations
GPCR: G-protein-coupled receptor; GEFs: guanine nucleotide exchange factors; SDF1α: stromal cell-derived factor 1 α (); PT: pertussis toxin; CXCR4: chemokine receptor type 4.
Supplementary Material
Figure S1.
Acknowledgements
This study was supported by NIAID/NIH intramural funds. We thank Dr. Bryan Fleming (NCI) for editorial assistance.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Jin T, Xu X, Hereld D. Chemotaxis, chemokine receptors and human disease. Cytokine. 2008;44:1-8 doi:10.1016/j.cyto.2008.06.017
2. Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME. et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50-6 doi:10.1038/35065016
3. Afonso PV, Janka-Junttila M, Lee YJ, McCann CP, Oliver CM, Aamer KA. et al. LTB4 is a signal-relay molecule during neutrophil chemotaxis. Developmental cell. 2012;22:1079-91 doi:10.1016/j.devcel.2012.02.003
4. Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu Rev Immunol. 2012;30:69-94 doi:10.1146/annurev-immunol-020711-075011
5. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678-89 doi:10.1038/nri2156
6. Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nature reviews Molecular cell biology. 2004;5:626-34 doi:10.1038/nrm1435
7. Murphy PM. International Union of Pharmacology. XXX. Update on chemokine receptor nomenclature. Pharmacological reviews. 2002;54:227-9
8. Fujiwara I, Remmert K, Piszczek G, Hammer JA. Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1970-9 doi:10.1073/pnas.1313738111
9. Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Developmental cell. 2009;17:310-22 doi:10.1016/j.devcel.2009.08.012
10. Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453-65
11. Dong X, Mo Z, Bokoch G, Guo C, Li Z, Wu D. P-Rex1 is a primary Rac2 guanine nucleotide exchange factor in mouse neutrophils. Current biology: CB. 2005;15:1874-9 doi:10.1016/j.cub.2005.09.014
12. Cote JF, Vuori K. GEF what? Dock180 and related proteins help Rac to polarize cells in new ways. Trends Cell Biol. 2007;17:383-93 doi:10.1016/j.tcb.2007.05.001
13. Li Z, Hannigan M, Mo Z, Liu B, Lu W, Wu Y. et al. Directional sensing requires G beta gamma-mediated PAK1 and PIX alpha-dependent activation of Cdc42. Cell. 2003;114:215-27
14. Li H, Yang L, Fu H, Yan J, Wang Y, Guo H. et al. Association between Galphai2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nature communications. 2013;4:1706. doi:10.1038/ncomms2680
15. Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC. et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nature cell biology. 2002;4:574-82 doi:10.1038/ncb824
16. Reddien PW, Horvitz HR. The engulfment process of programmed cell death in caenorhabditis elegans. Annual review of cell and developmental biology. 2004;20:193-221 doi:10.1146/annurev.cellbio.20.022003.114619
17. Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Developmental cell. 2001;1:491-502
18. Grimsley CM, Kinchen JM, Tosello-Trampont AC, Brugnera E, Haney LB, Lu M. et al. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. The Journal of biological chemistry. 2004;279:6087-97 doi:10.1074/jbc.M307087200
19. Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annual review of cell and developmental biology. 2005;21:247-69 doi:10.1146/annurev.cellbio.21.020604.150721
20. Katoh H, Negishi M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 2003;424:461-4 doi:10.1038/nature01817
21. Cote JF, Motoyama AB, Bush JA, Vuori K. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nature cell biology. 2005;7:797-807 doi:10.1038/ncb1280
22. Yan J, Mihaylov V, Xu X, Brzostowski JA, Li H, Liu L. et al. A Gbetagamma effector, ElmoE, transduces GPCR signaling to the actin network during chemotaxis. Developmental cell. 2012;22:92-103 doi:10.1016/j.devcel.2011.11.007
23. Millius A, Weiner OD. Chemotaxis in neutrophil-like HL-60 cells. Methods Mol Biol. 2009;571:167-77 doi:10.1007/978-1-60761-198-1_11
24. Saini V, Staren DM, Ziarek JJ, Nashaat ZN, Campbell EM, Volkman BF. et al. The CXC chemokine receptor 4 ligands ubiquitin and stromal cell-derived factor-1alpha function through distinct receptor interactions. The Journal of biological chemistry. 2011;286:33466-77 doi:10.1074/jbc.M111.233742
25. Patel M, Pelletier A, Cote JF. Opening up on ELMO regulation: New insights into the control of Rac signaling by the DOCK180/ELMO complex. Small GTPases. 2011;2:268-75 doi:10.4161/sgtp.2.5.17716
26. Lu M, Kinchen JM, Rossman KL, Grimsley C, Hall M, Sondek J. et al. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Current biology: CB. 2005;15:371-7 doi:10.1016/j.cub.2005.01.050
27. Lu M, Kinchen JM, Rossman KL, Grimsley C, deBakker C, Brugnera E. et al. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nature structural & molecular biology. 2004;11:756-62 doi:10.1038/nsmb800
28. Komander D, Patel M, Laurin M, Fradet N, Pelletier A, Barford D. et al. An alpha-helical extension of the ELMO1 pleckstrin homology domain mediates direct interaction to DOCK180 and is critical in Rac signaling. Molecular biology of the cell. 2008;19:4837-51 doi:10.1091/mbc.E08-04-0345
29. Sevajol M, Reiser JB, Chouquet A, Perard J, Ayala I, Gans P. et al. The C-terminal polyproline-containing region of ELMO contributes to an increase in the life-time of the ELMO-DOCK complex. Biochimie. 2012;94:823-8 doi:10.1016/j.biochi.2011.11.014
30. Patel M, Margaron Y, Fradet N, Yang Q, Wilkes B, Bouvier M. et al. An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Current biology: CB. 2010;20:2021-7 doi:10.1016/j.cub.2010.10.028
Author contact
![]() Corresponding author: tjinnih.gov
Corresponding author: tjinnih.gov

 Global reach, higher impact
Global reach, higher impact