Impact Factor
ISSN: 1837-9664
J Cancer 2012; 3:19-31. doi:10.7150/jca.3.19 This volume Cite
Research Paper
Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients
Centre for Biotechnology, Brock University, St. Catharines, ON, L2S 3A1, Canada.
Received 2011-8-14; Accepted 2011-11-2; Published 2011-12-9
Abstract
MicroRNAs (miRNA) are small endogenously expressed non-coding RNAs that negatively regulate expression of protein-coding genes at the translational level. Accumulating evidence, such as aberrant expression of miRNAs, suggests that they play a role in the development of cancer. They have been identified in various tumor types, demonstrating that different sets of miRNAs are usually deregulated in different cancers. To identify the miRNA signatures specific for Hepatitis C virus (HCV)-associated Hepatocellular carcinoma (HCC), miRNA expression profiling of 32 HCC post-HCV infected, 74 HCV-positive and 12 control individuals was carried out using whole genome expression profiling. Differential expression of two individual miRNAs between control and high risk HCV patients was detected and found to possibly target genes related to HCC development and progression. The sensitivity and specificity of miR-618 for detecting HCC among HCV-positive individuals was found to be 64% and 68%, respectively. Whereas, the sensitivity and specificity of miR-650 were 72% and 58%, respectively. Additionally, the sensitivity and specificity for miR-618/650 in tandem were 58% and 75%, respectively. These predictive values are greatly improved compared to the traditional α-feto protein (AFP) level-based detection method. The proposed HCC miRNA signatures may therefore be of great value for the early diagnosis of HCC, before the onset of disease in HCV-positive patients. The significance of this approach is amplified by the use of urine as a sample source as it offers a non-invasive approach for developing screening methods that can reduce mortality rates.
Keywords: Liver Cancer, Hepatitis C Virus, Urine, MicroRNA, Biomarkers.
Introduction
Hepatocellular carcinoma (HCC) is one of the leading causes of deaths worldwide and is responsible for 500,000 deaths per year (1). Individuals chronically infected with hepatitis B or C virus (HBV, HCV) are at high risk for the development of HCC, with disease progression occurring persistently over many years (2). Hepatitis B virus (HBV) and Hepatitis C virus (HCV) are the major risk factors for the development of HCC. Worldwide, approximately 370 million individuals are HBV positive and 130 million HCV positive (3). The development of HCC is also associated with other risk factors including gender, age and dietary factors (such as Aflatoxin B1 and alcohol) (4).
The major potentially curative form of therapy is still surgical resection, however only 10% of patients were at operable stages upon disease discovery. This is mainly due to the absence of reliable tools for early diagnosis and thus most patients were at a late stage either for surgical solutions or for effective treatment (5). The widely used serological tumor markers for HCC, α-fetoprotein (AFP) and des-γ-carboxy prothrombin (DCP), lack specificity and sensitivity. AFP specificity and sensitivity are 75% and 68%, respectively, whereas elevated DCP activity is only present in 44-47% of HCCs less than 3 cm in size (6, 7, 8). Most chronically infected patients remain asymptomatic for many years and the long latency between infection and development of HCC provides an important window of time during which individuals can be monitored for disease progression and intervention could be efficient (9). Therefore, the development of non-invasive biomarkers with high sensitivity and specificity that can be used for large-scale clinical investigations would be highly beneficial.
Various molecular alterations, leading to dysregulation of well-known molecular pathways in carcinogenesis, take place in the cancerous nodules and appear in HCCs (10, 11). Some of these pathways have recently been discovered as being regulated by microRNAs (miRNAs). MiRNAs are short noncoding RNAs (~22 nucleotides) that are believed to play an important role in the regulation of gene expression and hence control many biological processes (12). Since the discovery of miRNAs in 1993 in the nematode Caenorhabditis elegans (13), 470 miRNAs have been described in the miRBase database v9.15 and 265 potential miRNAs have also been identified in a RAKE analysis study (14, 15).
The production of a mature miRNA occurs in a stepwise manner, starting with a large RNA Pol II transcript that is processed by the sequential action of several proteins/enzymes of the RNAi pathway, resulting in the generation of a short double stranded RNA molecule (16). The “effector” (or antisense) strand of this short dsRNA molecule is incorporated into and guides the RNA-induced silencing complex (RISC) to its target mRNA (17). Recognition of the target is facilitated by partial (or in rare cases full) sequence complementarity between the miRNA and its target mRNA. Binding of the RISC to mRNA may result in translational repression if base pairing between the miRNA and its target mRNA is imperfect (as is the case for most of the miRNA mediated silencing), or mRNA target degradation, in case of full complementarity. MiRNA expression profiles seem to be tissue-specific and tumor-specific (18).
Half of the known miRNAs are localized inside or close to fragile sites, in minimal sites of loss of heterozygosity and at familiar breakpoints associated with cancer (19). Several recent studies have verified that miRNA expression signatures may have a significant impact on the classification, diagnosis and prognosis of the disease in cancer patients (20, 21). A comprehensive analysis of the miRNA expression in different cancers showed a higher diagnostic and prognostic accuracy for the miRNA genetic fingerprint than for more than 16,000 mRNAs (21). Recently, the biological functions and targets of some miRNAs have been reported. MiR-375 regulates insulin secretion by modulating the expression of myotropin (22). Abnormal expression of the miR-155 precursors, mature miR-15 and miR-16 were described in malignant lymphomas (19, 23). Furthermore, the downregulation of miR-143 and miR-145 was reported in colorectal neoplasia (16). In addition, miR-372 and miR-373 neutralize p53-mediated cyclin-dependent kinase inhibition and hence allow tumor growth in the presence of wild-type p53 (24).
Understanding the pathogenesis of viral-associated HCC is important in developing effective means of prevention and treatment of this highly malignant form of cancer (25). Nowadays, the identification of a non-invasive biological biomarker that can be used to screen high-risk patients in order to enhance HCC detection, earlier intervention and increase the probability of successful treatment is urgently needed. In the present study, the miRNA expression profiles in urine samples collected from HCC post HCV-postitive group, HCV-postitive group as well as from healthy control group were analyzed. MiRNA profiling was firstly carried out on miRNA isolated from urine samples pooled from each group using the miRNA expression profiling assay panel (Illumina). Furthermore, we evaluated the deregulation of 5 miRNAs, revealed from our data, as putative non-invasive urinary biomarkers for screening high-risk patients for the early detection of HCC.
