Impact Factor
ISSN: 1837-9664
J Cancer 2013; 4(5):358-370. doi:10.7150/jca.6289 This issue Cite
Research Paper
Ku Protein Levels, Localization and Association to Replication Origins in Different Stages of Breast Tumor Progression
1. Goodman Cancer Research Center, McGill University, Montreal, Quebec, Canada H3G 1Y6;
2. Department of Biochemistry, McGill University, Montreal, Quebec, Canada H3G 1Y6.
Received 2013-3-20; Accepted 2013-5-23; Published 2013-5-26
Abstract
Human origins of DNA replication are specific sequences within the genome whereby DNA replication is initiated. A select group of proteins, known as the pre-replication (pre-RC) complex, in whose formation the Ku protein (Ku70/Ku86) was shown to play a role, bind to replication origins to initiate DNA replication. In this study, we have examined the involvement of Ku in breast tumorigenesis and tumor progression and found that the Ku protein expression levels in human breast metastatic (MCF10AC1a) cells were higher in the chromatin fraction compared to hyperplastic (MCF10AT) and normal (MCF10A) human breast cells, but remained constant in both the nuclear and cytoplasmic fractions. In contrast, in human intestinal cells, the Ku expression level was relatively constant for all cell fractions. Nascent DNA abundance and chromatin association of Ku70/86 revealed that the c-myc origin activity in MCF10AC1a is 2.5 to 5-fold higher than in MCF10AT and MCF10A, respectively, and Ku was bound to the c-myc origin more abundantly in MCF10AC1a, by approximately 1.5 to 4.2-fold higher than in MCF10AT and MCF10A, respectively. In contrast, similar nascent DNA abundance and chromatin association was found for all cell lines for the lamin B2 origin, associated with the constitutively active housekeeping lamin B2 gene. Electrophoretic mobility shift assays (EMSAs) performed on the nuclear extracts (NEs) of the three cell types revealed the presence of protein-DNA replication complexes on both the c-myc and lamin B2 origins, but an increase in binding activity was observed from normal, to transformed, to cancer cells for the c-myc origin, whereas no such difference was seen for the lamin B2 origin. Overall, the results suggest that increased Ku chromatin association, beyond wild type levels, alters cellular processes, which have been implicated in tumorigenesis.
Keywords: Breast cancer, Ku protein, replication origins, nascent DNA, chromatin association.
Introduction
Breast cancer continues to be a leading cause of death among women worldwide. To date, two highly penetrant breast cancer susceptibility genes have been identified (BRCA1 and BRCA2). Carriers of a germline mutation in these genes have a 10- to 20-fold increased breast cancer risk, although the rarity of these mutations suggests that they can only partly (15-20%) account for familial breast cancers, indicating that either additional mutations in low-penetrant genes, or low-penetrant mutations in other highly penetrant genes, must also be involved in increasing the risk of developing non-BRCA breast tumorigenesis. The fact that both Ku and the highly penetrant BRCA genes are implicated in double-strand DNA break (DSB) repair [1, 2], supports the hypothesis that DSB-initiated chromosomal instability may be implicated in breast carcinogenesis and prognosis. This was further supported by a recent study showing that BRCA1 is recruited to DSBs in two distinct ways, Ku86-dependently and -independently, respectively, participating in DSB repair [3]. Non-homologous end-joining (NHEJ) is the prevailing pathway for repair of DNA double-strand breaks (DSBs) in mammalian cells and is initiated by the association of the Ku70/86 heterodimer to the DNA ends [4]. The Ku heterodimeric complex is a ring-shaped structure that displays high affinity for open DNA ends and is the initial sensor of DSBs [5]. Ku functions in the cell nucleus as a caretaker to protect against genomic instability [6-8], and has been suggested to be a tumor suppressor for murine T cell lymphoma [9].
In normal cells, the principal role of Ku proteins is to mediate DNA repair, but they are also implicated in many other cellular processes, including telomere maintenance, anti-apoptosis, tumor suppression, and regulation of specific gene transcription [10-13], cell cycle control and DNA replication [14-19]. Ku participates in the origin recognition complex (ORC) assembly and/or stabilization onto replication origins [16, 19]. Ku proteins are present in subcellular structures other than the nucleus. For example, Ku70 has been shown to bind to the pro-apoptotic complex and inhibit BAX-mediated apoptosis in vitro by preventing its relocalization to the mitochondria [20]. Moreover, upregulation of Ku proteins has been associated with the progression of some types of tumors. For example, the levels of Ku86 (or Ku80) are positively correlated with that of anti-apoptotic Bcl-2 and B cell chronic lymphocytic leukemia [21]. Ku86 has also been reported to be upregulated in bladder, breast, and primary hepatocellular carcinomas, compared to adjacent non-tumor tissues [22, 23]. Recently, Ku86 has been identified as a new potential player within intracellular signalling leading to breast cancer cell metastasis [24].
One of the key players in NHEJ is DNA-dependent protein kinase (DNA-PK), a complex consisting of DNA-PK catalytic subunit (DNA-PKcs) and a regulatory heterodimer consisting of Ku70 and Ku86, which are constitutively expressed at relatively high levels in cells [25]. The Ku heterodimer binds to DNA ends with high affinity independently of DNA sequence [26]. Although, both Ku and DNA-PKcs bind independently to the DNA ends, the greater part of this function is performed by the Ku70/86 heterodimer, rather than DNA-PKcs itself [27]. Ku recruits DNA-PKcs to the DNA and its interaction between with it stimulates the catalytic potential of the DNA-PK complex [28].
In eukaryotic cells, initiation of DNA replication takes place from multiple replication origins on each chromosome. Control mechanisms exist to ensure that DNA replication occurs only once per cell cycle, and when such mechanisms fail, checkpoint responses are activated to monitor the genome integrity and inhibit replication until DNA damage has been repaired [29]. The heterodimeric Ku protein (Ku70/86) promotes initiation of DNA replication, by binding to replication origins in a cell cycle dependent manner, reaching a maximum in late G1 and decreasing as cells enter S phase [16, 17, 30, 31]. Ku also interacts with other DNA replication proteins, including HsOrc2, DNA polymerases α, δ, ε, PCNA, topoisomerase II, RFC, and RPA [32].
