Impact Factor
ISSN: 1837-9664
J Cancer 2010; 1:150-177. doi:10.7150/jca.1.150 This volume Cite
Review
Novel diagnostic biomarkers for prostate cancer
Department of Pathology and Laboratory Medicine, University of Tennessee Health Science Center, Memphis, TN, USA
Received 2010-9-14; Accepted 2010-10-4; Published 2010-10-6
Abstract
Prostate cancer is the most frequently diagnosed malignancy in American men, and a more aggressive form of the disease is particularly prevalent among African Americans. The therapeutic success rate for prostate cancer can be tremendously improved if the disease is diagnosed early. Thus, a successful therapy for this disease depends heavily on the clinical indicators (biomarkers) for early detection of the presence and progression of the disease, as well as the prediction after the clinical intervention. However, the current clinical biomarkers for prostate cancer are not ideal as there remains a lack of reliable biomarkers that can specifically distinguish between those patients who should be treated adequately to stop the aggressive form of the disease and those who should avoid overtreatment of the indolent form.
A biomarker is a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention. A biomarker reveals further information to presently existing clinical and pathological analysis. It facilitates screening and detecting the cancer, monitoring the progression of the disease, and predicting the prognosis and survival after clinical intervention. A biomarker can also be used to evaluate the process of drug development, and, optimally, to improve the efficacy and safety of cancer treatment by enabling physicians to tailor treatment for individual patients. The form of the prostate cancer biomarkers can vary from metabolites and chemical products present in body fluid to genes and proteins in the prostate tissues.
Current advances in molecular techniques have provided new tools facilitating the discovery of new biomarkers for prostate cancer. These emerging biomarkers will be beneficial and critical in developing new and clinically reliable indicators that will have a high specificity for the diagnosis and prognosis of prostate cancer. The purpose of this review is to examine the current status of prostate cancer biomarkers, with special emphasis on emerging markers, by evaluating their diagnostic and prognostic potentials. Both genes and proteins that reveal loss, mutation, or variation in expression between normal prostate and cancerous prostate tissues will be covered in this article. Along with the discovery of prostate cancer biomarkers, we will describe the criteria used when selecting potential biomarkers for further development towards clinical use. In addition, we will address how to appraise and validate candidate markers for prostate cancer and some relevant issues involved in these processes. We will also discuss the new concept of the biomarkers, existing challenges, and perspectives of biomarker development.
Keywords: diagnostic biomarkers, prostate cancer
1. Introduction
Epidemiology of prostate cancer
With an estimated 192,280 new cases in 2009, prostate cancer is one of the most commonly diagnosed malignancies in American men (1). It is also the second leading cause of cancer death in American males, exceeded only by lung cancer. An estimated 27,360 men will die from prostate cancer in 2009 (1).
Prostate cancer is a disease of mainly older men. An early observation reports that more than 65% of all prostate cancers are diagnosed in men over the age of 65 (2). Compared with the occurrences in the White population, the incidence of prostate cancer is approximately 60% higher in Black men, while native Japanese and Chinese populations have a low risk of incidence and mortality (3). Furthermore, African-American men generally are diagnosed with more advanced stages of prostate cancer and at an earlier age (4). Consequently, much effort is being placed on detecting prostate cancer in an early, curable stage to decrease the rate of mortality from this disease. Along with genetics, social and environmental factors (especially diet and lifestyle) may act as the determining factors, which may explain why some individuals are at higher risk for developing prostate cancer than are others. Nevertheless, in most cases, this disease can be treated effectively and even eradicated when the disease is detected at a very early stage (2).
Biomarkers
The National Cancer Institute defines a biomarker as “a biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process or of a condition or disease.” A biomarker may be objectively measured and evaluated as an indication of normal biologic processes, pathogenic processes, or pharmacologic responses to a particular treatment or condition (5-7). Biomarkers are widely used as analytical tools to assess biological parameters for a rapid and comprehensive therapeutic analysis. In addition, biomarker measures can further the development and evaluation of new therapies (8).
In pilot studies involving therapeutic candidates, biomarkers can be used as criteria for deciding on lead compounds for the third phase of clinical trials (8). They also help in the understanding of clinical pharmacology, and are essential in the planning of clinical trials, which strives to promptly and ultimately assess safety and effectiveness (9, 10) (Table 1 and 2). Biomarkers that represent highly sensitive and specific indicators of disease pathways are often used as substitutes for outcomes in clinical trials where they can be used to predict and evaluate the clinical risk and/or benefit of a treatment, which is the optimal objective of all therapeutic interventions (11).
Use of Cancer Biomarkers in Patient Care
| Use of Biomarker | Clinical Goal |
|---|---|
| Risk Stratification | Used in evaluating the probability of the occurrence or recurrence of cancers. |
| Chemoprevention | To determine and target the cellular and molecular mechanisms of carcinogenesis in preneoplastic tissues. |
| Screening | Used to recognize early-stage cancers in the general population and administer early treatment. |
| Diagnosis and Classification | Used to reliably determine and distinguish the presence and type of cancer. |
| Prognosis | Helps in estimating the likely outcome of the disease, without considering treatment, to establish the intensity of treatment. |
| Prediction of treatment | Anticipate the response to respective treatments and select the therapy with the highest probability of being effective in a particular patient. |
| Therapy Tracking and Post-treatment Surveillance | Used in assessing the effectiveness and adverse effects of a treatment and to provide early determination and treatment of recurrent disease. |
Biomarker Application in Drug Development
| Use of Biomarker | Drug Development Goal |
|---|---|
| Target Verification | Used to establish that a probable drug target executes a pivotal function in the physiology of the disease. |
| Early Compound Selection | Determine the most favorable compounds in terms of safety and efficacy. |
| Pharmacodynamic Assays | Used to ascertain the drug's effect on the body to establish a dosing regimen. |
| Patient Selection for Clinical Trials | Aids in patient selection based on disease subtype or likelihood of positive response versus adverse reaction. |
| Surrogate Endpoint in Drug Approval | Used for a quick assessment of the safety and efficacy of the therapy by using a short-term outcome (biomarker) instead of a long-term primary endpoint. |
Types of cancer biomarkers
Cancer biomarkers are usually classified into three categories: prognostic, predictive, and pharmacodynamic. Prognostic biomarkers predict the natural course of the cancer and to distinguish the tumor's outcome. They also help determine whom to treat, how aggressively to treat, and which candidates will likely respond to a given drug and the most effective dose. Predictive biomarkers evaluate the probable benefit of a particular treatment. Pharmacodynamic biomarkers assess the imminent treatment effects of a drug on a tumor and can possibly determine the proper dosage in the early stages of clinical development of a new anticancer drug (12).