Materials and Methods
Urine sample collection. Urine samples were collected from the general hospital at Alexandria University (Alexandria, Egypt) and the National Institute for Liver Diseases at Menoufia University (Shebeen El-Kome, Egypt) upon the approval of the Research Ethics Board at Brock University (St. Catharines, Canada). Urine samples were collected (50mL) in the presence of 5 mM EDTA (final concentration). All the samples were divided into 3 groups based on a questionnaire filled by the clinician responsible for each patient. The 3 groups participating in this study were divided as follows: 32 patients with HCC post-HCV infection, 74 patients with chronic HCV infection and 12 normal individuals (Table 1). Patients with other conditions such as HBV/HCV co-infection and Schistosomiasis were excluded. All three groups were examined by HCV 5' non-coding region reverse transcriptase PCR (5' NCR RT-PCR). The results confirmed that the HCC post-HCV and HCV-positive patients share the same etiological factor, HCV genotype 4 (data not shown).
Clinical pathological parameters of the patients involved in this study.
| Control group | HCC post-HCV group | HCV-positive group | |
|---|---|---|---|
| Number of patients | 12 | 32 | 74 |
| Age (mean ± SD) | 28 ±4 | 50 ±8 | 36 ±18 |
| HCV RNA* | -Ve | +Ve | +Ve |
| Sex | Male (6) Female (6) | Male (26) Female (6) | Male (50) Female (24) |
* All patients were confirmed to be HCV positive or negative by polymerase chain reaction for HCV NCR. Sequencing the PCR product confirmed the genotype of the HCV to be genotype 4.
Isolation of total RNA from urine. Total RNA was isolated from 3mL of urine using the Urine (Exfoliated cell) RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada). The isolation was performed using a modified protocol. Briefly, 3mL of urine was mixed with 350μL of lysis solution. The sample was vortexed for 15 seconds and the lysate was then transferred to an RNase-free microcentrifuge tube. To the lysate, 200μL of 95-100% ethanol was added and mixed by vortexing for 10 seconds. The lysate + ethanol were applied to the column. The column was then centrifuged for 1min at 10,000 rpm. After centrifugation, the flow-through was discarded and the spin column was re-assembled with the collection tube. The previous step was repeated until all the lysate had been loaded onto the column. The column was washed twice by applying 500μL wash solution to the column and centrifuging for 1min at 10,000 rpm. The column was spun for an additional minute at 14,000 rpm to ensure that the column was completely dry prior to RNA elution. The column was placed into a new 1.7mL elution tube (provided with the kit). To the column, 50μL of elution buffer was added and the column was spun down for 2min at 2,000 rpm followed by a 1min spin at 14,000rpm. The purified RNA sample was stored at -70°C till further analysis.
Capillary electrophoresis. The purified miRNAs were loaded onto an Agilent® RNA Nano 6000 chip and resolved on an Agilent® 2100 BioAnalyzer according to the manufacturer's instruction (Agilent Technologies).
MicroRNA whole-genome expression profiling. The differential miRNA expression profiling was performed using the miRNA expression profiling assay panel (Illumina, San Diego, CA, USA) at the SickKids genomic core facility (Toronto, ON, Canada). This assay is an adaptation of the proven DASL® (cDNA-mediated Annealing, Selection, Extension and Ligation) assay. Hybridization and scanning of miRNA expression bead array was performed according to the manufacturer's instructions (Illumina, San Diego, CA, USA).
Relative expression profiling for candidate miRNAs using RT-qPCR. Relative miRNA expression levels for the 5 candidate miRNAs (miR-625, miR-532, miR-618, miR-516 and miR-650) were analyzed among the HCC post HCV-positive group, HCV-positive group and the control group. Candidate miRNAs were reversibly-transcribed using 3µl of RNA template and 0.5µl gene-specific stem-loop RT primer (Table 2). The reaction was then made up with RNase-free water to 5μL and incubated for 5 mins at 70°C followed by a 10 minute incubation at 4°C. During the 4°C incubation, a mix of 10µl 5X first strand Buffer, 5µl of 0.1mM DTT, 2.5µl of 10mM dNTPs, 2µl SuperScript III (Invitrogen) and 25.5µl nuclease-free water were added to the initial reaction for a final reaction volume of 50µL. The final reaction was then incubated at 25°C for 10 min, 42°C for 90 min, 70°C for 15 min with a final hold at 4°C. For controls, minus RT reactions were set up (only for the control group) where the enzyme was replaced with nuclease-free water. All samples were analyzed using the iCycler iQ real time PCR detection system with iQ SYBR Green Supermix (BioRad Laboratories). For each sample, 3μl from its corresponding RT reaction were mixed in a 7.5μl iQ SYBR Green Supermix, 4.26μl nuclease-free water and 0.12μl each specific forward and reverse miRNA primer (Table 2) in a total of 15μl. All RT-qPCR experiments were carried out in triplicate and the expression levels were normalized to 5S rRNA levels. The relative expression level (fold change) for each candidate miRNA within each group was then calculated using the equation 2-∆Ct test / 2-∆Ct control.