In this study, we analyzed the levels and subcellular localization of Ku as well as its binding activity to the replication origins c-myc and lamin B2 in a set of isogenic human breast normal (MCF10A), hyperplastic (MCF10AT) and metastatic (MCF10AC1a) cell lines, as a model mimicking the stepwise malignant transformation observed clinically. The data show that although Ku protein expression is relatively constant in the cytoplasmic and nuclear fractions in all cell lines, its association with chromatin was higher in the metastatic cells, while it was less in the hyperplastic cells and even less in the normal ones. By comparison, constant levels of Ku expression were observed in the cytoplasmic, nuclear, and chromatin extracts in all intestinal cells. These data suggest that Ku is a specific chromatin binding protein in breast cells. Ku was also found to bind specifically to the replication origins, c-myc and lamin B2, as these origins were enriched in Ku immunoprecipitates compared to non origin-containing regions of the genome. With the c-myc origin, higher association of Ku was observed in the metastatic cells cells compared to hyperplastic and normal cells, while relatively constant binding was observed with the constitutively active lamin B2 origin in all cell lines. Overall the data suggest the Ku is expressed and bound to chromatin at higher levels in more advanced breast cancer compared to hyperplastic and normal breast cells and that certain origins are more active due to this increased association. These data support a role of Ku as an important genomic factor in breast tumor progression.
Materials and Methods
Cell culture
MCF10A, MCF10AT were grown in DMEM/F12 (Wisent, St-Bruno, Quebec, Canada) as previously described (Dentath et al., 2003). MCF10CA1a cells were cultured in DMEM/F12 supplemented with 5% horse serum (Invitrogen, Burlington, Ontario, Canada), and penicillin/ streptomycin (20µg/ml). Human HCT116 and Caco2 cells were cultured in DMEM supplemented with 10% FBS, 1% non-essential amino-acids and 1% penicillin/streptomycin solution. HIEC-7 cells were grown in DMEM containing 25 mM glucose and supplemented with 10% FBS, 19 mM NaHCO3, 20 mM HEPES, 10µg/ml insulin, 20ng/ml EGF, and 0.5 µg/ml hydrocortisone. All cells were maintained in a humified atmosphere of 5% CO2 at 37°C, and grown at a confluency of 80% for most experiments except, when the cells reached 30%-50% confluence, they were harvested for the isolation of nascent DNA, while upon reaching 100% confluence, they were serum-starved for 48h-72h and harvested for the isolation of genomic DNA.
Cell fractionation
Nuclear and cytoplasmic protein fractions were isolated using a NEPER® Nuclear and Cytoplasmic Extraction Reagent Kit (Pierce Biotechnology, Rockford, IL, USA). Purity of cytoplasmic and nuclear fractions was verified using antibodies against Tubuline (cytoplamic). For the chromatin fraction, cells were harvested from T-75 flasks, washed with ice-cold PBS, resuspended in 1 ml of lysis buffer A [10 mM HEPES-KOH, pH 7.9, 100 mM NaCl, 300 mM sucrose, 0.1% Triton X-100 and Complete Protease Inhibitor tablet (Roche Molecular Biochemicals, Laval, Quebec, Canada)], and the cells were incubated on ice for 10 minutes. After centrifugation at 2000 g for 3 minutes at 4°C, pellets were washed once more with ice-cold lysis buffer A and resuspended in lysis buffer B [10 mM HEPES-KOH, pH 7.9, 200 mM NaCl, 300 mM sucrose, 0.1% Triton X-100, 5 mM MgCl2 and Complete Protease Inhibitor tablet (Roche Molecular Biochemicals, Laval, Quebec, Canada)] containing 1000 U of DNase I (Wisent, St-Bruno, Quebec, Canada). Following incubation at room temperature for 30 minutes, the chromatin-enriched fraction was isolated in the supernatant after centrifugation at 2500 g for 5 minutes at 4°C. Protein contents were determined using a BCA protein detection kit (Pierce Biotechnology, Rockford, IL. USA).
Western Blot
Equal amounts of protein (5 µg) were diluted with loading buffer and boiled at 95°C for 5 min , then loaded onto 10% SDS-PAGE. After electrophoresis, gels were transferred to polyvinylidene diflouride membranes (Bio-Rad, Hercules, CA, USA) for 45 minutes at 4°C in transfer buffer. Membranes were blocked using Tris-buffered saline (TBS) plus 5% milk powder (blocking buffer) for 2 hr at room temperature, and then incubated with primary antibody at a 1:1,000 dilution in blocking buffer at 4°C overnight, followed by washes with TBS-T (TBS containing 0.15% Tween) and incubation with the secondary antibody for 1 hr at room temperature. The following antibodies were used: anti-Ku70 (sc-1486), anti-Ku86 (sc-9034), anti-Orc1 (sc-23887), anti-Orc2 (sc-32734), anti-Orc3 (sc-21862), anti-Orc4 (sc-20634), anti-Orc5 (sc-19728), anti-Orc6 (sc-32735), anti-MCM7 (sc-46687), anti-DNA-PKcs (sc-9051), anti-α Tubulin (sc-8035) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Ku70/80 (ab3180; Abcam), anti-actin (A 2066, Sigma). Membranes were then rinsed with TBS-T and water. After rinsing, the membranes were incubated with enhanced chemiluminescence (ECL) reagents for 5 min after which the membranes were sealed in plastic wrap and photographed using ECL hyperfilm to detect immunoreactive bands.
Immunoflorescence
For immunostaining with anti-Ku antibody, MCF10A, MCF10AT, and MCF10AC1a cells were plated on coverslips, rinsed with PBS and fixed with methanol for 10 minutes at -20°C. Then treated with 0.3% Triton X-100 in PBS for 15 minutes at room temperature. After treatment with blocking buffer (5% BSA) for 1 hour at RT, the cells were incubated with a mouse monoclonal anti-Ku86 antibody (1:50, sc-9034; Santa Cruz) over night at 4°C. After washing with PBS, cells were incubated with Alexa Fluor1 488- conjugate goat anti-rabbit antibody (A-11034, 1:500, Invitrogen, St-Bruno, ON, Canada). Co-staining with 0.1 mg/ml DAPI for 1 minute at RT was also performed to allow visualization of nuclear DNA. Fluorescence data were collected using a Nikon Eclipse E800 microscope and analyzed with the Nikon ACT-1 software.
Isolation of genomic DNA
Genomic DNA was extracted using the GenElute Mammalian Genomic DNA Miniprep Kit (Sigma, Oakville, ON, Canada), as per manufacturer's instructions. Typically, 10ng of genomic DNA was used to assess the copy number per haploid genome by real-time PCR. Genomic DNA from at least two different cellular populations for each cell line was prepared and the experiment was performed at least twice in triplicate for each data point.
Isolation of nascent DNA
Nascent DNA was prepared using a modified λ exonuclease digest method [33] with the following modifications, the λ exonuclease digested samples were heated at 100_C for 3min, then immediately subjected to electrophoreses on a 2% agarose gel. DNA was visualized by staining with 0.02% (w/v) methylene blue (Sigma) and the origin-containing nascent DNA, ranging between 350bp to 1000bp in size was excised from the gel, purified with the Sephaglas BandPrep Kit (GE Healthcare, Piscataway, NJ, USA), as per manufacturer's instructions, and resuspended in TE. Typically, 5µl of nascent DNA was used to assess origin activity by real-time PCR. Nascent DNA from at least two different cellular populations for each cell line was prepared and the experiment was performed at least twice in triplicate for each data point.