Instead of analyzing the tumor cells themselves, the molecular composition of a tumor can be indirectly characterized by analyzing blood samples and searching for variations in serum proteins, thereby improving the precision of screening and curtailing the need for invasive diagnostic procedures. Some difficulties were encountered initially in an attempt to reproduce these cancer-specific serum proteins. With advances in our ability to measure quantitatively, collect standardized samples, and resolve the problems of reduced sensitivity in detection, confidence in the results of this approach has risen (12). Measurements from biomarkers can be used to adjust empirical results of clinical trials by establishing a relationship between the effects of interventions on molecular/cellular pathways and clinical responses, thereby providing a way for scientists to comprehend mechanistically the differences in clinical response that may be affected by uncontrolled variables (5).
2. BIOLOGY AND STAGING OF ROSTATE CANCER
Biology of prostate cancer
Located under the bladder and in front of the rectum, the prostate is a small, soft gland with the urethra running directly through it (2). Androgens regulate the prostate gland as the major stimulus for cell division in prostatic epithelium (13). Although androgens are regarded as major contributors to prostatic carcinogenesis, there is little direct evidence to demonstrate that androgens cause prostate cancer. In part because of the lack of easily measurable hormonal events in men, there is insufficient evidence to establish an indirect role for androgens relative to the cause of the disease (14).
Prostate cancer occurs when the rate of cell division surpasses cell death, leading to uncontrolled tumor growth. Subsequent to the initial transformation event, further mutations of a multitude of genes, including the genes for p53 and retinoblastoma, can result in tumor progression and metastasis (15). More than 95% of prostate cancers are adenocarcinomas that arise from prostatic epithelial cells (16). Of these cases, 70% occur in the peripheral zone, 15-20% in the central zone, and 10-15% in the transitional zone. The majority of cancer cells are multifocal and influenced simultaneously by numerous regions of the prostate gland, indicating that prostate cancer is probably the result of clonal and nonclonal tumors (15). The cells from these tumors can metastasize through the lymphatic system and the bloodstream if untreated and allowed to grow. Arriving at their final destination, the tumor cells lodge and grow secondary tumors, resulting in a dramatic decline in the cure rates for the disease. The presence of these prostate cancer cells in another site, such as bone, does not change its classification to bone cancer-for instance. The new tumor is still considered to be prostate cancer (2).
There is an architectural and cytological similarity between prostate cancers identified clinically and those detected incidentally at autopsy, although differences do exist in numerous pathologic features. Compared to the clinically identified cancers, incidentally found cancers are usually small, sufficiently or moderately differentiated, and confined to the prostate (17-21). In addition, unsuspected prostate cancers found at the time of cystoprostatectomy for the treatment of bladder cancer are similar to autopsy cancers (20). Seventy-eight percent of unexpected prostate cancers found in cystoprostatectomy specimens are small, confined to the prostate, and moderately to well differentiated, compared with only 9% of the clinically detected cancers with such features (19). Twenty-nine percent of clinically found cancers are advanced as compared with none of the cystoprostatectomy cancers (22).
Prostate cancer staging systems
Stage and grade classification of a tumor is frequently complemented by the biomarker expression when biologically targeted therapeutics are discussed. The stage of the cancer indicates certain aspects of the cancer such as the tumor size, depth of its penetration, extent to which the cancer has spread, and to which organ(s) it has metastasized and invaded, as well as its effect on the organ(s) in relation to the stage (23). The stage at diagnosis of cancer is considered the most important indicator regarding survival of the patient. Stage is also very important because the required therapy is directly related to and frequently varied based on it (24).
Two main classification systems are used to stage tumors: the Jewett system (stages A through D) described in 1975 and since modified (25) and the TNM system adopted in 1997 by the American Joint Committee on Cancer (AJCC) and the International Union against Cancer. In 2002, the TNM classification system was further revised by the AJCC (26). These systems can reveal nonpalpable tumors by identifying an increase in serum prostate-specific antigen (PSA) level or an aberrant transrectal ultrasound image. These systems can also categorize patients based on tumor detection technique and distinguish nonpalpable prostate cancers (those detected during transurethral resection) from palpable ones (those detected by digital rectal examination) (27).
The TNM staging system is based on the extent of the tumor size and grade (T), detection in the lymph nodes (N), and any other possible metastasis (M) (28). It corresponds to one of five stages of the traditional staging system (a progression of the cancer from Stage 0 to Stage IV), but it has the advantage of revealing more detail by separating designations for the primary tumor, regional nodes, and distant metastases via more specific alphanumeric subcategories. An added number or letter is used to specify the size or extent of the tumor and the extent of spread (2, 27) (Table 3). The staging system is important and essential, however insufficient it is by itself. Other significant variables that may contribute to the evaluation include the grade; PSA level; DNA ploidy; nuclear morphometry; and a number of cellular, molecular, genetic, and environmental factors (29).
Knowledge of the stage of disease facilitates determining how aggressively to treat the disease and how likely the available treatment options will eliminate the disease (2). Although it can be difficult to accurately stage the cancer, incorrect staging can result in improper treatment and substantial decrease in the patient's chance of survival (23). The results from some common tests like digital rectal examination (DRE), serum PSA test, or transrectal ultrasound (TRUS) can reveal the probability of the incidence of prostate cancer. Any positive cases from these tests are usually followed by biopsy and histological examination for verification. Several other tests, including X-rays, MRIs, CT scans, and bone scans, can then be used to determine the stage of cancer and to detect any localized cancers outside the prostate (2). Despite the fact that the staging systems can reveal the extent of disease, the test results cannot be used independently to ascertain the stage of the disease, to select the best treatment options, or to envisage outcomes because they are not capable of detecting very small groups of cancer cells (30).
