Impact Factor
ISSN: 1837-9664
J Cancer 2016; 7(4):462-466. doi:10.7150/jca.13566 This issue Cite
Research Paper
Newcastle Disease Virus Hemagglutinin Neuraminidase as a Potential Cancer Targeting Agent
1. Department of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang, Selangor Darul Ehsan, Malaysia;
2. Department of Microbiology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Serdang, Selangor Darul Ehsan, Malaysia;
3. Institute of Bioscience, Universiti Putra Malaysia, Serdang, Selangor Darul Ehsan, Malaysia.
Received 2015-8-16; Accepted 2015-12-3; Published 2016-1-29
Abstract
The hemagglutinin-neuraminidase (HN) protein of Newcastle disease virus (NDV) with its immunotherapeutic activities and sialic acid binding abilities is a promising cancer adjuvant. The HN was surfaced displayed on Lactococcus lactis and its cancer targeting ability was investigated via attachment to the MDA-MB231 breast cancers. To surface display the HN protein on the bacterial cell wall, HN was fused to N-acetylmuraminidase (AcmA) anchoring motif of L. lactis and expressed in Chinese hamster ovary cells. The expressed recombinant fusion proteins were purified and mixed with a culture of L. lactis and Lactobacillus plantarum. Immunofluorescence assay showed the binding of the recombinant HN-AcmA protein on the surface of the bacterial cells. The bacterial cells carrying the HN-AcmA protein interacted with the MDA-MB231 breast cancer cells. Direct and fluorescent microscopy confirmed that L. lactis and Lb. plantarum surface displaying the recombinant HN were attached to the breast cancer MDA-MB231 cells, providing evidence for the potential ability of HN in targeting to cancer cells.
Keywords: Hemagglutinin-neuraminidase, Cancer targeting, Lactococcus lactis, Surface display.
Introduction
Newcastle disease virus (NDV) is a paramyxovirus with potential oncolytic and apoptotic properties. It is able to replicate selectively inside cancer cells [1, 2]. Its heamaglutinin-neuraminidase (HN) protein is a multifunctional protein. It plays a pivotal role in the pathogenesis, immunogenicity and attachment of the virus [3, 4]. It is involved in mediating the attachment of the virus to sialic acid-containing cellular receptors. This protein consists of two domains associated with the hemagglutinin and neuraminidase activities which mediate the recognition as well as cleavage of the sialic acid containing receptors [5].
Several studies highlight the potential adjuvant activity of the HN in cancer therapy [6-8]. The HN protein can exert T cell co-stimulatory function as well as boost innate immunity [8, 9]; HN can stimulate dendritic cells to produce high levels of interferon-α and activate macrophages as well as stimulate the tumor necrosis factor-related apoptosis-inducing ligand immune response [9, 10]. In addition, HN has the ability to stimulate the NK cell activity and expedite adhesive interactions of lymphocytes with infected tumor cells [11]. Altogether, these characteristics make HN an adjuvant of interest in cancer immunotherapy.
In the present study, we report the targeting ability of the HN against MDA-MB231breast cancer cells. To achieve this, the binding domain of AcmA, anchor motif of L. lactis, was used to surface display the HN protein onto the bacterial cell surface which was afterward subjected to interact with MDA-MB231 breast cancer cell. The FreeStyle ™ MAX CHO Expression based system was chosen to be used to produce and display the antigen-anchor fusion proteins on the cell surface of L. lactis.
Materials and Methods
Bacterial strain, cell line and growth media
The L. lactis NZ9000 and recombinant L. lactis NZ9000 carrying GFP [12] were propagated in M17 containing 0.5% (w/v) glucose at 30oC. The FreeStyle™ CHO-S® cells were cultured in FreeStyle™ CHO expression medium supplemented with L-glutamine to a final concentration of 8 mM at 37oC in 5% CO2 atmosphere.
Construction of HN-acmA cassette
PCR was used to amplify the coding region of HN and acmA gene. The acmA (from genomic DNA of L. lactis MG1363; Accession no: LLU17696) and HN (from pCite2a-HN plasmid, Virology lab, Department of Microbiology, Universiti Putra Malaysia) DNA fragments were amplified and digested with XhoI before being ligated to make a surface display cassette. Then the HN-acmA cassette and pcDNA 3.1 expression vector were digested with BamHI and XbaI (Fermentas, USA). Ligation was achieved by using T4 DNA ligase (Fermentas, USA). The ligation mixture was purified using the Wizard® PCR Preps DNA Purification Systems (Promega, USA) before the recombinant plasmids were transformed into FreeStyle™ CHO-S® cells.