Real-time PCR quantification analyses
PCRs were carried out in a total volume of 20ml with 5ml of genomic, nascent, or immunoprecipitated DNA, using the LightCycler (Roche Diagnostics) as previously described [31]. The sequences and amplification conditions for all primer sets are shown in Table I. Genomic DNA (0.01, 0.1, 1, and 10 ng) from MCF10A cells was used to generate the standard curves needed for quantification of the PCR products. A negative control without template DNA was included with each set of reactions. PCR products were also resolved on 2% agarose gels, visualized with ethidium bromide and photographed with an Eagle Eye apparatus (Speed Light/BT Sciencetech-LT1000). No extraneous bands were generated with any of the primer sets.
Chromatin Immunoprecipitation
Immunoprecipitation (IP) was carried out as previously described [17, 31], with the following modifications: Briefly, sheared chromatin lysates were pre-cleared by incubation with 50 μl of protein A/G agarose (Roche Molecular Biochemicals, Laval, QC, Canada) for 1 h at 4°C to reduce background caused by non-specific adsorption to the beads, then cleared chromatin lysates were incubated overnight at 4°C with either 20 µg of anti-Ku70/86 (ab3108, clone 162, Abcam) or normal anti-rabbit IgG (7074, Cell Signaling) with constant rotation. Protein A/G agarose (50 µl) was added and incubated at 4°C for 2 h. The pelleted beads were washed successively twice with 1 ml of lysis buffer A [10mM HEPES-KOH, PH 7.9, 100 mM NACL, 300mM sucrose, 0.1% Triton X-100 and completed protease inhibitor tablet (Roche Molecular Biochemicals)] for 15 min each at 4°C, followed by 1 ml of WB1 (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 0.1% NP40, 0.05% sodium deoxycholate, complete protease inhibitor tablet), 1 ml of WB2 (50 mM Tris-HCl pH 7.5, 0.1% NP40, 0.05% sodium deoxycholate, complete protease inhibitor tablet) and 1 ml of sterile ice-cooled TE. The beads were resuspended in 200 µl TE/1% SDS, incubated at room temperature (rt) for 15 min and centrifuged at 3000 r.p.m. for 1 min at rt. Half of the supernatant was then incubated overnight at 65°C to reverse the cross-links, followed by 100 µg of proteinase K at 55°C for 2 h. The DNA was purified using QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) and eluted in 100 µl TE. The remaining half of the supernatant was boiled for 10 min in SDS-PAGE loading buffer and subjected to electrophoresis on a 5% stacking/10% separating SDS-PAGE gel for western blot analysis.
Names, sequences and amplification conditions of primers used for real-time PCR quantification of DNA with the LightCycler (Roche Diagnostics). `F' and `R' designate the forward and reverse primers, respectively. The size of the PCR products in base pairs (bp) and the annealing temperature used in the PCR cycling conditions in °C is also indicated.
| Primer name | Sequence (5'-3') | Amplicon size (bp) | Annealing Temp. (C°) |
|---|---|---|---|
| LB2-F | GGCTGGCATGGACTTTCATTTCAG | 232 | 66 |
| LB2-R | GTGGAGGGATCTTTCTTAGACATC | ||
| LB2C1-F | GTTACCAGTCAGGCGCATGGGCC | 240 | 66 |
| LB2C1-R | CCATCAGGGTCACCTCTGGTTCC | ||
| Myc11-F | TATCTACACTAACATCCCACGCTCTG | 192 | 62 |
| Myc11-R | CATCCTTGTCCTGTGAGTATAAATCA | ||
| Myc1-F | TCG TCTCAACCTCAGCACTGGTGACA | 248 | 60 |
| Myc1-R | GACTTTGCTGTTTGCTGTCAGGCT |
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed to assess binding of Ku protein with specific origin sequences, c-myc and lamin B2. Nuclear extracts (NE) from MCF10A, MCF10AT, MCF10AC1a cells were prepared with the NE-PER nuclear extraction reagent (Pierce Biotechnology, Rockford, IL, USA). A non-radioactive method was employed whereby the 3′-end double-strand DNA oligonucleotides of the Myc11 and LB2 probes were labeled with biotin (Pierce). The binding reactions contained 10 μg of NE, 0.5X binding buffer, 10 mm Tris, pH 7.5, 50 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, 0.05% Nonidet P-40, 2.5% glycerol, 1 μg of poly(dI-dC), and 2 nm of biotin-labeled DNA. The reactions were incubated at room temperature for 20 min. The competition reactions were performed by adding 5 and 20-fold excess unlabeled Myc11 and LB2 probes to the reaction mixture. Following separation on a 6% non-denaturing gel at 100 V for 3 h in a 100 mm Tris-borate-EDTA buffer. The samples were transferred to a positively charged nylon membrane. The membrane was UV-cross linked, and the biotin end-labeled probes were detected with streptavidin-HRP using a luminol enhancer solution as described in LightShift chemiluminescent electrophoretic mobility shift assay kit (Pierce Biotechnology, Rockford, IL, USA). Supershift experiments, to determine the binding specificity, were done using antibodies to Ku70 (sc-1486, Santa Cruz) and Ku86 (sc-1484, Santa Cruz).
Results
Ku70 and Ku86 expression in chromatin, nuclear, and cytoplasmic extracts
Ku70/86 protein subunits were detected by Western blot analysis on chromatin, nuclear, and cytoplasmic extracts from all cell lines (Fig. 1). In human breast metastatic cells (MCF10AC1a), the abundance of Ku was approximately two-fold higher in the chromatin fraction compared to the hyperplastic (MCF10AT) and normal (MCF10A) breast cells, whereas it was relatively constant in both nuclear and cytoplasmic fractions (Fig. 1A, upper panel). In contrast, analysis of Ku expression in intestinal cells (Caco2, HCT116, and HIEC7) revealed no significant changes in any of the cellular fractions (Fig. 1A, lower panel). In all human breast cell lines, the Ku70 subunit detectable levels were consistently lower than those of the Ku86 subunit (Fig. 1A, upper panel), while in human intestinal cells the detectable levels of both Ku subunits were similar (Fig. 1A, lower panel). Immunofluorescence analyses revealed a nuclear localization of Ku in all three human breast cell lines (Fig. 1B), in agreement with previous reports describing a nuclear localization of the Ku proteins [34].