List of oligonucleotides used in this project.
| Name | Sequence |
|---|---|
| SL-miR-625 | 5'GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATA CGACAGGACT 3' |
| miR-625 (Forward) | 5' AGGGGGAAAGTTCTATAG-3' |
| SL-miR-532 | 5'GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATA CGACACGGTC 3' |
| miR-532 (Forward) | 5' CATGCCTTGAGTGTAGGA 3' |
| SL-miR-618 | 5'GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATA CGACACTCAG 3' |
| miR-618 (Forward) | 5' AAACTCTACTTGTCCTTCTG 3' |
| SL-miR-516 | 5'GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGA CAAAGTG 3' |
| miR-516 (Forward) | 5' CATCTGGAGGTAAGAAGCAC 3' |
| SL-miR-650 | 5'GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATAC GACGTCCTG 3' |
| miR-650 (Forward) | 5' AGGAGGCAGCGCTCTCAG 3' |
| 5S rRNA (Forward) | 5' GCCATACCACCCTGAACG 3' |
| 5S rRNA (Reverse) | 5' AGCCTACAGCACCCGGTATT 3' |
Results
Micro-RNA Microarray Expression Profiles among HCC-post HCV positive, HCV positive and control group
Micro-RNA was isolated using the miRNA purification Kit (Norgen Biotek, Thorold, ON, Canada) from urine samples pooled from the three groups participating in this project (i.e. HCC post HCV-positive group, HCV-positive group and control group). The quality and the quantity of the isolated miRNA were tested using the Agilent RNA 6000 Kit. The RNA concentrations in the HCC post HCV-positive group, HCV-positive group and the control group were 82ng/μL, 148ng/μL and 30ng/μL, respectively.
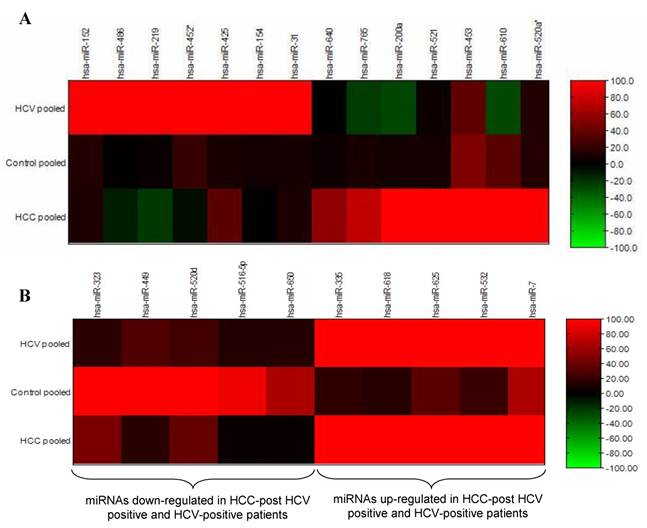
Differential expression analyses of miRNA isolated from pooled urine samples was performed using the illumina MicroRNA Expression Profiling Assay, which includes 470 miRNAs described in the miRBase database v9.15 and 265 potential miRNAs identified in a RAKE analysis study (14, 15). The signal intensity of each gene in the HCC-post HCV positive group and the HCV positive group was normalized against its equivalent in the control group (Figure 1). After calculating the fold change in the expression of different miRNAs in the HCC-post HCV positive group and in the HCV positive group relative to the control group, miR-152, miR-486, miR-219, miR452*, miR-425, miR-154 and miR-31 were significantly up-regulated only among chronic HCV patients. Furthermore, miR-640, miR-765, miR-200a, miR-521 and miR-520* were significantly up-regulated only in the HCC-post HCV positive group, relative to their expression (or lack thereof) in the control group. MiR-453 and miR-610 were expressed relatively weakly in the control group, but their expression was markedly increased in the HCC-post HCV positive group. It should also be noted that miR-486 and miR-219 (up-regulated among the HCV-positive group) were found to be down-regulated in the HCC-post HCV positive group.
The opposite was true for the expression levels of miR-765, miR200a and miR-610, which were significantly up-regulated in the HCC-post HCV group but were down-regulated in the HCV positive group. Moreover, miR-323, miR-449, miR-502d, miR-92b, miR-516-5p and miR-650 were found to be commonly down-regulated in both the HCC-post HCV positive group and in the HCV positive group, relative to the control group. Finally, miR-335, miR-618, miR-625, miR-532, miR-7 were found to be commonly up-regulated in both the HCC-post HCV positive group and in the HCV positive group, relative to the control group. Scanning the TargetScan and MiRanda databases (26) revealed that the commonly up-regulated miRNAs (miR-625, miR-532 and miR-618) were likely playing a significant role in the development of HCC among high risk HCV positive patients. The methyl-CpG binding domain protein 2 gene (MBD2) is likely to be targeted by miR-625. MiR-532 likely targets ligase I, DNA, and the ATP-dependent gene (LIG 1), whereas, the putative target of miR-618 was found to be the low density lipoprotein-related protein 12 (LRP12).
Heat map of miRNA expression in HCC-post HCV positive, HCV positive and control group. A) MiRNA up-regulated in HCC-post HCV positive and HCV positive group. B) MiRNA de-regulated in both HCC post HCV-positive and HCV-postitive group relative to the control group. The signal intensity from each miRNA tested in either the HCC or the HCV groups was normalized against its equivalent in the control group.
Validation of miR-625, miR-532, miR-618, miR-516-5p and miR-650 Deregulation among HCC-post HCV positive Patients using RT-qPCR
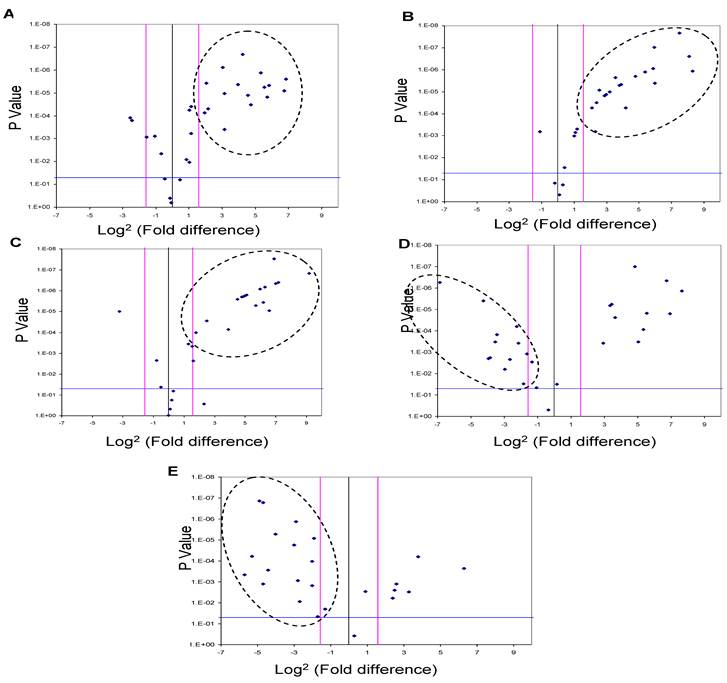
Five of the miRNAs, three up-regulated (miR-625, miR-532 and miR-618) and two down-regulated (miR-516-5P and miR-650), whose levels were significantly different in the urine pooled from the HCC post HCV-positive group relative to the control group, were studied as potential HCC biomarker(s) since their putative targets have a role in cancer development. In order to evaluate their potential as biomarkers, the relative expression levels of these miRNAs were examined among patients in the HCC-post HCV positive group, using RT-qPCR. Each gene was normalized against the 5S rRNA for both the HCC post HCV-positive and the control group. The relative expression of each miRNA was calculated as a fold difference of expression, using the equation 2-∆Ct (target miRNA) HCV / 2-∆Ct (target miRNA) control. Differences in the expression levels of miRNAs between the groups were considered significant when there was a three or more fold difference in expression and the p value was less than 0.05 (Figure 2).