The TNM Staging System
| Primary Tumor (T) | Early Stage | Advanced Stage |
|---|---|---|
| TX: Primary tumor cannot be evaluated | ||
| T0: No evidence of primary tumor | ||
| T1: Although the tumor is present, it is clinically not palpable or visible by imaging. It may have been detected by needle biopsy, after finding a raised PSA level | ||
| T1a: Found incidental to other surgery; tumor was incidentally found in less than 5% of prostate tissue resected (for other reasons) | ||
| T1b: Found incidental to other surgery; present in 5% or more of tissue | ||
| T1c: Identified by needle biopsy performed a result of an elevated serum PSA | ||
| T2: Tumor confined within prostate, the tumor can be palpated on examination, but has not spread outside the prostate | ||
| T2a: the tumor is in half or less than half of one of the prostate gland's two lobes | ||
| T2b: the tumor is in more than half of one lobe, but not both | ||
| T2c: The tumor is in both lobes but is still inside the prostate gland | ||
| T3: Tumor extends through prostate capsule | ||
| T3a: the tumor has spread through the capsule on one or both sides | ||
| T3b: the tumor has invaded one or both seminal vesicles | ||
| T3c Extends into seminal vesicles | ||
| T4: The tumor has spread into other body organs nearby, such as the rectum or bladder | ||
| T4a: Invades bladder neck, external sphincter, or rectum | ||
| T4b: Invades muscles and/or pelvic wall | ||
| Regional Lymph Nodes (N) | ||
| NX: Regional lymph nodes cannot be evaluated | ||
| N0: No regional lymph node involvement; no cancer cells found in any lymph nodes | ||
| N1: One positive lymph node smaller than 2 cm across, there has been spread to the regional lymph nodes | ||
| N2: More than one positive lymph node Or one that is between 2 and 5cm across | ||
| N3: Any positive lymph node that is bigger than 5 cm across | ||
| Distant Metastasis (M) | ||
| MX: Distant metastasis cannot be evaluated | ||
| M0: No distant metastasis | ||
| M1: there is distant metastasis | ||
| M1a: the cancer has spread to lymph nodes beyond the regional ones | ||
| M1b: the cancer has spread to bone | ||
| M1c: the cancer has spread to other sites |
The TNM staging system based primarily on the anatomical extent of disease, which considers the tumor size or depth (T), lymph node spread (N), and presence or absence of metastases (M). The TNM system is used as a standard for staging and predicting survival, choice of early treatment, and stratification of patients in clinical trials.
3. SCREENING FOR PROSTATE CANCER
Prostate cancer generally does not present any symptoms until it becomes locally advanced or metastatic disease. Therefore, in the past, efforts at screening and early detection have used all available tools for diagnosis in asymptomatic patients before the presentation of symptoms (14). The detection and management of prostate cancer is controversial, especially regarding screening and therapy choice after diagnosis. For example, a patient can be diagnosed late in life with a low-grade prostate cancer that may not have any impact on the quality or length of his life, while a younger man with a high-grade lesion can have an advanced disease and die within 5 years because of the disease's aggressive progression. This intriguing observation demonstrates the unusual biological heterogeneity of prostate cancer and demands distinctive classification (14).
Nonetheless, a greater number of patients are now diagnosed at an earlier stage thanks to the advanced tools for prostate cancer diagnosis that has improved considerably in recent years. Just as the screening and early diagnosis techniques for cervical and breast cancer have been shown to successfully reduce the death rates, respectively, from these cancers, screening for prostate cancer has successfully accomplished the same goal (31). Some of the major techniques used in assessing prostate cancer in its early stage are the DRE, PSA blood test, and TRUS (31, 32).
DRE is regarded as a basic tool for screening and early detection of prostate cancer and is estimated to have about a 59% overall accuracy (33). Despite its seemingly poor sensitivity, DRE is a routine method for prostate cancer screening because it often detects cancers missed by other tests (34). Its main advantage is that it may detect cancer in some men with normal PSA levels and whose tumors are small and well differentiated in most cases (33). An additional advantage of DRE is that it is a relatively inexpensive procedure that is normally well tolerated, and it can be used to investigate other abnormal conditions of the prostate, such as benign prostatic hyperplasia (BPH) (35). The main limitation of DRE is that most palpable cancers are not early cancers, and many clinically important cancers are located in regions of the gland that are distant and thus evasive to digital palpation (34). A population-based case-control study on men who died as a result of prostate cancer reported that DRE screening might have prevented as many as 50 to 70% of the deaths from the disease (36). However, controversial results from two other studies revealed that there is no evidence that men who died as a result of prostate cancer were less likely to have received the screening compared to those who survived (37, 38). Based on studies investigating the sensitivity and specificity of DRE and its role in the early detection of prostate cancer, the majority of experts agree that detection is less likely when using DRE independently as opposed to DRE in combination with other predictors (39).
Serum prostate-specific antigenPSA, discovered in 1971, is considered the most important biomarker for detecting, staging, and monitoring cancer of the prostate in its early stage (40-47). PSA is a member of the family of human kallikrein proteases with a molecular mass of approximately 30 kDa and chymotryptic-like activity. In serum, PSA is bound primarily by α1-antichymotrypsin (ACT), an endogenous protease inhibitor, and also by another similar inhibitor, α2-microglobulin (A2M). PSA was initially thought to be solely synthesized by epithelial cells of the prostate and thus was used as a biomarker for diagnosing and managing prostate cancer (48). However, PSA has also been found in a variety of human normal and tumor cell lines and in biological fluids synthesized by numerous cells, although mainly by prostatic epithelial cells (46, 49).
PSA testing was initially used for monitoring prostate cancer patients. After it was commercially introduced, it became extensively used for screening and diagnosing the disease. The noticeable increase in prostate cancer incidence rates in the United States, which started in the late 1980s and peaked in 1992, is believed to be in accordance with the time period when PSA testing was introduced (22). Like DRE, PSA testing is a relatively inexpensive procedure and has high patient acceptance. The main advantage of PSA testing is its superior sensitivity. The main disadvantage of the test is that it is not very specific because common pathological conditions such as BPH and prostatitis can also cause moderately to conspicuously abnormal test results. These false-positive results may lead to further diagnostic evaluation, increasing costs and use of more invasive procedures. Conversely, efforts to prevent such overdiagnosis that may result from the high number of false-positives may lead to delayed treatment for the aggressive, potentially life-threatening cancers (22).
In an effort to find ways of improving specificity, several variations on the basic PSA test have been proposed. For example, the free PSA ratio, which may be a more specific test, compares the amount of free PSA circulating in the blood (unbound) to the amount attached to other blood proteins (50). Furthermore, PSA levels are normally elevated in older men relative to younger men regardless of the absence or presence of cancer. Therefore, a continuous rise in PSA level over time from a relatively low level may be more indicative of cancer than a moderately increased PSA that is stagnant (51). Higher PSA values have also been observed in African American men with newly diagnosed prostate cancer when compared with newly diagnosed Caucasian men (52). Studies have shown that African American men have notably larger cancer volumes even within clinical stage category at diagnosis. Thus, special efforts at screening are necessary to minimize the discrepancy. It may be practical, therefore, to start testing at younger ages in African American men in an effort to detect tumors earlier when they are still confined. Some researchers recommend age- and race-adjusted PSA values for detecting cancer, with lower PSA limits for African American men (53-54).
Serum PSA value can independently predict a pathological stage. However, the serum PSA level alone may not adequately predict pathological stage because the relationship between pathological stage and serum PSA varies by tumor grade, volume, and site of origin (55-57). Nevertheless, comparative studies have demonstrated that PSA and its related testing can increase the detection rate of prostate cancer in men with no symptoms (31, 58). It has also been shown that the stage distribution of cancers detected through PSA screening was much more favorable than that which occurred in the population without PSA screening.