Expression of HN-acmA cassette
Protein expression in FreeStyle™ CHO-S® cells
The transformation of suspension FreeStyle™ CHO-S cells was done according to the manufacture's instruction (Invitrogen™, Life Technologies; USA). Five days prior to post transfection, the transfected FreeStyle™ CHO-S® cells were subcultured in culture medium containing 100 µg/ml geneticin. After approximately two passages, the culture was monitored microscopically for the presence of viable cells. The positively transfected cells were harvested by centrifuge at 2500 ×g at 4oC for 5 min; and the cells were resuspended in the appropriate amount of CytoBuster (Novagen, USA), at room temperature for 5 min. The mixture was then centrifuged for 10 min at 16000 ×g to separate the cell from the soluble protein.
Purification and Western blot analysis of recombinant protein
The Ni-NTA purification system was used to purify the expressed His-tagged recombinant protein according to the manufacture's instruction (Novagen, USA). The purified proteins were separated by SDS-PAGE before they were transferred to a polyvinylidene fluoride (PVDF) membrane for incubation with primary antibody (mouse Anti-NDV; Abcam) followed by secondary antibody (Goat Anti-mouse IgG HRP). The hemagglutination titer test was performed according to OIE Manual of Standard Diagnostic Tests [13].
Binding of the fusion proteins to L. lactis and Lb. plantarum
Lb. plantarum and L. lactis were grown overnight from single colonies in MRS and GM17, respectively. The overnight cultures were then inoculated into fresh media and grown to OD600 of 0.6 at 37oC and 30oC, respectively. The cells were then harvested at 2000 ×g for 5 min and the cell pellet was resuspended in 600 µl of the fresh media. The bacterial suspension was then mixed with 200 µl of the purified protein (2 mg/ml) and incubated at 30oC for 2 h. The cells were pelleted at 2000 µg for 5 min and then washed with 1 ×PBS, 3 times. The binding capability of the fusion proteins was analyzed by immunofluorescence staining using mouse anti NDV antibody as primary and FITC conjugated anti mouse antibodies raised in rabbit as secondary antibodies according to [14].
Attachment of bacteria to breast cancer MDA-MB231 cell line
The adhesion between the recombinant L. lactis which surface displayed HN-AcmA and MDA-MB231 breast cancer cells was visualized under florescent microscopy. A cell suspension of 1 × 105 cells was grown in complete RPMI medium in six-well tissue culture plates. When the cells reached 80% confluency, the medium was completely removed and the cells were fed with RPMI medium lacking antibiotics for 24 h. The cells were then washed three times with sterile phosphate-buffered saline. An aliquot of 2 ml of DMEM (without serum and antibiotics) was added to each well and incubated at 37oC for 30 min. Then, L. lactis carrying GFP (1 × 109 cfu) surface displaying HN-AcmA were resuspended in 1 ml RPMI medium (without serum and antibiotics) before they were added to the above wells. The plates were incubated at 37°C in 5% CO2 -95% air for 45 min. The monolayers were washed five times with sterile PBS. The adhesion was visualized under both direct and fluorescent microscope.
Results
Amplification, construction and expression of fusion HN-acmA cassette
The HN-acmA cassettes were generated by insertion of an XhoI restriction site at the C and N terminus of HN and acmA, respectively (Figure 1a). The production of HN-acmA in FreeStyle™ CHO-S® cells was studied by SDS-PAGE and Western blotting after purification. The 76 kDa expressed HN-AcmA was detected using SDS-PAGE and Western blotting after purification, respectively.
Immunofluorescent staining
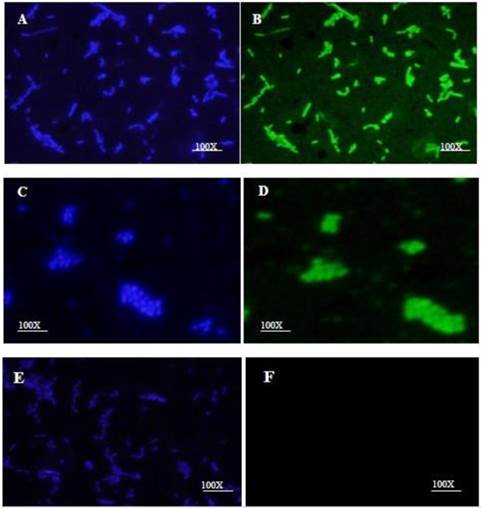
The HN-AcmA protein was purified on Ni2+ affinity columns and mixed with a culture of Lb. plantarum and L. lactis NZ9000. Immunofluorescence microscopy verified the binding of the fusion proteins on the cell wall surface of bacteria. Both lactobacilli (Fig. 2B) and lactococci (Fig. 2D) cells which were incubated with HN-AcmA exhibited bright fluorescence on the cell surface, indicating the presence of the fusion proteins on the cell wall surface. The control bacterial cells showed no fluorescence (Fig. 2F).