MiR-625, miR532 and miR-618 were found to be significantly up-regulated in 56%, 62.5% and 72% of HCC-post HCV positive patients, respectively (i.e. 18, 20 and 23 of 32 patients). Moreover, miR-516-5p and miR-650 were found to be significantly down-regulated in 50% and 72% of HCC post HCV-positive patients, respectively (i.e. 16 and 23 of 32 patients).
A Volcano Plot graphs the log2 for the fold difference in A) miR-625, B) miR-532, C) miR-618 D) miR-516-5p and E) miR-650 expression versus its p value from the t-test among HCC-post HCV positive group. The black line indicates a fold-change in gene expression of 1. The pink lines indicate the desired fold change in miRNA expression threshold (2). The blue line indicates the desired threshold for the p value of the t-test (P < 0.05). Samples with a significant miRNA deregulation (dotted oval) were chosen at a fold difference ≥ 3 and p < 0.05.
Screening the Expression of miR-618 and miR-650 among High-Risk HCV positive patients using qRT-PCR
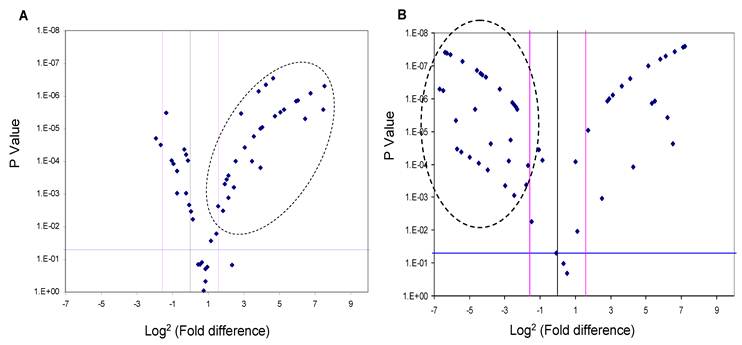
Since the deregulation of expression of miR-618 and miR-650 appeared to be a characteristic event among HCC patients, with 72% of HCC-post HCV positive patients exhibiting this deregulated pattern of expression, the expression levels of miR-618 and miR-650 were examined among the 74 HCV positive patients using RT-qPCR in order to evaluate the potential of using these aberrant miRNA expression signatures as an HCC biomarker. MiR-618 was significantly (three or more fold difference in expression levels and a p value of the student t-test lower than 0.05) up-regulated in 35 of the 74 HCV positive patients (Figure 3A), while, miR-650 was significantly down-regulated in 42 of the 74 HCV positive patients (Figure 3B).
A Volcano Plot graphs the log2 for the fold difference in A) miR-618 and B) miR-650 expression versus its p value from the t-test among HCV positive group. The black line indicates a fold-change in gene expression of 1. The pink lines indicate the desired fold-change in gene expression threshold (3). The blue line indicates the desired threshold for the p value of the t-test (P < 0.05). Samples with a significant miRNA deregulation (dotted oval) were chosen at a fold change ≥ 3 and p < 0.05.
Diagnostic Value of miR-618 and miR-650 for the Detection of HCC among High-Risk HCV infected Patients
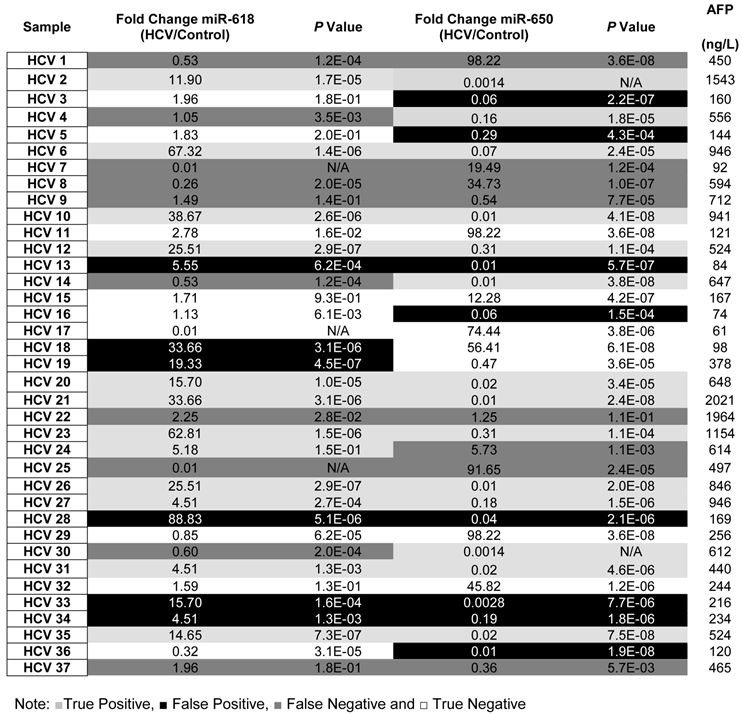
To evaluate the potential of using aberrant expression of miR-618 and miR-650 for diagnostic purposes, the medical records, specifically AFP levels and CT scan results, of all HCV-positive patients were updated two years after initial sample collection. It should be noted that all HCV positive patients were diagnosed as being HCC-free (AFP level lower than 400ng/L and a negative CT scan) at the time of initial sample collection. The specificity, sensitivity and the diagnostic accuracy of miR-618 up-regulation and miR-650 down regulation (separately or in tandem) for diagnosing HCC were then determined, based on the updated medical records of HCV patients. The following criteria were applied: patients with AFP levels of 400ng/L or more, with a positive CT scan, and exhibiting significant deregulation of either miR-618 or miR-650 expression, or deregulation of both, were considered true positives; patients whose AFP levels were 400ng/L or more, with a positive CT scan, and not exhibiting significant deregulation of any of the miRNAs (either separately or in tandem) were considered false negatives; patients with AFP levels lower than 400ng/L, with a negative CT scan, and exhibiting significant deregulation of miR-618 or miR-650 expression, or deregulation of both, were considered false positives; and finally, samples with AFP levels lower than 400ng/L and not exhibiting significant deregulation of either miR-618 or miR-650 expression, or deregulation of both, were considered true negatives.