Serum PSA testing is vital not only in screening and early detection, but it has also been found to be essential in diagnosing localized prostate cancer. PSA testing is now a standard application clinically for staging and monitoring prostate cancer (46). The prevalent acceptance of PSA screening has increased the diagnoses of prostate cancer at an earlier stage and age (59) and has reduced the likelihood by half that a new case will be localized and by 33% that a new case will be metastatic. Furthermore, because PSA screening has become routine, the occurrence of prostate cancer in men over 70 years has declined (60-62). However, despite a decrease in the incidence of prostate cancer since 1992, an apparent increase in the prevalence has continued (63). The possible explanations for this may be the combined effects of the continuous rise in the life expectancy of the US population (64) and the inclination toward early detection. Consequently, the prolonged lifespan and increased number of survivors living with the diagnosis of prostate cancer is leading to an increase in the cost of treating the disease (65).
The most important adverse effect of prostate cancer screening is overdiagnosis and overtreatment (66). Overdiagnosis refers to detecting prostate cancer through PSA testing that would otherwise not have been diagnosed in the person's lifetime. A randomized study of screening for prostate cancer was initiated in the early 1990s to assess the outcome of screening for PSA on death rates from prostate cancer. The researchers concluded that PSA-based screening reduced the death rate from prostate cancer by 20%, but the screening was associated with an increased risk of overdiagnosis (67). Previous studies had already demonstrated that risks incurred by either screening/diagnosis (68, 69) or resulting treatment (70-76) of prostate cancer were both substantial. Further study revealed that, for every patient who benefits from PSA diagnosis-initiated treatment, 47 patients undergo unnecessary biopsy and other treatments because of false-positive PSA test results (67). After 7 to 10 years of follow-up, the rate of death from prostate cancer was very low and did not differ significantly between the two groups used in the study. Also, prostate cancer screening offered no reduction in death rate after 7 years and no apparent indication of benefit among 67% of the subjects who completed 10 years of follow-up (67). Although PSA testing as a reliable biomarker for prostate cancer diagnosis remains controversial today, researchers have reported that overdiagnosis rates are about 29% for Whites and 44% for Blacks, suggesting that most of the cancers detected through PSA testing would have been diagnosed in the lifetime of the patient and that PSA screening detects mainly cancers with a high clinical consequence to patients (22).
The use of PSA for prostate cancer screening has led to a great increase in the number of men undergoing TRUS (77), a procedure in which a probe that sends out high-energy sound waves is inserted into the rectum against the prostate gland to image the entire gland (2). Areas of the gland with varying morphology frequently produce different images. The advantage of TRUS is its high sensitivity, and, thus, it plays a very important role in early detection. Besides being a screening test, TRUS can also be used to guide needle biopsies of the prostate gland for diagnostic purposes (22). However, it has poor specificity when used as the sole screening for modality.
Research conducted in the early 1990s revealed that PSA combined with DRE is the most effective screening and early detection modality for prostate cancer (32, 78-81). As screenings became more prevalent, a study in the late 1980s concluded that the occurrence of prostate cancer significantly increased and that PSA testing was associated with the acceleration of the overall occurrence of prostate cancer (81-84). Research revealed that conducting extensive PSA testing led to a decline in the mortality rate while the rate of incident cases continued to rise (85-87). Some studies have cast doubts over the relationship between the decline in prostate cancer mortality and the increase of PSA screening (88-90). This controversy may be partly result from the fact that PSA is more prostate specific than cancer specific, which leads to a consequent increase in the rate of false-positive results (91).
Several researchers have proposed that, before PSA testing was available, the decrease in death due to prostate cancer could have resulted from the increase in early detection with DRE, as shown by earlier stage at diagnosis and by increasing rates of surgery for localized prostate cancer in the decade prior to the start of widespread PSA testing. Another possible explanation for the decrease in prostate cancer mortality is that the tendency to classify prostate cancer as the underlying cause of death has shifted (92). Some researchers have argued in the alternative that the survival rates that are specific to the stage of the cancer imply that the swift reduction in mortality followed by a large increase in incidence may be a result of the large amount of high-grade prostate cancers detected before metastasis. Consequently, it is reasonable to consider the decrease in the incidence rate of advanced disease as predictive of a subsequent decrease in prostate cancer mortality rates rather than considering the trend in the context of the estimated average lead time gained (85).
The American Cancer Society National Prostate Cancer Detection Project reported that 5 years following an annual testing by PSA, DRE, and TRUS, 92% of cancers detected were localized to the prostate, compared to 66% in a contemporary national database covering men the same age (93). Prostate cancer screening appears effective when one considers the population mortality trends related to this disease; prostate cancer mortality began to decline in the United States after several years of steadily growing death rates (94). Several reports have argued that PSA testing may not to be responsible for reduced mortality observed within the first 10 years following the onset of widespread the testing (95). Some studies compared the efficiency of PSA testing alone or in combination with DRE and TRUS. As discussed earlier, despite any conclusive evidence showing that screening asymptomatic men and treating those with early stage disease improved survival, serum PSA screening has been generally accepted and still remains as a commonly used diagnostic biomarker for prostate cancer (96). Furthermore, although overdiagnosis may result from prostate cancer screening, most of the risk occurs in men in whom the cancer would not have been detected if they had not been screened (67).
4. CRITERIA FOR SELECTING/ IDENTIFYING PROSTATE CANCER BIOMARKERS
Criteria as a biomarker
A biomarker must be shown to correlate with an interested outcome, such as disease progression, recurrence, or survival, if it is to be seen as useful for diagnosing and monitoring a disease. Several analyses with different variables should demonstrate that the biomarker could predict the relevant stage or grade irrespective of the characteristics frequently accessible. For statistical implication to be assessed, these tests ought to be performed on a set of cases with adequate ending data and at a sufficient number of incidents. With the help of tissue microarrays, this process is becoming significantly more efficient (97).
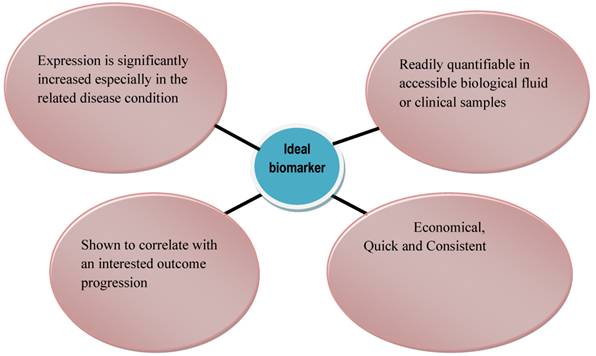
The selection of a biomarker should have a biological or therapeutic basis or, at minimum, the biomarker should indicate a reliable correlation with the presence, characteristics, or aggressiveness of the cancer. Also, there should be an evaluation of the strength of the marker in relation to the outcome of the disease, which, together with other factors, should be carried out as an independent predictor in a multivariable assay (98). An ideal biomarker should be quick, consistent, economical, and quantifiable in an accessible biological fluid or clinical sample (e.g., plasma, urine, or prostatic fluid) that is readily interpretable by a clinician (99, 100) (Fig. 1). Its expression should be significantly increased (or decreased) in the related disease condition, and no overlap should exist in the levels of biomarker between healthy control subjects and untreated patients.