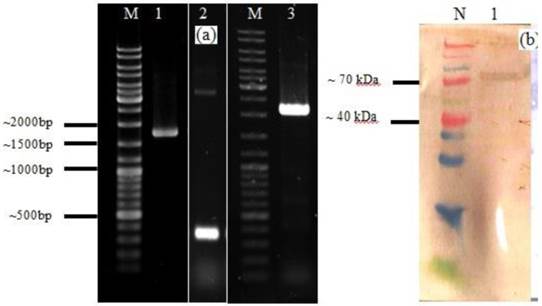
A. Agarose gel electrophoresis analysis of PCR product of HN of NDV, acmA, and HN-acmA Cassette; Lane M: GeneRuler DNA ladder mix; Lanes 1: PCR product of HN of NDV, which is around ~ 1750 bp. Lanes 2: PCR product of acmA, which is around ~ 366 bp. Lanes 3: PCR product of HN-acmA, which is around ~ 2100 bp. B. SDS-PAGE and Western blot analysis of the purified recombinant HN-AcmA protein ; Lane N: Spectra Multicolor Broad Range Protein Ladder; Lane 1: pCDNA:HN-AcmA; Expression was confirmed through Western blot by using specific polyclonal antibody directed against NDV.
Immunofluorescent micrograph of the binding of the HN-AcmA expressed in CHO cell to Lb. plantarum and L. lactis. A & B immunofluorescent micrograph of Lb. plantarum cells surface displayed by recombinant HN-AcmA. C& D immunofluorescent micrograph of L. lactis cells surface displayed by recombinant HN-AcmA. D& F control: L. lactis cells. The cells were immunostaining with anti-NDV antibody and DAPI and were visualized under two filter of DAPI (A, C, E) and FITC (B, D, F). The cells were observed under ×100 objective.
Attachment of HN surface displayed L. lactis to MDA-MB 231 breast cancer cell line: A&C interaction of L. lactis cells surface displayed by recombinant HN-AcmA with MDA-MB 231 breast cancer cell; (A) Direct microscopy (C) Fluorescent microscopy; N= nucleus; B= Bacteria. D&F Control: Attachment of L. lactis to MDA-MB 231 breast cancer cell line: (D) Direct microscopy (F) Fluorescent microscopy. Cells were photographed with a light microscope under ×40 objective.
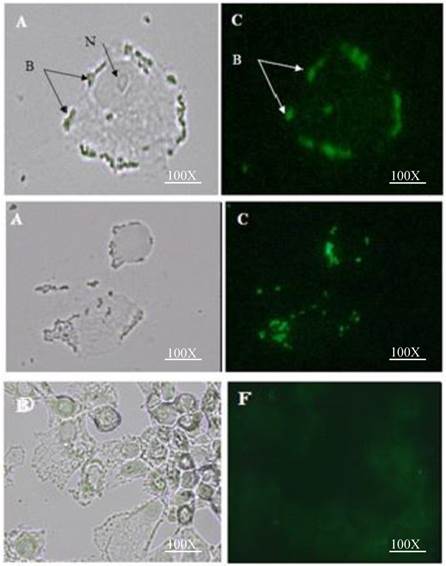
Bacterial attachment to breast cancer cell line
The attachment efficiency of the purified HN-AcmA surfaced displayed on L. lactis to the MDA-MB231 breast cancer cells was visualized using direct and fluorescent microscope (Figure 3). When the L. lactis were surface displayed with HN-AcmA (Fig 3; A and C), they acquired the ability to be attached to the surface of the MDA-MB231 breast cancer cells. However, the normal L. lactis cells (Fig 3; D and F) did not illustrate the binding capability to these cancer cells. In addition the HN surfaced displayed L. lactis induced apoptosis in the cancer cells.