Copy number of c-myc and lamin B2 chromosomal sites
Prior to assessing the activity of two replication origins used in this study, namely the origins associated with the c-myc [35] and lamin B2 [36] loci, we analyzed the copy number of these replication origins and their respective control (non-origin-containing) regions, as previously described [37]. Briefly, equal amounts of genomic DNA from each cell line were amplified by quantitative real-time PCR. The results show that all the regions of interest, namely, the origin-containing LB2 and Myc11 regions, and their respective control (non-origin-containing) regions, LB2C and Myc1, were present at one copy per haploid genome (Fig. 2A and B), permitting comparison of their origin activity.
Nascent DNA abundance
To analyze the activity of the c-myc [38] and lamin B2 [36, 39] replication origins, an analysis of the nascent DNA abundance was performed, as previously described [33]. The sequence and annealing temperature of the real time PCR primers used for each region is described in Table I. The human origin-containing regions (Myc11 and LB2) and their respective origin-lacking regions (Myc1 and LB2C) were used as positive and negative controls, respectively. Nascent DNA was prepared by the λ-exonuclease digested DNA method, as described in Materials and Methods and in [33, 38]. Nascent DNA abundance revealed that the c-myc origin activity in metastatic (MCF10AC1a) cells was approximately 2.5 to 4-fold greater than that in the hyperplastic (MCF10AT) and normal (MCF10A) ones, respectively (Fig. 3A). In contrast, the activity of the lamin B2 origin was similar in all cell lines (Fig. 3B), in agreement with previous findings [31, 33, 37].
Quantitative Real Time PCR (QPCR) of immunoprecipitated DNA
To analyse whether the DNA bound protein that was immunoprecipitated with anti-Ku antibodies after cross-linking with formaldehyde was bound to origin-containing sequences, and to quantify this association, quantitative real time PCR was performed, with the use of four sets of primers to amplify the two replication origin regions (Myc 11 and LB2), and their respective non-origin control regions (Myc1 and LB2C). This was compared with DNA obtained by immunoprecipitation with an anti-IgG antibody, used as control. Sonicated non-imunoprecipitated DNA was used to generate the standard curve. The efficiency for all graphs ranged between 1.96 and 2, giving a good linear correlation, and the slopes of all standard curves were similar (ranging from -3.22 to -4.23), indicating similar amplification efficiencies for all primer sets, thus permitting quantitative comparisons between different primer sets. Regarding Ku's DNA binding activity, in agreement with the results obtained with the EMSA and nascent abundance assays, the association of immunoprecipitated Ku70/86 with Myc11 origin sequences in breast cancer cells (MCF10AC1a) showed approximately 2.5- and 4.2-fold higher abundance by comparison to transformed (MCF10AT) and normal (MCF10A) breast cells, respectively (Fig. 4A), while, with LB2 containing origin sequences, no significant replication origin differences were found between the amount of amplified DNA in the three cell lines (Fig. 4B), in agreement with previous results found using different transformed/tumor and normal cells [40].
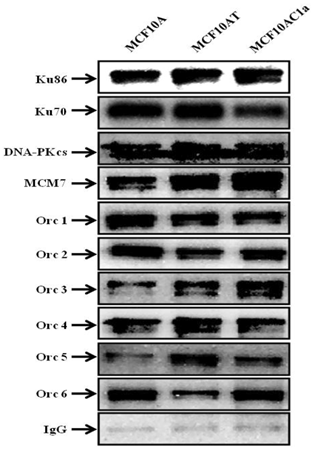
Furthermore, co-immunoprecipitations of Ku with other DNA repair/replication proteins were performed, using anti-Ku70/86, anti-DNA-PKCs, anti-Orc1-6, and anti-MCM7 antibodies (Fig. 5). The results revealed the interaction between Ku and all the above-mentioned proteins in all the cell lines used in this study.
Expression levels and localization of Ku protein in normal (MCF10A), transformed (MCF10AT) and cancer (MCF10AC1a) breast cell lines. A) Western blot analysis of Ku70 and Ku86 heterodimer in cytoplasmic, nuclear, and chromatin fractions. 5µg of protein extracts from each cell line were immunoblotted for Ku86 and Ku70. Actin was used as a loading control. Tubulin was used to verify the quality of nuclear and cytoplasmic preparation. B) Immunofluorescence Staining of DNA with DAPI. Cells were grown to a confluency of 80%, and then incubated with anti-Ku86 antibody followed by incubation with Fluor® 488-conjugate goat anti-rabbit antibody along with 0.1 mg/ml of DAPI. Cell images were captured with fluorescence microscopy using 60x1.4 objective and 488/364 nm laser wave lengths to detect Fluor®488 and DAPI, respectively. The Ku86 is white and DAPI is grey. The image above is a representation of one of three experiments performed.
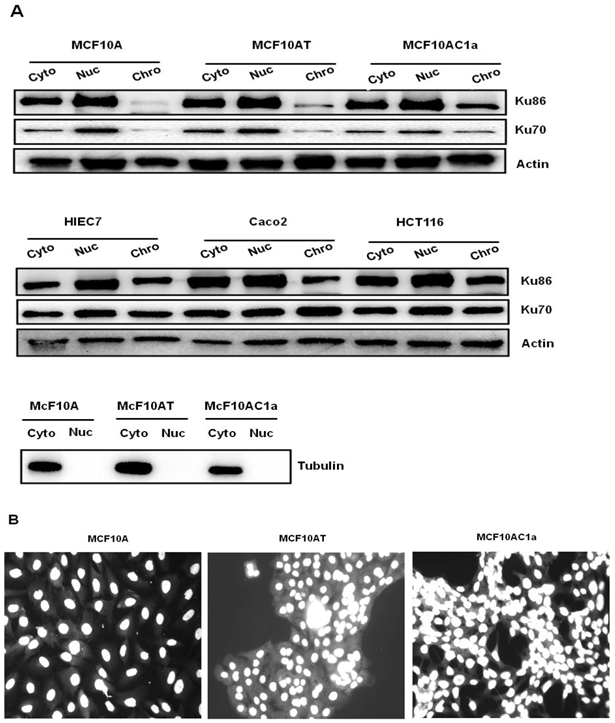
Copy number per haploid genome at the lamin B2 and c-myc chromosomal loci in MCF10A, MCF10AT, MCF10AC1a cells. Histogram plots of copy number/haploid genome at the lamin B2 (A), and c-myc (B) chromosomal loci in the MCF10A, MCF10AT, MCF10AC1a cell lines. The results were normalized by making MCF10A equal to one copy/haploid genome for all primer sets. The location and sequence information of the primers used for the amplification of the origin-containing regions (LB2, and Myc 11; grey bars) and their respective non-origin-containing control regions (LB2C1, and Myc 1; white bars) are denoted in Table I. Each primer set amplifies a region of approximately 150-300bp in size. The error bars represent the average of at least two experiments performed in triplicate and one standard deviation.