By applying the above mentioned criteria (see also Table 3), the sensitivity and the specificity of miR-618 up-regulation for detecting HCC among HCV-positive patients, based on AFP levels and CT scan results, were 64% and 68%, respectively with a 66% diagnostic accuracy. Whereas, the sensitivity and the specificity of miR-650 down-regulation for detecting HCC among HCV-positive patients were 72% and 58%, respectively with a 65% diagnostic accuracy. Additionally, the sensitivity and the specificity of tandem deregulation of miR-618 and miR-650 expressions for detecting HCC were 58% and 75%, respectively with a diagnostic accuracy of 69%. There was no correlation between the relative expression of miR-618 or miR-650 and AFP levels.
Discussion
MicroRNAs (MiRNAs) are negative gene regulators that have been shown to control many crucial biological processes, such as cellular proliferation, cellular differentiation and apoptosis. Not surprisingly, miRNAs play a critical role in the initiation and progression of human cancer (27). Recent evidence indicates that miRNAs can act as both tumor suppressors and oncogenes (or oncomirs) (28). Half of the known miRNAs are localized inside or close to fragile sites, sites of loss of heterozygosity and at familiar breakpoints associated with cancer (19). Several recent studies reported that miRNA expression signatures may have a significant impact on the classification, diagnosis and prognosis of cancer (20, 21). A comprehensive analysis of miRNA expression in different cancers showed a higher diagnostic and prognostic accuracy than expression profiling of 16,000 mRNAs (21).
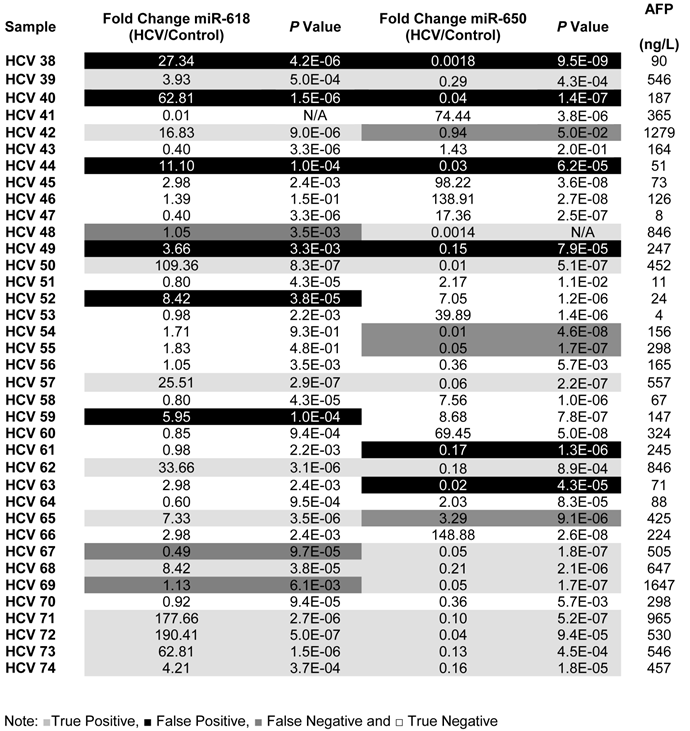
A list showing the fold change, p values for the expression of miR-618, miR-650 as well as the AFP levels for each patient in the Testing set.
Understanding the pathogenesis of viral-associated HCC is important for developing effective means of prevention and treatment of this highly malignant form of cancer (25). Currently, the identification of a non-invasive biological biomarker that can be used to screen high-risk patients is urgently needed to enhance HCC detection, resulting in earlier intervention and increasing the probability of successful treatment. Recent studies have already identified several miRNAs whose expression is deregulated in viral hepatitis infections and in HCC (29). Although important information was gathered about the hepatocarcinogenic effect of these deregulated miRNAs and the viral-host interactions that lead to the viral pathogenic effect, none of these studies evaluated the potential of using deregulated miRNA expression patterns to predict the development and progression of HCC among high-risk HCV infected patients.
The aim of our biomarker discovery approach was to investigate the potential of using deregulation of miRNA expression in HCC-post HCV infected patients for the early diagnosis of HCC in high risk groups. The expression levels of miRNAs isolated from urine were studied using the Illumina MicroRNA Expression Profiling Assay. To analyze the global differences in miRNA expression patterns between the two test groups participating in this study and the control group, the miRNAs isolated from individuals belonging to the same group were pooled together and analyzed. After analysis, four different categories of differentially expressed miRNAs were identified. The first category of miRNAs were up-regulated only in the HCV positive group, relative to the control group (miR-152, miR-486, miR-219, miR452*, miR-425, miR-154 and miR-31). The second category of miRNAs were up-regulated only in the HCC-post HCV positive group, relative to the control group (miR-640, miR-765, miR-200a, miR-521 and miR-520a*). The miRNAs identified in these two categories are important for studying HCV pathogenicity, HCV-host interaction, and the hepatocarcinogenic effect of miRNA as a consequence of HCV infection. Unfortunately, these miRNAs can not be used as HCC biomarkers because they were not commonly deregulated in the HCC-post HCV positive group and the HCV positive group.