Discussion
Targeted therapies are cornerstone of precision anticancer development. One promising strategy for targeting is the receptor-mediated system in which a tumor recognition moiety is connected through a smart linker to target the cancer [15]. The cancer cell aberrantly expresses or overexpresses glycan on the surface which is different than their normal counterparts [16, 17]. Finding ligands tailored to the abnormality of this glycan with higher affinity can be explored as a targeting agent.
Sialic acids and their derivatives which are abundant at the terminal positions of the cancer glycoproteins play a pivotal role in cancer biology [18]. The high expression of sialic acids has been shown to contribute to cancer cell protection from apoptosis, progression of metastasis, and enhanced resistance to therapy [19]. In breast cancers, MUC1 expression level is accompanied by greatly altered glycosylation [20, 21]. These O-glycans on the surface of MUC1 in cancer cells are shorter, less complex and more sialylated [22, 23]. Considering the detrimental effects of hyper-sialylation, strategies to take advantage of these alterations as a targeting biomarker as well as strategies to reduce unbalanced sialic acid expression might have a dual benefit as a targeting and therapeutic agent. This research investigates the tumor targeting capability of the HN protein in the context of its subcellular compartmentalization on the surface of L. lactis using AcmA.
The non-recombinant surface display system that mimics the surface antigen of pathogens was used in this study [24]. In this system the target protein is first produced as a fusion protein in a high level expression system and then bound non-covalently onto the cell surface of L. lactis by simply mixing them with live cells. To achieve this, the CHO expression system was used to express high levels of the anchored antigen fusion proteins which were subsequently purified and displayed onto the cell wall surface of L. lactis through attachment from the exterior. Immunofluorescent staining was used to confirm the efficient binding of the fusion proteins onto the cell wall surface. The tumor attachment study was utilized to investigate whether the surface displayed HN on L. lactis has a targeting capacity to the cancer cells. Based on adhesion, percentage of adhesion to MDA-MB231 breast cancer cells was high in L. lactis strains which surface displayed HN compared to normal L. lactis (Figure 3). In addition in concordance to the published studies by Ghirici et al, the HN show that have the ability to induce apoptosis on the cancer cells [25]. The data presented here demonstrates that L. lactis surface-expressed HN protein may provide new insight as the targeting agent to target vaccine trough cancer cells. To our knowledge, this is the first study demonstrating that recombinant HN-AcmA can be expressed in mammalian (CHO) expression system as well as targeting capabilities of HN protein to the cancer cells. This system would be helpful in the future to allow for breakthrough discovery for a novel way of expressing fusion proteins harboring viral epitopes to be displayed on the surface of L. lactis and Lb. plantarum as well as new insight in cancer targeting.
Abbreviations
HN: hemagglutinin-neuraminidase; NDV: Newcastle disease virus.
Acknowledgements
We would like to thank the Ministry of Science, Technology and Innovation, Malaysia and Malaysian Genome Institute (MGI) for funding this study under the grant number 02-01-04-SF1273. We also thank Kees Leenhouts for the kind gift of plasmids pNZ8048 and L. lactis hosts NZ9000.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fournier P, Schirrmacher V. Oncolytic Newcastle disease virus as cutting edge between tumor and host. Biology. 2013;2:936-75
2. Prestwich RJ, Harrington KJ, Pandha HS, Vile RG, Melcher AA, Errington F. Oncolytic viruses: a novel form of immunotherapy. Expert Rev Anticancer Therapy. 2008:1581-8
3. Huang Z, Panda A, Elankumaran S, Govindarajan D, Rockemann DD, Samal SK. The hemagglutinin-neuraminidase protein of Newcastle disease virus determines tropism and virulence. Journal of virology. 2004;78:4176-84
4. Zaitsev V, von Itzstein M, Groves D, Kiefel M, Takimoto T, Portner A. et al. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. Journal of Virology. 2004;78:3733-41