In the general population, the levels of biomarker should not vary widely so that the severity and prognosis of the disease can be predicted based on large deviations of the biomarker from the reference values in the control population (98, 101). Furthermore, within the general population, the abundance or activity of an ideal biomarker should be similar in subjects (99). In this way, it provides a great advantage for clinical diagnosis and monitoring of disease activity. It would also provide a correlation between subjects with a disease and those with other deadly conditions occurring in the context of a particular disease for which the biomarker will be examined (98, 99). One critical factor that determines the selection of a candidate biomarker is the caliber of scientific and clinical results such as (i) linking the gene or protein function to the biology of the disease, (ii) relating the candidate biomarker to the presence of the disease, (iii) variations in stage, (iv) reaction to therapy, and (v) overall survival; all of which will back the possible efficacy of the candidate biomarker. For prostate cancer biomarkers used in early detection or monitoring of disease, the model candidate should be prostate specific and able to differentiate between normal BPH, prostatic intraepithelial neoplasia, and cancerous prostate tissues (100).
Validation of biomarker
Biomarkers are essential factors in clinical and biological research. Identifying a new candidate biomarker is followed by a thorough operational evaluation to validate its application in the clinical setting (99). Biomarkers that have been scientifically scrutinized must pass several proposed practical tests prior to being accepted for clinical practice. Five conceptual phases of biomarker development have been suggested (102-103): (i) preclinical exploratory, (ii) clinical assay and validation, (iii) retrospective longitudinal, (iv) prospective screening, and (v) cancer control.
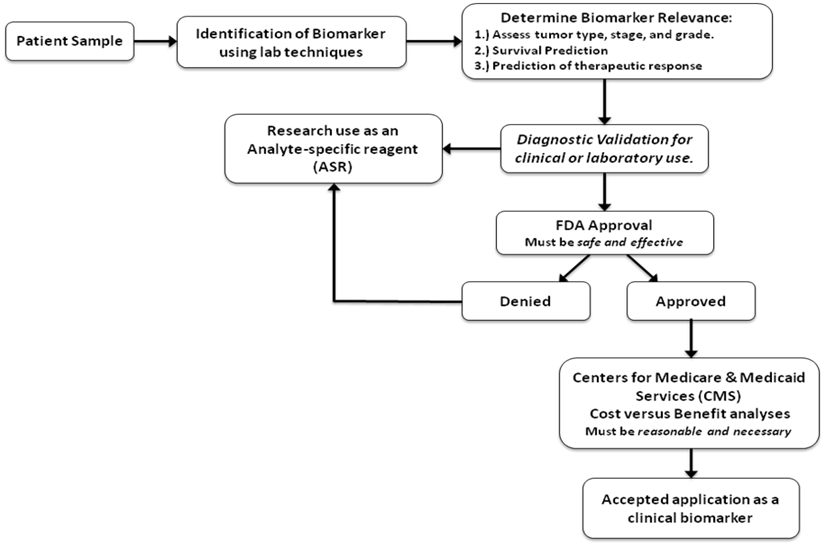
The first step involves identifying the biomarker and evaluating it for a specific clinical indication. Prior to submitting it for US Food and Drug Administration (FDA) approval, analytical and clinical confirmation must be carried out (Fig. 2). Alternatively, if the marker is intended solely for research, it may not require FDA approval. After approval by the FDA, the Center for Medicaid and Medicare Services (CMS) might conclude that the biomarker is essential for improving patient care (104). However, a number of challenges are involved in the process, and the majority of candidate markers are still in the premature phases of development. Thus far, clinical studies are usually reflective, and the few promising studies that have been conducted have frequently produced conflicting results (104).
Characteristics of an Ideal Biomarker
Steps involved in the validation of a biomarker. The initial step involves identifying the biomarker, followed by assessing its relevance to the particular information sought. A diagnostic validation for its clinical use is done, and, if the results are positive, it is submitted to the FDA for approval. If approval is denied, it may go back to the lab to be used in research as an analyte-specific reagent. An approval, on the other hand, paves the way for it to go to the Center for Medicaid and Medicare Services (CMS). It may go directly to the CMS and boycott the FDA if it is for research purposes only [104].
5. BIOMARKERS FOR THE DIAGNOSIS AND PROGNOSIS OF PROSTATE CANCER
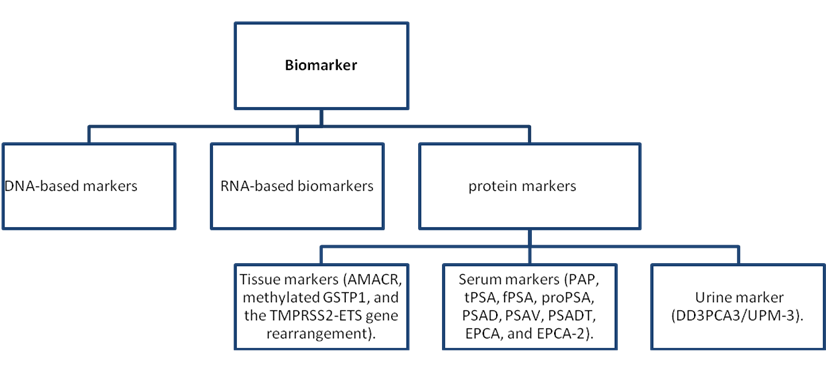
As described earlier, a biomarker is in general an analyte that signifies the presence and/or degree of a biological process, which in itself is frequently directly linked to the clinical expressions and result of a particular disease (99). Biomarkers have been shown to possess many important applications including use as a diagnostic tool to identify patients with a disease or abnormal condition, a tool for staging disease or classifying the extent of disease, an indicator of disease prognosis, and for predicting and monitoring clinical response to an intervention (105). Cancer early detection markers indicate the presence of an early cancer or that cancer will occur with nearly 100% certainty within a very short time interval. This is in contrast to screening markers (22). Biomarkers for the diagnosis and prognosis of prostate cancer include DNA-based markers, RNA-based biomarkers, and protein markers (see Fig. 3 for examples).