The expression of ten miRNAs was commonly deregulated in both the HCC-post HCV group and the HCV positive group. Of these ten deregulated miRNAs, five miRNAs were significantly down-regulated (miR-323, miR-449, miR-520d, miR-516-5p and miR-650), while the other five miRNAs were up-regulated (miR-335, miR-618, miR-625, miR-532, and miR-7), relative to the control group. The common deregulation of these miRNAs represented possible HCC biomarker candidates.
It was decided that three commonly down-regulated miRNAs (miR-323, miR-449 and miR-520d) and two commonly up-regulated miRNAs (miR-335 and miR-7) would not be investigated as possible HCC biomarker candidates. The decision for not using miR-7 and miR-335 as biomarker candidates was based on their putative targets and a review of relevant literature. MiR-7 is thought to be an endocrine miRNA, highly expressed in the human pancreatic islets (30). Since HCV is known to infect the pancreatic cells (31), the presence of miR-7 in both the HCC-post HCV positive group and the HCV positive group may have resulted from the destruction of pancreatic cells as part of the cellular immune response against HCV infection. As for miR-335, it reportedly inhibits tumor metastasis through regulating a set of genes whose expression is associated with a risk of distal metastasis, especially in breast cancer (32). It is, therefore, not specific enough to be used as an HCC biomarker.
Similarly to miR-7 and miR-335, three commonly down-regulated miRNAs (miR-323, miR-449 and miR-520d) were excluded from this study based on specificity factors. The transforming growth factor-α (TGF-α) is thought to be the putative target of miR-323. Growth factors are generally secreted by tumor cells in an autocrine fashion and are considered important factors in tumorigenesis (33). TGF-α has been reported to be over-expressed in various tumor tissues, and is likely to be associated with the occurrence of lymph nodes, liver metastases and short survival (34). TGF-α is also over-expressed in colorectal carcinoma and is associated with poor prognosis of this cancer (35). It is likely that expression of miR-323 down-regulates the expression of TGF-α; consequently, the down-regulation of miR-323 leads to the uncontrolled over-expression of TGF-α.
As expected, up-regulation of the putative targets of miR-449 and miR-520d, Stathmin 1 and Matrin 3, respectively, was previously reported in the Proteomic-based biomarker approach. As a reminder, Matrin 3 has been identified by the HPLC-MS/MS analysis in proteins pooled from the HCV positive group, whereas Stathmin 1 was commonly present in both the HCC-post HCV positive group and the HCV positive group. Stathmin 1 over-expression in HCC patients was associated with higher vascular invasion and considered to be an important prognostic factor for HCC, which may serve as a useful marker to predict early tumor recurrence (ETR) (36). Matrin 3 is an RNA-binding protein, reported to be one of the cellular proteins that interacts with the 3' UTR of HCV and that plays a role in HCV replication and HCV-associated pathogenesis (37). Theoretically, the down-regulation of both miR-449 as well as miR-520d should be followed by the over-expression of their putative targets (i.e. Stathmin 3 and Matrin 3) and this was confirmed by the data generated from the proteomic-based approach.
Thus, we focused on 5 miRNAs (miR-516-5p, miR-650, miR-625, miR-532 and miR-618) as possible HCC biomarker candidates among high-risk HCV patients. Based on the TargetScan and MiRanda databases, miR-516-5p, miR-650, miR-625, miR-532 and miR-618 were found to possibly target genes related to HCC development and progression.
We reviewed the putative targets of the chosen miRNAs closely in order to explain the significance of the deregulation of their expression in the context of HCC. The putative target of miR-650 is TRAF4. TRAF4 is likely to play an important role in carcinogenesis as it was suggested that its over-expression could participate in and/or promote tumorigenesis (38). Accordingly, we hypothesized that its over-expression was a result of the down-regulation of its regulator, miR-650. MiR-650 is, therefore, a possible tumor-suppressor gene and, thus, a potential candidate HCC biomarker among the high-risk HCV positive population.
The putative target of miR-516-5p is Glypican 3 (GPC3). The serum level of GPC3 is significantly elevated in patients with HCC in comparison to healthy individuals or patients with nonmalignant liver diseases (39). It has already been suggested that GPC3 could be used as a marker for HCC (40), therefore, the down-regulation of miR-516-5p, which is a theoretical regulator of GPC3 expression, could also be a potential HCC biomarker among high-risk HCV positive patients.
The putative target of the up-regulated miR-625 is methyl-CpG-binding domain 2 (MBD2). MBD2 is one of five known MBD containing proteins (41) that specifically bind to any methylated gene and mediate its transcriptional repression (42). Given the epigenetic role of MBD proteins in regulating gene expression, they may also be involved in cancer development by affecting the expression of cancer-related genes (43). MBD2 has been reported to bind specifically to methylated gene promoters and recruit histone deacetylases and chromatin remodeling proteins. Consequently, the binding of these factors leads to the alteration of the chromatin structure that may be resistant to transcriptional machinery, causing repression of gene expression (44). In accordance with our data which showed that a large number of tumor-suppressor genes were methylated (“silenced”) among HCC-post HCV positive patients, the up-regulation of miRNA-625 may serve to reduce aberrant methylation events of TSGs by targeting the MBD2 protein and, thus, decrease the tumor growth rate. Therefore, miR-625 may have a tumor-suppressor role.
The putative target of miR-532 is the DNA ligase I (LIG 1), which plays an important role in DNA replication. It has been reported that DNA ligase I is responsible for the majority of DNA ligase activity in proliferating cells compared to non-dividing or resting cells (45, 46). In 2001, Sun et al., reported the presence of high DNA ligase I activity in tumor tissues compared to normal tissues. This activity may improve the survival of tumor cells, especially in the presence of anti-cancer drugs, which directly or indirectly damage DNA. It was also postulated that DNA ligase I could be used as a unique target for the design of new anti-cancer agents (47). Therefore, we assumed that the up-regulation of miR-532 is a natural anti-cancer response that can target and suppress DNA ligase I over-expression and, thus, can be used as an HCC biomarker candidate.