5. Takimoto T, Taylor GL, Connaris HC, Crennell SJ, Portner A. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. Journal of Virology. 2002;76:13028-33
6. Ji H-f, Chi B-r, He D-y, Li C, Hu N-n, Wang K. et al. Antitumor effects of Newcastle disease virus hemagglutinin-neuraminidase used as a molecular adjuvant. Chemical Research in Chinese Universities. 2013;29:270-4
7. Ni J, Schirrmacher V, Fournier P. The hemagglutinin-neuraminidase gene of Newcastle Disease Virus: A powerful molecular adjuvant for DNA anti-tumor vaccination. Vaccine. 2010;28:6891-900
8. Ni J, Galani IE, Cerwenka A, Schirrmacher V, Fournier P. Antitumor vaccination by Newcastle Disease Virus Hemagglutinin-Neuraminidase plasmid DNA application: Changes in tumor microenvironment and activation of innate anti-tumor immunity. Vaccine. 2011;29:1185-93
9. Piao B-g, Li X, Sun L-l, Kan S-f, Liu L, Huang H-y. et al. Activity of T Cells Stimulated by Hemagglutinin-neuraminidase of Newcastle Disease Virus in vivo. Chemical Research In Chinese Universites. 2011;27:455-60
10. Zeng J, Fournier P, Schirrmacher V. Induction of interferon-α and tumor necrosis factor-related apoptosis-inducing ligand in human blood mononuclear cells by hemagglutinin-neuraminidase but not F protein of Newcastle disease virus. Virology. 2002;297:19-30
11. Jarahian M, Watzl C, Fournier P, Arnold A, Djandji D, Zahedi S. et al. Activation of natural killer cells by Newcastle disease virus hemagglutinin-neuraminidase. Journal of Virology. 2009;83:8108-21
12. Baradaran A, Sieo CC, Foo HL, Illias RM, Yusoff K, Rahim RA. Cloning and in silico characterization of two signal peptides from Pediococcus pentosaceus and their function for the secretion of heterologous protein in Lactococcus lactis. Biotechnology Letters. 2013;35:233-8
13. Manual OT. Avian influenza. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 2012
14. Richter E, Wick G. Fluoro-immuno-cytoadherence (FICA): A new method for the identification and enumeration of antigen-binding cells. Zeitschrift für Immunitätsforschung: Immunobiology. 1977;152:351-62
15. Mohanty C, Das M, R Kanwar J, K Sahoo S. Receptor mediated tumor targeting: an emerging approach for cancer therapy. Current Drug Delivery. 2011;8:45-58
16. Reis CA, Osorio H, Silva L, Gomes C, David L. Alterations in glycosylation as biomarkers for cancer detection. Journal of Clinical Pathology. 2010;63:322-9
17. Meany DL, Chan DW. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin Proteomics. 2011:8
18. Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Annals of the New York Academy of Sciences. 2012;1253:16-36
19. Büll C, Boltje TJ, Wassink M, de Graaf AM, van Delft FL, den Brok MH. et al. Targeting aberrant sialylation in cancer cells using a fluorinated sialic acid analog impairs adhesion, migration, and in vivo tumor growth. Molecular Cancer Therapeutics. 2013;12:1935-46
20. Freire-de-Lima L. Sweet and sour: the impact of differential glycosylation in cancer cells undergoing epithelial-mesenchymal transition. Frontiers in oncology. 2014:4
21. Taylor-Papadimitriou J, Burchell J, Miles D, Dalziel M. MUC1 and cancer. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 1999;1455:301-13
22. Sihlbom C, van Dijk Härd I, Lidell ME, Noll T, Hansson GC, Bäckström M. Localization of O-glycans in MUC1 glycoproteins using electron-capture dissociation fragmentation mass spectrometry. Glycobiology. 2009;19:375-81
23. Singh R, Bandyopadhyay D. MUC1: a target molecule for cancer therapy. Cancer biology & therapy. 2007;6:481-6
24. Raha A, Varma N, Yusoff K, Ross E, Foo HL. Cell surface display system for Lactococcus lactis: a novel development for oral vaccine. Applied Microbiology and Biotechnology. 2005;68:75-81
25. Ghrici M, El Zowalaty M, Omar AR, Ideris A. Induction of apoptosis in MCF-7 cells by the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus Malaysian strain AF2240. Oncology reports. 2013;30:1035-44
Author contact
![]() Corresponding author: Khatijah Yusoff (kyusoffedu.my) KY, Department of Microbiology, Faculty of Biotechnology & Biomolecular Science, Universiti Putra Malaysia, 43400 Serdang, Selangor DE, Malaysia.
Corresponding author: Khatijah Yusoff (kyusoffedu.my) KY, Department of Microbiology, Faculty of Biotechnology & Biomolecular Science, Universiti Putra Malaysia, 43400 Serdang, Selangor DE, Malaysia.

 Global reach, higher impact
Global reach, higher impact