A lot of the biomarkers used currently were discovered unexpectedly while others were detected through the use of basic reasoning with understanding of the fundamental biochemical defect. Biomarkers are used to observe the natural course of the condition and to assess the effectiveness of treatments. Also, they may help in the development of set therapeutic objectives, thereby presenting standards for judging success in the management of chronic diseases (99). For some time, biomarkers have served as an indicator of the presence of a particular disease and reflected the activity of a given condition either during its natural course or in response to a given therapeutic intervention. In addition, they may be useful for prognostic purposes in the outcome of diseases, with particular attention on the quantitative biomarkers that demonstrate a relationship with the clinical manifestation of the disease and that have an effect on quality of life, risk of complications, or survival. Surrogate biomarkers have a significant function in disease monitoring after accepted treatments are introduced. Surrogates are particularly important for those treatments that are uncommon, such as cases in which the direct study has proved to be very difficult because of the limited number of patients and varying expression of their primary illness or in which the efficiency of the treatment must justify the high cost (99).
A general classification of biomarkers based on their description [104].
The management of prostate cancer has undergone several dramatic changes as a result of the evolution of biomarkers used in screening, detecting, and predicting the disease (106, 107). Human prostatic acid phosphatase (PAP) (or serum acid phosphatase (AP) was reportedly the first serum biomarker for prostate cancer. Gutman and his colleagues observed in the 1930s that patients with prostate cancer metastasized to bone had elevated levels of PAP activity at the site of metastasis and high serum levels of the protein (108-110). This finding effectively established the value of serum acid phosphatase activity as an aid in diagnosing metastatic carcinoma of the prostate and consequently as a biomarker for prostate cancer progression and reaction to androgen deprivation therapy of prostate cancer that had metastasized (97, 110). The total serum AP comprises a mixture of phosphatases from most tissues of the body (111). Men with high preoperative PAP had a greater chance of developing lymph-node-positive disease and metastases than did their counterparts with normal PAP (112). Posttreatment PAP, as determined by other researchers, could predict outcome when combined with other clinical factors (113).
Although AP, with an elevated level in more than 70% of patients (114), was linked early with prostate cancer that had spread, both AP and prostate-specific AP (PAP, its subtype) are not sensitive enough for screening. Patients with localized cancer frequently display normal levels, and neither PAP nor AP show sufficient sensitivity to be used as a reliable biomarker for recurrence or response to systemic therapy. Sudden variations in PAP and AP have been observed, which has led to questioning whether this enzyme is a legitimate biomarker in cancer diagnosis. Furthermore, the use of ACP has been reduced because of the development of PSA screening, which is a more sensitive and specific tumor marker (22).
PSA was later discovered as a biomarker for prostate cancer following the discovery of serum PAP. The PSA test was first used in the field of forensic science as a marker for human semen (41), and it was first purified in the late 1970s from prostate extracts (43). Later studies established that PSA could be quantified in serum, and its serum levels were high in men with prostate cancer (44). The prostate gland produces PSA, and the test measures PSA levels in the blood (serum). Because PSA is from the body and can be used in disease detection, it is often referred to as a biological marker or a tumor marker. Both prostate cancer and benign prostatic conditions (e.g., BPH) can increase PSA levels from a normally low level to an elevated state in the blood (2). PSA can be present in a free form or complexed with α1-antichymotrypsin or α2-macroglobulin in circulation (115). Patients with cancer can be distinguished from those with BPH based on the percentage of free to total PSA in the serum. For those with an elevated level of PSA, particularly with a PSA range between 4 and 10 ng/ml, patients are more likely to have prostate cancer when the free PSA is less than 20-25% of the total serum PSA level. ProPSA, the precursor form of PSA, may serve as an additional indicator in differentiating cancers from benign processes (22). Moreover, in a group of patients examined with PSA levels between 2.5 and 4 ng/ml, the ratio of proPSA to free PSA showed more specificity in detecting cancers when compared to the detection rate by free PSA alone (116).
Serum PSA was initially used for screening men with an existing diagnosis of prostate cancer (117) and was regarded as an ideal marker for identifying recurring disease subsequent to treatment. The fact that levels of serum PSA remained undetectable demonstrated the absence of recurrent disease among men who underwent radical prostatectomy (118). PSA gradually replaced serum PAP, which was considered inferior to PSA, for prostate cancer screening, staging, and prognostication (119). However, PAP is once again attracting some attention because of the fact that several studies have shown that it is a good prognostic marker for patients with aggressive disease who went through local therapy and are at high risk for distant relapse (120). However, PAP has no role as a diagnostic screening tool (22).
In 1994, PSA was officially approved for prostate cancer screening by the FDA, and 4.0 ng/ml was set as the upper limit of normal range (see time line in Fig. 4). Following its prevalent use for identifying the incidence of prostate cancer, PSA became, and is still, the most frequent method of detecting prostate cancer and has resulted in a considerable stage migration. The observed decline in mortality rate both in the United States and around the world has been partially attributed to the ongoing screening based on PSA levels (121). However, there are still some significant controversies over PSA screening because no study has successfully shown any significant correlation between such screening and a decline in mortality rate. Further research has facilitated significant modification in using and understanding serum PSA, despite the lack of direct evidence. PSA is useful not only for detecting prostate cancer but also for drawing a parallel between its levels at diagnosis and more advanced stages (122).
Timeline for Early Prostate Cancer Biomarkers for Diagnosis
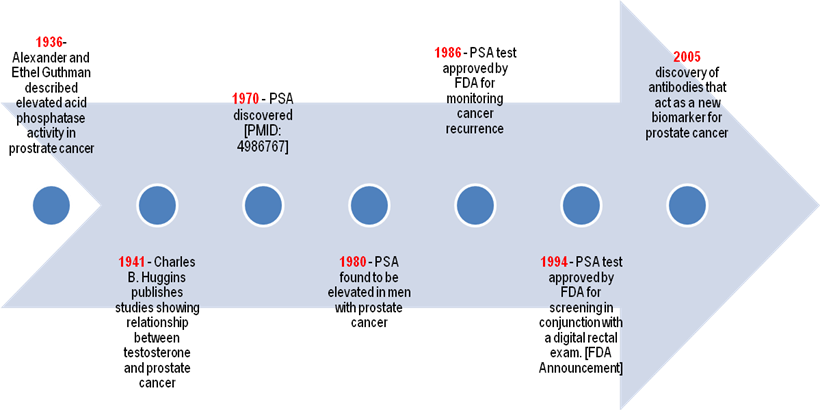
Several clinicians currently use a threshold range of 2.5-3.0 ng/ml for proposing prostate biopsy, because the prognosis of prostate cancer is better when the disease is identified at lower PSA levels (123). However, limitations in the use of PSA for screening prostate cancer are becoming noticeable. Many men, as they grow older, suffer from various nonmalignant processes, including BPH and prostatitis that often lead to serum PSA increase that results in a limited specificity of PSA testing for cancer detection (105). On the contrary, the high levels of PSA can be muddled up by certain alterations in the prostate and by clinical procedures. Overall, an increase in the levels of PSA is not as specific as it is sensitive for prostate cancer diagnosis. According to Thompson et al., prostate cancer has been detected in about 15% of men with normal or very low levels of total PSA, thereby making it difficult to reliably rule out the possibility of cancer at any PSA level (124). This has resulted in a call for reevaluating the approach to diagnose prostate cancer and a search for new prostate cancer biomarkers (46, 124-127).