As for miR-618, it was found to possibly target the Low-Density Lipoprotein Receptor-Related Protein 12 (LPR12), which was firstly identified by Qing et al. in 1999. LPR 12 was found to be expressed in normal tissues and was decreased in corresponding malignant tissues (48). Several studies have demonstrated that LPR 12 can act as a tumor suppressor gene as tumor formation/growth was not reduced when it was down-regulated (49). Since LPR 12 was found to be the putative target of miR-618, miR-618 is thought to function as an oncogene. The up-regulation of miR-618 could suppress LPR 12 expression and lead to the progression of HCC. Therefore, the up-regulation of mir-618 was evaluated as a possible HCC biomarker.
Based on the above-mentioned correlations between the observed deregulated miRNAs and HCC progression and development, we decided to evaluate the deregulations of expression of miR-625, miR-532, miR-618, miR-516-5p and miR-650 as non-invasive biomarkers for the early diagnosis of HCC among high-risk HCV positive patients. Initially, we found that miR-625, miR-532 and miR-618 were up-regulated in 56%, 62.5% and 72% of HCC-post HCV positive patients, respectively. In addition, miR-516-5P and miR-650 were found to be down-regulated in 50% and 72% of HCC-post HCV positive patients, respectively. Interestingly, the down-regulation of miR-516-5p among 50% of the HCC-post HCV-positive patients agrees with the diagnostic value of its putative target, Glypican3, which can correctly identify HCC with a sensitivity of 50% (40). Importantly, these results suggested that the deregulation of both miR-618 and miR-650 is a characteristic miRNA signature of HCC among HCV positive patients (72%). This is in accordance with their putative roles as regulators of genes involved in HCC progression and development. MiR-618 up-regulation suppresses the expression of the tumor-suppressor gene (LPR12) and miR-650 down-regulation leads to the over-expression of TARF4.
To evaluate the potential of using miR-618 and/or miR-650 as urinary miRNA-based biomarkers for the early detection of HCC among high-risk HCV positive patients, we studied their expression among HCV positive patients who were not diagnosed as HCC patients at the time of initial sample collection. The expression levels of miR-618 and miR-650 among HCV positive patients, relative to the control group, were evaluated using RT-qPCR. MiR-618 was significantly (≥ 3 fold difference of expression and p value < 0.05) up-regulated in 35 of the 74 HCV positive patients, whereas miR-650 was significantly down-regulated in 42 of the 74 HCV positive patients. The patients who were exhibiting either significant up-regulation of miR-618 or significant down-regulation of miR-650 were suspected as HCC patients. In order to evaluate the diagnostic value of the deregulation of these two miRNAs for identifying HCC among chronic HCV positive patients, the medical records of the HCV infected patients, specifically their AFP levels and CT scan results, were updated two years after initial sample collection. The updated records showed that AFP levels were elevated in 35 HCV infected patients, all of whom also tested positive for HCC by CT scans. Only one patient, #7, had a normal AFP level, but was diagnosed with HCC by CT scan. In total, 36 patients were identified as having developed cancer.
Of the 35 HCV positive patients who showed significant up-regulation of miR-618, 23 were confirmed as HCC positive based on the updated medical records. Furthermore, 26 of the 42 HCV patients who showed significant down-regulation of miR-650 were confirmed as HCC positive. The sensitivity and specificity of miR-618 for detecting HCC among HCV positive patients were 64% and 68%, respectively, with an overall diagnostic accuracy of 66%. The Positive predictive value (PPV) of miR-618 for correctly identifying true HCC positive patients was 66% and the Negative predicative value (NPV) for correctly identifying true negative HCC patients was 67%. Whereas, the sensitivity and specificity of miR-650 for detecting HCC among HCV-positive patients were 72% and 58%, respectively, with an overall diagnostic accuracy of 65%. The PPV of miR-650 for correctly identifying true HCC positive patients was 62% and the NPV for correctly identifying true negative HCC patients was 69%. Additionally, the sensitivity and specificity for miR-618/ miR-650 tandem were 58% and 75%, respectively, with an overall diagnostic accuracy of 69%. The PPV of miR-618/miR-650 for correctly identifying true HCC positive patients was 68% and the NPV for correctly identifying true negative HCC patients was 70%.
Despite the poor predictive values of our findings, the sensitivity of miR-650 and the specificity of the mir-618/miR-650 combination were greatly improved compared to the AFP level-based detection method (sensitivity of 68% and specificity of 75%) (50).The proposed HCC miRNA signatures may be of great value for the early diagnosis of HCC, before the onset of the disease, among high-risk HCV infected patients. The significance of this approach is amplified by the sample source (urine) from which these miRNA signatures were extracted. The advantage of using urine as a sample source for studying diseases is that it offers a non-invasive approach for developing screening tools that can reduce worldwide mortality rates.