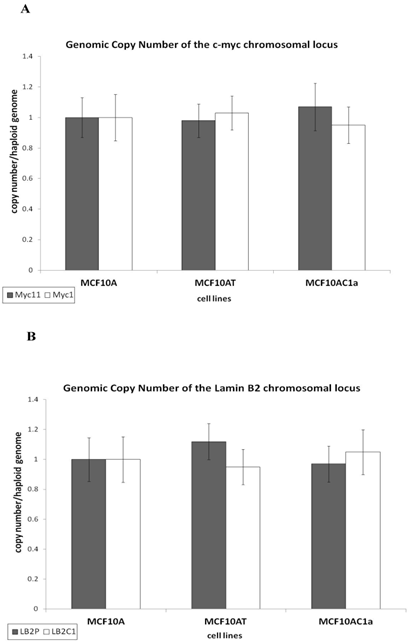
Comparative analysis of the nascent DNA abundance at the lamin B2 and c-myc chromosomal loci in MCF10A, MCF10AT, MCF10AC1a cells. Histogram plots of the quantification by real-time PCR of nascent DNA abundance (ng) at the lamin B2 (A), and c-myc (B) chromosomal loci in the MCF10A, MCF10AT, MCF10AC1a cell lines. The location and sequence information of the primers used for the amplification of the origin-containing regions (grey bars) and the non-origin-containing control regions (white bars) are as described in Table I. The error bars represent the average of at least two experiments performed in triplicate and one standard deviation.
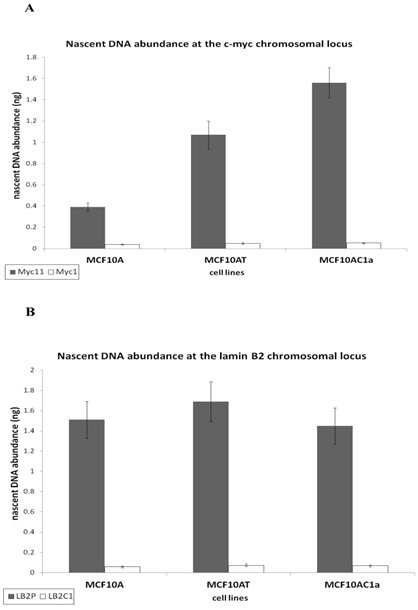
Real-time PCR analyses of immunoprecipitated DNA with anti-Ku70/86 antibody at c-myc (A) and lamin B2 (B) replication origins in MCF10A, MCF10AT, and MCF10AC1a cells. For each immunoprecipitate, the fold enrichment of the both origin-containing (Myc11 and LB2), and origin-lacking (Myc1 and LB2C) was determined. IgG was used as negative control. Each bar represents the average of three experiments, each done in duplicate.
Coimmunoprecipitation of Ku, DNA-PKcs, MCM7, and Orc1-6 proteins in chromatin immunoprecipitated fractions from MCF10A, MCF10AT, and MCF10AC1a cells. Chromatin fractions were immunoprecipitated with anti-Ku70/86 antibody, or IgG (control), as indicated above. The immunoprecipitates were immunoblotted for Ku86, Ku70, DNA-PKcs, MCM7, Orc1-6 proteins using their corresponding antibodies.
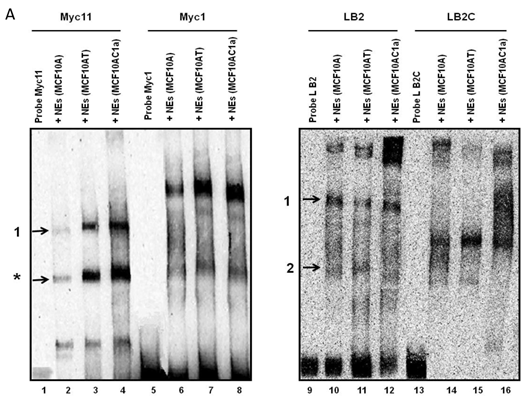
Ku 70/86 presence within complexes formed on replication origin sequences
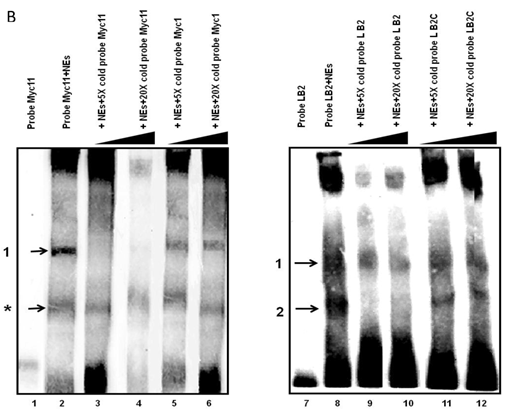
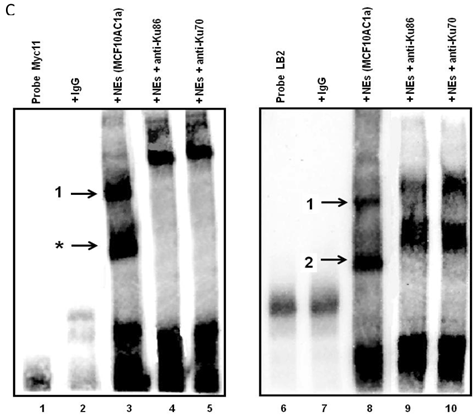
Electrophoretic mobility shift assays (EMSAs) were performed to compare the DNA binding activity of the Ku70/86 heterodimeric complex between the three cell types under study, namely breast cancer (MCF10AC1a), transformed (MCF10AT), and normal (MCF10A) breast cells. Basic EMSA revealed the formation of specific protein-DNA complexes on both biotin-labeled probes (Myc11 and LB2) in all cell lines (Fig. 6A). With the c-myc origin probe (Myc 11), higher binding was observed on origin probes in MCF10AC1a cells compared to MCF10A and MCF10AT cells, while relatively equal binding was observed for all cell lines, when the lamin B2 origin probe was used. Competitive EMSAs inhibited complex formation in the presence of increasing amounts of cold-specific competitor probes for both Myc11 and LB2 sequences (Fig. 6B, lanes 3, 4, 9, and 10), but, in the case of LB2, complex 1 was inhibited to a lesser extent than complex 2, which was completely inhibited, even at 5X-molar excess of competitor. In contrast, no inhibition of complex formation was found by increasing amounts of non-specific competitor probes (Myc1 and LB2C) (Fig. 6B, lanes 5, 6, 11, and 12). Supershift EMSAs were performed using antibodies against Ku70 and Ku86, after the incubation of Myc11 and LB2 probes with NEs from MCF10AC1a cancer cells. The results (Fig. 6C), show that addition of anti-Ku70 and anti-Ku86 antibodies to the Myc11 origin (lanes 4 and 5), resulted in a shift of both the origin-specific (complex labeled as 1) and non-specific (complex labeled as *) protein-DNA complexes, as evidenced by the appearance of higher molecular weight bands compared to both of the complexes obtained in lane 3 (Fig. 6C). Addition of the same antibodies to the reactions with the LB2 origin probe resulted in a supershift of both the specific complexes formed [Fig. 6C, compare complexes 1 and 2 (lanes 9 and 10) to complexes in lane 8]. Addition of IgG to Myc11 and LB2 probes (Fig. 6C, lanes 2 and 7, negative controls), did not affect the mobility of the complex, confirming specificity of the antibody. Overall, the results obtained with the supershift EMSAs indicated the presence of Ku protein in all formed complexes.