6. CANDIDATE MARKERS FOR PROSTATE CANCER
One of the established objectives of cancer research is to identify the molecular mechanisms that dictate the initiation and progression of the disease and then to ascertain those molecular markers associated with the cancer to target cancer cells with specifically designed drugs. Like all respective biomarkers correlated to their respective cancers, prostate biomarkers exhibit some or all of these abilities: detect the presence of prostate cancer, monitor and relay its progression, prognosticate the possibility of a recurrence, predict the response to therapy, and foresee whether the patient will be disease-free and survive. Clinicians rely on biomarkers to detect cancer at premature stages or prior to tumor metastasis, when the efficacy of therapeutic drugs is greater.
The need for effective prostate cancer biomarkers is therefore urgent and great, and the search for them has been a priority of researchers for years. In the last decade, PSA has been widely used as a useful tool for screening prostate cancer. However, PSA and other established biomarkers are still not ideal, as they lack diagnostic specificity and prognostic value and lead to a high rate of false-positives. Consequently, the lack of specific and sensitive biomarkers for early detection of prostate cancer calls for investigating novel and existing biomarkers and developing new approaches to identify and validate more accurate diagnostic and prognostic biomarkers for prostate cancer.
Current advancements in proteomics, tissue microarray, DNA microarray, immunohistochemical staining, and other biotechnologies have paved the way and have significantly increased the pace at which novel biomarkers are being discovered and developed. Using these methodologies, researchers have reported several biomarkers with great potential, and they are currently undergoing further investigation for validation. Although the existing method of discovery usually identifies several candidate markers in each investigation, only a few of them ever make it through clinical validation.
We will discuss and list (Table 4) some of the biomarkers that have a substantial amount of supportive data that are biologically and clinically persuasive for them to be further developed and other potential candidates still being investigated. A few of the recent candidates that have generated some excitement for their potential as biomarkers for prostate cancer are also discussed. Needless to say, this list is by no means exhaustive, and it will keep growing with the help of the advance of newer/better technologies in molecular analysis.
Description of the Biological Function of Selected Serum Markers
| Serum Marker | Description/Type | Biological Function | Purpose |
|---|---|---|---|
| Chromogranin-A | Pro-hormone peptide released by neuroendocrine cells | Uncertain definite function. Possesses calcium-binding abilities and may act through paracrine and autocrine manners. | Prognosis |
| Neuron-specific enolase | Isomer of the glycolytic enzyme 2-phospho-D-glycerate hydrolase released by neuroendocrine cells | Uncertain definite function. Possibly serves as paracrine and autocrine factor. | Prognosis |
| Human kallikrein 2 | Serine protease with trypsin-like substrate specificity | Splits pro-PSA to create PSA | Diagnosis |
| Urokinase-type plasminogen activator system | Serine protease and transmembrane receptors | Converts plasminogen to plasmin | Diagnosis (fragments) and prognosis |
| Interleukin-6 | Cytokine | Implicated in hematopoiesis and the immune response through mediation of B-cell differentiation and the acute-phase inflammatory response | Prognosis |
| Transforming growth factor-β | Cytokine | Involved in cellular proliferation, cellular chemotaxis, cellular differentiation, angiogenesis, humoral immunity, cell-mediated immunity, and wound healing | Prognosis |
| Prostate membrane-specific antigen | Type II integral membrane glycoprotein with cell surface carboxypeptidase function | Possesses folate hydrolase function. Also is involved in the cell stress reaction, signal transduction, cell migration, and nutrient uptake. May possess questionable receptor function. | Diagnosis |
| Prostate-specific cell antigen | Glycosyl phosphatidylinositol-anchored cell surface glycoprotein | Known cell surface marker. Perhaps involved in several stem cell activities involving proliferation or signal transduction. | Prognosis |
| α-Methylacyl-CoA racemase (autoantibodies) | Peroxisomal and mitochondrial racemase | Engaged in bile acid synthesis, stereoisomerization, and β-oxidation of branched-chain fatty acids | Diagnosis |
| Early prostate cell antigen-1, -2 | Nuclear matrix protein | May be involved in early prostate carcinogenesis; however, has uncertain contribution to nuclear morphology | Diagnosis |
| GSTP1 hypermethylation | CpG island hypermethylation of DNA encoding the protein, glutathione S-transferase π | Hypermethylation of GSTP1 inhibits transcription. GSTP1 usually acts by conjugation of oxidant and electrophilic carcinogens to glutathione to inactivate them | Diagnosis |
| Testosterone | Steroid hormone | Acts in the natural growth and support of the prostate gland and seminal vesicles. Many actions on sexual development and anabolism. Also involved in endocrine signal transduction. | Prognosis |
| Estrogen | Steroid hormone | Many effects on female sexual development. Also acts in the control of sperm development and in endocrine signal transduction. | Prognosis |
| Sex hormone-binding globulin | Serum glycoprotein-binding protein | Adheres to and carries testosterone and estradiol. Also involved in endocrine signal transduction. | Prognosis |
| Caveolin-1 | Integral membrane protein | Works to regulate cholesterol metabolism and cellular transformation and is engaged in transducing cell-to-cell signals | Prognosis |
| E-cadherin | Calcium-dependent cell adhesion protein | Plays major role as a cellular adhesion molecule in cell-to-cell adhesion of secretory tissues | Prognosis |
| β-Catenin | Adhesion protein (80-kDa fragment isolated in prostate cancer) | Aggregates with cadherin to regulate the formation of adherent junctions between cells | Prognosis |
| MMP-9 | Zinc-dependent endogenous protease | Acts in cell migration through and degradation of the ECM and in cell-cell adhesion. | Prognosis |
| Tissue inhibitor of MMPs (TIMP 1, 2) | Protease inhibitor | Prevents synthesis of ECM | Prognosis |
| Hepatocyte growth factor | Polypeptide growth factor (secretory protein of fibroblasts) | A cellular growth, motility, and morphogenic factor. Also, involved in cell scattering and angiogenesis. | Diagnosis/ prognosis |
| MIC-1 | Cytokine (TGF-β superfamily) | Uncertain role, but may induce apoptosis | Diagnosis/ prognosis |
| Cytokine macrophage MIF | Cytokine (secreted by lymphocytes) | Modulates inflammation and the immune response. Activates cellular proliferation and angiogenesis, while inhibiting some tumor-suppressor genes. | Diagnosis |