Acknowledgements
The authors would like to thank the Egyptian government and the Egyptian Cultural and Educational Bureau in Canada for their financial support of this project. The authors will also like to thank the staff of Norgen Biotek for their technical support.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223- 1232
2. Steel LF, Mattu TS, Mehta A, Hebestreit H, Dwek R, Evans AA, London WT, Block T. A proteomic approach for the discovery of early detection markers of hepatocellular carcinoma. Dis Markers. 2001;3:179-189
3. Moradpour D, Blum HE. Pathogenesis of Hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2005;17:477- 483
4. Colombo M, Sangiovanni A. Etiology, natural history, and treatment of hepatocellular carcinoma. Antiviral Res. 2003;60:145- 150
5. He QY, Zhu R, Lei T, Ng MY, Luk JM, Sham P, Lau GK, Chiu JF. Toward the proteomic identification of biomarkers for the prediction of HBV related hepatocellular carcinoma. J Cell Biochem. 2008;3:740-752
6. Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin Liver Dis. 2001;5:145-159
7. El-Houseini ME, Mohammed MS, Elshemey WM, Hussein TD, Desouky OS, Elsayed AA. Enhanced detection of hepatocellular carcinoma. Cancer Control. 2005;4:248-53
8. Soga K, Watanabe T, Aikawa K, Toshima M, Shibasaki K, Aoyagi Y. Serum des-gamma-carboxyprothrombin level by a modified enzyme immunoassay method in hepatocellular carcinoma: clinical significance in small hepatocellular carcinoma. Hepatogastroenterology. 1998;45:1737-1741
9. Steel LF, Mattu TS, Mehta A, Hebestreit H, Dwek R, Evans AA, London WT, Block T. A proteomic approach for the discovery of early detection markers of hepatocellular carcinoma. Dis Markers. 2001;3:179-189
10. Lee JS, Thorgeirsson SS. Comparative and integrative functional genomics of HCC. Oncogene. 2006;25:3801- 3809
11. Lemmer ER, Friedman SL, Llovet JM. Molecular diagnosis of chronic liver disease and hepatocellular carcinoma: the potential of gene expression profiling. Semin Liver Dis. 2006;26:373- 383
12. Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580- 587
13. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843- 854
14. Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, Plasterk RH. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375- 1377
15. Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, van Zonneveld AJ, Mano H, Plasterk R, Cuppen E. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289- 1298
16. Michael MZ, O'Connor SM, Van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882- 891
17. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776- 780
18. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257- 2261
19. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro - RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524-15529
20. Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189- 198
21. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834- 838
22. Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226-230
23. Van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, Poppema S. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20-28
24. Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169- 1181
25. Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346
26. Griffiths-Jones S, Grocock RJ, Van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140- D144
27. Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60- 12
28. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269
29. Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537-2545
30. Bravo-Egana V, Rosero S, Molano RD, Pileggi A, Ricordi C, Domínguez-Bendala J, Pastori RL. Quantitative differential expression analysis reveals miR-7 as major islet microRNA. Biochem Biophys Res Commun. 2008;366:922-926
31. Yan FM, Chen AS, Hao F, Zhao XP, Gu CH, Zhaov LB, Yang DL, Hao LJ. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol. 2000;6:805-811
32. Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147-152
33. Bianco R, Troiani T, Tortora G, Ciardiello F. Intrinsic and acquired resistance to EGFR inhibitors in human cancer therapy. Endocr Relat Cancer. 2005;12:159-171
34. De Jong KP, Stellema R, Karrenbeld A, Koudstaal J, Gouw AS, Sluiter WJ, Peeters PM, Slooff MJ, De Vries EG. Clinical relevance of transforming growth factor alpha, epidermal growth factor, p53, and Ki67 in colorectal liver metastases and corresponding primary tumors. Hepatology. 1998;28:971-979
35. Liang JL, Wan DS, Pan ZZ, Zhou ZW. Correlations of c-erb-B2, EGFR and TGF-alpha expression to recurrence of Dukes'A and B colorectal carcinoma. Ai Zheng. 2007;26:647-651
36. Yuan RH, Jeng YM, Chen HL, Lai PL, Pan HW, Hsieh FJ, Lin CY, Lee PH, Hsu C. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression and is associated with tumour progression, early recurrence and poor prognosis in hepatocellular carcinoma. J Pathol. 2006;209:549-558
37. Harris D, Zhang Z, Chaubey B, Pandey VN. Identification of cellular factors associated with the 3'-nontranslated region of the hepatitis C virus genome. Mol Cell Proteomics. 2006;5:1006-1018
38. Camilleri-Broët S, Cremer I, Marmey B, Comperat E, Viguié F, Audouin J, Rio MC, Fridman WH, Sautès-Fridman C, Régnier CH. TRAF4 overexpression is a common characteristic of human carcinomas. Oncogene. 2007;26:142-147
39. Nakatsura T, Yoshitake Y, Senju S, Monji M, Komori H, Motomura Y, Hosaka S, Beppu T, Ishiko T, Kamohara H, Ashihara H, Katagiri T, Furukawa Y, Fujiyama S, Ogawa M, Nakamura Y, Nishimura Y. Glypican-3, overexpressed specifically in human hepatocellular carcinoma, is a novel tumor marker. Biochem Biophys Res Commun. 2003;306:16-25
40. Wright LM, Kreikemeier JT, Fimmel CJ. A concise review of serum markers for hepatocellular cancer. Cancer Detect Prev. 2007;31:35-44
41. Berger J, Bird A. Role of MBD2 in gene regulation and tumorigenesis. Biochem. Soc. Trans. 2005;33:1537-1540
42. Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538-6547
43. Schlegel J, Guneysu S, Mennel HD. Expression of the genes of methyl-binding domain proteins in human gliomas. Oncol Rep. 2002;9:393-395
44. Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131-1137
45. Lindahl T, Branes DE. Mammalian DNA ligases. Annu Rev Biochem. 1992;269:251-281
46. Jessberger R, Schar P, Robins P, Ferrari E, Riwar B, Hubscher U. Regulation of DNA metabolic enzymes upon induction of preB cell development and V(D)J recombination: up-reglation of DNA polymerase δ. Nucleic Acids Res. 1997;25:289-296
47. Sun D, Urrabaz R, Kelly S, Nguyen M, Weitman S. Enhancement of DNA ligase I level by gemcitabine in human cancer cells. Clin Cancer Res. 2002;8:1189-1195
48. Qing J, Wei D, Maher VM, McCormick JJ. Cloning and characterization of a novel gene encoding a putative transmembrane protein with altered expression in some human transformed and tumor-derived cell lines. Oncogene. 1999;18:335-342
49. Battle MA, Maher VM, McCormick JJ. ST7 is a novel low-density lipoprotein receptor-related protein (LRP) with a cytoplasmic tail that interacts with proteins related to signal transduction pathways. Biochemistry. 2003;42:7270-7282
50. El-Houseini ME, Mohammed MS, Elshemey WM, Hussein TD, Desouky OS, Elsayed AA. Enhanced detection of hepatocellular carcinoma. Cancer Control. 2005;4:248-253
Author contact
![]() Corresponding author: Moemen AK Abdalla, Centre for Biotechnology, Brock University, St. Catharines, ON, L2S 3A1, Canada. E-mail: demerdashmcom. Telephone: 905-650-9065
Corresponding author: Moemen AK Abdalla, Centre for Biotechnology, Brock University, St. Catharines, ON, L2S 3A1, Canada. E-mail: demerdashmcom. Telephone: 905-650-9065

 Global reach, higher impact
Global reach, higher impact