EMSA revealing the binding of Ku protein to the c-myc and lamin B2 replication origin sequences. A) Basic EMSA was performed to analyze the binding of proteins present in the nuclear extract of MCF10A, MCF10AT, and MCF10AC1a cells with Myc11 origin probe (lanes 2-4), Myc1 control probe (lanes 6-8), LB2 origin probe (lanes 10-12), and LB2C control probe (lanes 14-16), as described in Materials and Methods. Complexes 1 and 2 (indicated by arrows) represent specific protein-DNA complexes (formed in the presence of Myc11 and LB2 origin probes), while complexes labeled with an asterisk (indicated by arrows) represent a non-specific protein complex (formed in the presence of Myc1 control probe). B) Competitive EMSAs using MCF10AC1 cell nuclear extracts (NEs) and increasing amounts of cold Myc11 (origin) (lanes 3 and 4) and LB2 (origin) (lanes 9 and 10) probes, and Myc1 (non-origin) and LB2C (non-origin) control probes (lanes 5, 6, 11 and 12). C) Supershift EMSAs using MCF10AC1a cells, showing that the addition of anti-Ku70 and anti-Ku86 antibodies supershiftted the DNA-protein complexes (arrows) formed on the Myc11 (lanes 4 and 5) and LB2 (lanes 9 and 10) origins. Free probe without protein extracts (lanes 1 and 6) and IgG with probe (lanes 2 and 7) were used as negative control for both probes. All experiments were performed in triplicate and gave rise to reproducible results.
Discussion
Ku was originally reported to be a nuclear protein, consistent with its functions as a subunit of DNA-PKcs. Several studies, however, have reported the cytoplasmic or cell surface localization of Ku proteins in various cell types [41-44]. In the present study, we examined the subcellular localization of Ku proteins in breast and intestinal cell lines. Our results confirmed the nuclear localization of Ku and showed that the abundance of the Ku protein in the chromatin fraction was greater in breast metastatic cancer (MCF10AC1a) cells compared to hyperplastic (MCF10AT) and normal (MCF10A) breast cell lines, whereas, no changes were observed in both the cytoplasmic and nuclear extracts. In contrast, the abundance of Ku in the intestinal cell lines examined (Caco2, HCT116, and HIEC7) remained relatively constant in all fractions. These results suggest that Ku proteins are specific to breast cancer progression and indicate that in intestinal cancer cells, tumoregenesis is not associated with Ku protein expression. Many studies have suggested a key role of Ku in cancer development and progression, acting as either tumor suppressor gene or as oncogene [45]. Low levels of Ku have been linked to increased genomic instability and tumorigenesis (due to its role in DSB repair) [46], while its increased expression has been associated with resistance to apoptosis (due to the anti-apoptotic role of Ku70) [20, 47] and hyperproliferation (consistent with its role in DNA replication). Indeed, Ku expression and/or function is known to be deregulated in breast tumors [22, 48] and in several other human cancers [49-52]. Those studies suggest that a delicate balance exists in Ku expression, as overexpression of Ku promotes oncogenic phenotypes, including hyperproliferation and resistance to apoptosis, whereas, low expression of Ku leads to genomic instability and tumorigenesis, and it may contribute to either condition [46]. As our results with the breast metastatic cancer (MCF10AC1a) cells suggest, development of cancer tumorigenesis is associated with Ku overexpression, and are in agreement with previous reports showing that low protein expression of the Ku70/86 heterodimer predicted good response to radiotherapy in early breast cancer [48].
Also in agreement with previous results, where a high replication origin activity was seen in tumor/transformed cell compared to normal ones [31, 33, 37, 40], the breast metastatic cancer (MCF10AC1a) cells had the highest replication origin activity at the c-myc locus, by comparison to the normal (MCF10A) and hyperplastic (MCF10AT) breast cells (Fig. 3A). A possible explanation for this observation is that the c-myc origin in MCF10AC1a cells may be activated in more cells per population by comparison to the origins corresponding to MCF10AT and MCF10A cells, as also seen with this origin in other cell types (HeLa, NSF, WI38 and WI38 (SV40) [31]. Alternatively, this might be due to origin interference, as other initiation sites may be present in the vicinity of the origins on MCF10AT and MCF10A chromosomes [53], resulting in a broader and flatter peak by comparison to the origin associated with MCF10C1a cells.
In the present study we have also investigated Ku's origin binding activity, which is crucial for its role in maintaining genomic stability, DNA repair and DNA replication. As previously reported, Ku is involved in DNA replication, and directly interacts with replication origins in vitro and in vivo [25, 54-56]. In vivo studies have also indicated the binding of Ku to specific regulatory elements [16, 17, 19, 30, 57-61]. As previously shown, Ku70/86 DNA-binding activity could indicate the cellular capacity to maintain genomic integrity and could be used as a marker of DNA repair capability [62]. A number of studies, using closed DNA minicircles or plasmids that lack free ends, have shown that Ku also exhibits sequence-specific DNA binding to regulatory DNA elements effecting gene transcription and DNA replication [13, 54]. The basic EMSAs used here (Fig. 6A) to analyze protein-DNA origin interaction in the three human breast (normal, transformed and breast cancer) cell lines, revealed the formation of complexes being more prominent in the transformed/tumor cells compared to the normal ones for the Myc11 probe, but staying constant for the LB2 probe, both of which are origin-containing, but the latter is associated with a constitutively expressed housekeeping gene. These results suggest that during progression of breast cancer the c-myc origin seems to be bound more readily by a specific protein complex compared to less transformed and normal cells. Analysis of protein-DNA origin interactions by competitive EMSAs indicated an origin-specific association (Fig. 6B), while supershift EMSAs, using anti-Ku70 and anti-Ku86 antibodies, confirmed that the specific complexes formed in both replication origins contain Ku proteins (Fig. 6C). Furthermore, co-immunoprecipitation analyses showed the interaction of Ku70/86 with DNA repair and replication complexes, revealing that DNA-PKcs, MCM7, and Orc1-6 proteins are part of the observed complexes (Fig. 5). Previous co-immunoprecipitation studies have shown that the Ku heterodimer associates specifically with the human HsOrc-2,-3,-4, and -6 subunits and with DNA-PKcs [16, 32, 63]
In conclusion, this study demonstrates a differential expression and chromatin association of the Ku protein in normal, transformed and tumor breast cell lines. The results show a gradual increase in overexpression and chromatin association of Ku from normal cells, compared to transformed ones, and finally to tumor cells, suggesting that increased expression and chromatin bound levels of the Ku protein are implicated in the process of breast tumor proliferation. Furthermore, these findings suggest that deregulation of Ku may be involved in the step-wise progression to the development of breast cancer.
Acknowledgements
This research was supported by grants from the Cancer Research Society and The Natural Sciences and Engineering Research Council of Canada (NSERC).
Competing interests
The authors declare that they have no competing interests.
References
1. Powell SN, Kachnic LA. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene. 2003;22:5784-91 doi:10.1038/sj.onc.1206678
2. Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA repair. 2007;6:1018-31 doi:10.1016/j.dnarep.2007.02.020
3. Wei L, Lan L, Hong Z, Yasui A, Ishioka C, Chiba N. Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Molecular and cellular biology. 2008;28:7380-93 doi:10.1128/MCB.01075-08
4. Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA repair. 2006;5:1042-8 doi:10.1016/j.dnarep.2006.05.026
5. Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutation research. 2001;480-481:37-50
6. Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H. et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. The EMBO journal. 1998;17:5497-508 doi:10.1093/emboj/17.18.5497
7. Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE. et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510-4 doi:10.1038/35006670
8. Roth DB, Gellert M. New guardians of the genome. Nature. 2000;404:823-5 doi:10.1038/35009180
9. Li GC, Ouyang H, Li X, Nagasawa H, Little JB, Chen DJ. et al. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Molecular cell. 1998;2:1-8
10. Dvir A, Stein LY, Calore BL, Dynan WS. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. The Journal of biological chemistry. 1993;268:10440-7
11. Kuhn A, Gottlieb TM, Jackson SP, Grummt I. DNA-dependent protein kinase: a potent inhibitor of transcription by RNA polymerase I. Genes & development. 1995;9:193-203
12. Labhart P. DNA-dependent protein kinase specifically represses promoter-directed transcription initiation by RNA polymerase I. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2934-8
13. Giffin W, Torrance H, Rodda DJ, Prefontaine GG, Pope L, Hache RJ. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265-8 doi:10.1038/380265a0
14. Park SJ, Ciccone SL, Freie B, Kurimasa A, Chen DJ, Li GC. et al. A positive role for the Ku complex in DNA replication following strand break damage in mammals. The Journal of biological chemistry. 2004;279:6046-55 doi:10.1074/jbc.M311054200
15. Zannis-Hadjopoulos M, Sibani S, Price GB. Eucaryotic replication origin binding proteins. Frontiers in bioscience: a journal and virtual library. 2004;9:2133-43
16. Sibani S, Price GB, Zannis-Hadjopoulos M. Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry. 2005;44:7885-96 doi:10.1021/bi047327n
17. Sibani S, Price GB, Zannis-Hadjopoulos M. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/- cells. Journal of cell science. 2005;118:3247-61 doi:10.1242/jcs.02427
18. Shimura T, Martin MM, Torres MJ, Gu C, Pluth JM, DeBernardi MA. et al. DNA-PK is involved in repairing a transient surge of DNA breaks induced by deceleration of DNA replication. Journal of molecular biology. 2007;367:665-80 doi:10.1016/j.jmb.2007.01.018
19. Rampakakis E, Di Paola D, Zannis-Hadjopoulos M. Ku is involved in cell growth, DNA replication and G1-S transition. Journal of cell science. 2008;121:590-600 doi:10.1242/jcs.021352
20. Sawada M, Sun W, Hayes P, Leskov K, Boothman DA, Matsuyama S. Ku70 suppresses the apoptotic translocation of Bax to mitochondria. Nature cell biology. 2003;5:320-9 doi:10.1038/ncb950
21. Klein A, Miera O, Bauer O, Golfier S, Schriever F. Chemosensitivity of B cell chronic lymphocytic leukemia and correlated expression of proteins regulating apoptosis, cell cycle and DNA repair. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2000;14:40-6
22. Pucci S, Mazzarelli P, Rabitti C, Giai M, Gallucci M, Flammia G. et al. Tumor specific modulation of KU70/80 DNA binding activity in breast and bladder human tumor biopsies. Oncogene. 2001;20:739-47 doi:10.1038/sj.onc.1204148
23. Seimiya M, Tomonaga T, Matsushita K, Sunaga M, Oh-Ishi M, Kodera Y. et al. Identification of novel immunohistochemical tumor markers for primary hepatocellular carcinoma; clathrin heavy chain and formiminotransferase cyclodeaminase. Hepatology. 2008;48:519-30 doi:10.1002/hep.22364
24. Lagadec C, Romon R, Tastet C, Meignan S, Com E, Page A. et al. Ku86 is important for TrkA overexpression-induced breast cancer cell invasion. Proteomics Clinical applications. 2010;4:580-90 doi:10.1002/prca.200900148
25. Mimori T, Hardin JA, Steitz JA. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. The Journal of biological chemistry. 1986;261:2274-8
26. Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes & development. 1999;13:916-34
27. Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:525-30
28. Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131-42
29. Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & development. 2001;15:2177-96 doi:10.1101/gad.914401
30. Novac O, Matheos D, Araujo FD, Price GB, Zannis-Hadjopoulos M. In vivo association of Ku with mammalian origins of DNA replication. Molecular biology of the cell. 2001;12:3386-401
31. Di Paola D, Zannis-Hadjopoulos M. Comparative analysis of pre-replication complex proteins in transformed and normal cells. J Cell Biochem. 2012;113:1333-47 doi:10.1002/jcb.24006
32. Matheos D, Ruiz MT, Price GB, Zannis-Hadjopoulos M. Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication. Biochimica et biophysica acta. 2002;1578:59-72
33. Di Paola D, Price GB, Zannis-Hadjopoulos M. Differentially active origins of DNA replication in tumor versus normal cells. Cancer research. 2006;66:5094-103 doi:10.1158/0008-5472.CAN-05-3951
34. Koike M, Shiomi T, Koike A. Dimerization and nuclear localization of ku proteins. The Journal of biological chemistry. 2001;276:11167-73 doi:10.1074/jbc.M010902200
35. Leffak M, James CD. Opposite replication polarity of the germ line c-myc gene in HeLa cells compared with that of two Burkitt lymphoma cell lines. Molecular and cellular biology. 1989;9:586-93
36. Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G. et al. Fine mapping of a replication origin of human DNA. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7119-23
37. Di Paola D, Rampakakis E, Chan MK, Zannis-Hadjopoulos M. Differential chromatin structure encompassing replication origins in transformed and normal cells. Genes & cancer. 2012;3:152-76 doi:10.1177/1947601912457026
38. Tao L, Dong Z, Leffak M, Zannis-Hadjopoulos M, Price G. Major DNA replication initiation sites in the c-myc locus in human cells. J Cell Biochem. 2000;78:442-57
39. Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M. et al. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023-6
40. Di Paola D, Rampakakis E, Chan MK, Arvanitis DN, Zannis-Hadjopoulos M. Increased origin activity in transformed versus normal cells: identification of novel protein players involved in DNA replication and cellular transformation. Nucleic acids research. 2010;38:2314-31 doi:10.1093/nar/gkp1192
41. Prabhakar BS, Allaway GP, Srinivasappa J, Notkins AL. Cell surface expression of the 70-kD component of Ku, a DNA-binding nuclear autoantigen. The Journal of clinical investigation. 1990;86:1301-5 doi:10.1172/JCI114838
42. Grawunder U, Finnie N, Jackson SP, Riwar B, Jessberger R. Expression of DNA-dependent protein kinase holoenzyme upon induction of lymphocyte differentiation and V(D)J recombination. European journal of biochemistry / FEBS. 1996;241:931-40
43. Dalziel RG, Mendelson SC, Quinn JP. The nuclear autoimmune antigen Ku is also present on the cell surface. Autoimmunity. 1992;13:265-7
44. Reeves WH. Antibodies to the p70/p80 (Ku) antigens in systemic lupus erythematosus. Rheumatic diseases clinics of North America. 1992;18:391-414
45. Gullo C, Au M, Feng G, Teoh G. The biology of Ku and its potential oncogenic role in cancer. Biochimica et biophysica acta. 2006;1765:223-34 doi:S0304-419X(06)00002-3 [pii] 10.1016/j.bbcan.2006.01.001
46. Feldmann E, Schmiemann V, Goedecke W, Reichenberger S, Pfeiffer P. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic acids research. 2000;28:2585-96
47. Amsel AD, Rathaus M, Kronman N, Cohen HY. Regulation of the proapoptotic factor Bax by Ku70-dependent deubiquitylation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5117-22 doi:0706700105 [pii] 10.1073/pnas.0706700105
48. Soderlund Leifler K, Queseth S, Fornander T, Askmalm MS. Low expression of Ku70/80, but high expression of DNA-PKcs, predict good response to radiotherapy in early breast cancer. International journal of oncology. 2010;37:1547-54
49. Luk JM, Su YC, Lam SC, Lee CK, Hu MY, He QY. et al. Proteomic identification of Ku70/Ku80 autoantigen recognized by monoclonal antibody against hepatocellular carcinoma. Proteomics. 2005;5:1980-6 doi:10.1002/pmic.200401084
50. Korabiowska M, Voltmann J, Honig JF, Bortkiewicz P, Konig F, Cordon-Cardo C. et al. Altered expression of DNA double-strand repair genes Ku70 and Ku80 in carcinomas of the oral cavity. Anticancer research. 2006;26:2101-5
51. Mazzarelli P, Parrella P, Seripa D, Signori E, Perrone G, Rabitti C. et al. DNA end binding activity and Ku70/80 heterodimer expression in human colorectal tumor. World journal of gastroenterology: WJG. 2005;11:6694-700
52. Parrella P, Mazzarelli P, Signori E, Perrone G, Marangi GF, Rabitti C. et al. Expression and heterodimer-binding activity of Ku70 and Ku80 in human non-melanoma skin cancer. Journal of clinical pathology. 2006;59:1181-5 doi:10.1136/jcp.2005.031088
53. Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Molecular biology of the cell. 2006;17:5337-45 doi:10.1091/mbc.E06-04-0298
54. Ruiz MT, Matheos D, Price GB, Zannis-Hadjopoulos M. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Molecular biology of the cell. 1999;10:567-80
55. Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367-78 doi:10.1038/nrm1367 nrm1367 [pii]
56. Rampakakis E, Arvanitis DN, Di Paola D, Zannis-Hadjopoulos M. Metazoan origins of DNA replication: regulation through dynamic chromatin structure. J Cell Biochem. 2009;106:512-20 doi:10.1002/jcb.22070
57. Knuth MW, Gunderson SI, Thompson NE, Strasheim LA, Burgess RR. Purification and characterization of proximal sequence element-binding protein 1, a transcription activating protein related to Ku and TREF that binds the proximal sequence element of the human U1 promoter. The Journal of biological chemistry. 1990;265:17911-20
58. Falzon M, Fewell JW, Kuff EL. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. The Journal of biological chemistry. 1993;268:10546-52
59. Camara-Clayette V, Thomas D, Rahuel C, Barbey R, Cartron JP, Bertrand O. The repressor which binds the -75 GATA motif of the GPB promoter contains Ku70 as the DNA binding subunit. Nucleic acids research. 1999;27:1656-63
60. Schild-Poulter C, Matheos D, Novac O, Cui B, Giffin W, Ruiz MT. et al. Differential DNA binding of Ku antigen determines its involvement in DNA replication. DNA and cell biology. 2003;22:65-78 doi:10.1089/104454903321515887
61. Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK. et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798-802 doi:10.1126/science.1127196
62. Hwang BJ, Smider V, Chu G. The use of electrophoretic mobility shift assays to study DNA repair. Methods Mol Biol. 1999;113:103-20 doi:10.1385/1-59259-675-4:103
63. Wang H, Zeng ZC, Perrault AR, Cheng X, Qin W, Iliakis G. Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic acids research. 2001;29:1653-60
Author contact
![]() Corresponding author: Dr. Maria Zannis-Hadjopoulos, 3655 Promenade Sir William Osler, Rm. 707, Montreal, QC, Canada H3G 1Y6. Tel: 514-398-3536. Fax: 514-398-6769. E-mail: maria.zannisca.
Corresponding author: Dr. Maria Zannis-Hadjopoulos, 3655 Promenade Sir William Osler, Rm. 707, Montreal, QC, Canada H3G 1Y6. Tel: 514-398-3536. Fax: 514-398-6769. E-mail: maria.zannisca.

 Global reach, higher impact
Global reach, higher impact