| hK11 | Serine protease (human kallikrein superfamily) | Has an uncertain function. Acts like trypsin but, depending on the tissue or body compartment in which it is present, may possibly have many different functions. | Diagnosis |
| Progastrin-releasing peptide (ProGRP 31-98) | Neuropeptide | Split to form GRP. GRP acts in the regulation of metabolism, behavior, smooth muscle activity, some exocrine and endocrine operations, and cellular chemotaxis. | Prognosis |
| Apolipoprotein A-II (8.9 kDa isoform) | Lipoprotein (abundant in HDL) | Effects plasma free fatty acid levels via operating in lipid metabolism and transport | Diagnosis |
| 50.8-kDa protein | Unknown, identified by mass spectrometry | Uncertain function but possibly is parallel to the action of vitamin D-binding protein | Diagnosis |
| ILGF-1, -2 | Growth hormone-dependent polypeptides | In the prostate gland, both modulate cellular proliferation, differentiation, and apoptosis. Also, acts in endocrine signal transduction. | Diagnosis |
| Leptin | Adipocyte-derived peptide | In metabolism, modulates hunger, energy use, and fat metabolism and is also known to induce angiogenesis | Diagnosis |
| Endoglin (CD105) | Homodimeric transmembrane glycoprotein | Controls TGF-β superfamily signaling pathway and therefore subsequently affects angiogenesis, cellular propagation, apoptosis, cell adhesion, and cell movement | Prognosis |
| EGFR family (c-erbB-1 (EGFR), c-erbB-2 (HER2/neu), c-erbB-3 (HER3) and c-erbB-4 (HER4)) | Transmembrane glycoprotein receptors | Transduce signals for multiple growth factors | Diagnosis and prognosis |
| TSP-1 | Homotrimeric extracellular matrix glycoprotein | Inhibits angiogenesis by inhibiting cell development, movement, and propagation and is also an effector molecule for the tumor suppressor gene p53 | Diagnosis |
| VEGF | Dimeric, heparin-binding protein | An important endothelial cell growth factor that controls angiogenesis and augments vascular permeability | Prognosis |
| Huntingtin-interacting protein 1 (autoantibodies) | Cytoplasmic clathrin-binding protein | Acts in growth factor receptor transport. Also, transforms fibroblasts by lengthening the half-life of growth factor receptors. | Diagnosis |
| Prostasome (autoantibodies) | Prostatic secretory granules and vesicles composed of a lipid bilayer membrane and composite protein content | Consist of proteins that act in numerous enzymatic reactions, transport, structure, GTP activity, molecular chaperoning, and signal transduction | Diagnosis |
| ZAG | Glycoprotein | Induces lipid decline in adipocytes and therefore is implicated as possibly acting in cachexia | Diagnosis |
| CGRP | Neuropeptide | Vasodilatation and possibly regulation of protease secretion | Prognosis |
| PSP94 | Nonglycosylated secretory peptide | In all probability acts as a growth and calcium regulator, apoptosis inducer, and an inhibitor of FSH. | Diagnosis |
| Other methylated genes including RASSF1α, APC, RARB2 and CDH1 | Hypermethylated DNA encoding for various peptides | Hypermethylation predictably inactivates gene transcription | Diagnosis |
Adapted from reference (159).
a-Methylacyl Coenzyme A Racemase (AMACR)
AMACR is an enzyme localized to the peroxisome and involved in fat metabolism and has been identified to function as a growth promoter, independent of androgens, in prostate cancer (128, 129). By using various experimental methods and different prostate cancer specimens, the AMACR gene has been shown to be overexpressed in prostate cancer tissue at the mRNA and protein levels and making it a highly specific tissue biomarker currently used to aid in the pathological diagnosis (130-132).
AMACR has been reported to be involved in the crucial role in peroxisomal b-oxidation of branched chain fatty acid molecules (133). When prostate cancer tissues were compared with normal controls, a 9-fold increase in mRNA levels of AMACR was discovered in 88% of the sample prostate cancer tissues (134). This finding has prompted other researchers to propose the possibility of analyzing the levels of AMACR from urine to detect prostate cancer (134). Another potential use of AMACR includes analyzing and interpreting specimens of prostate needle biopsy (and results) that are usually diagnostically challenging (131). Immunodetectable serum autoantibodies generated in response to the AMACR tumor-associated antigen may also be useful in preliminary diagnosis, especially if combined with PSA screening. A considerably more enhanced sensitivity and specificity in prostate cancer patients with mid-range PSA levels have been observed with AMACR antibodies than that with PSA. This demonstrates that AMACR can be useful in discriminating control subjects from those with prostate cancer (135).
Some of the limitations of AMACR as a biomarker include the possibility of humoral response and production of endogenous AMACR antibody as a result of certain cancers other than prostate in patients suffering from autoimmune diseases (97). In addition, AMACR levels have also been observed to be commonly increased in patients with other urological disorders like BPH. However, the diagnostic capability for characterizing organ-confined and metastatic prostate cancer was increased by adding the AMACR test to serum PSA testing (136). Therefore, a new promising and noninvasive screening test for prostate cancer is to use quantitative reverse transcriptase polymerase chain reaction (RT-PCR) to identify the ratio of AMACR-to-PSA transcript (129). Nevertheless, further testing is under way to assess and possibly validate the prospective use of this serum biomarker.
Glutathione S-transferase P1 (GSTP1)
Glutathione S-transferase π is an example of a biomarker that has been extensively studied in prostate cancer, primarily as a tissue marker. GSTs are a ubiquitous family of multifunctional enzymes that conjugate reactive substrates with reduced glutathione (GSH) and are involved in detoxification (137, 138). Their role in protecting the cells from oxidative attack (137), and thereby being upregulated in the presence of free radicals, makes them a prime candidate for consideration as a cancer biomarker. The GSTP1 gene has been observed to be unmethylated in all normal human tissues and BPH, but hypermethylated in specimens of prostate cancer tissues (138, 139). Hypermethylation of the GSTP1 promoter is a common change that occurs during carcinogenesis and is regarded to be a main characteristic of prostate carcinogenesis (140).
GSTP1 has been shown to be acutely sensitive in detecting the presence of prostatic intraepithelial neoplasia and prostrate cancer, thereby distinguishing patients with these diseases from patients with BPH (138, 141-145). Moreover, the increased hypermethylation of the GSTP1 gene in neoplastic events can consistently distinguish between prostate cancer and BPH (146). With the help of PCR, the methylated GSTP1 gene has also been detected in the urine of men who have undergone prostate biopsy. This implies the possible additional use of this biomarker in risk-stratification of men undergoing prostate biopsy (147).
GSTPI displays several good characteristics that make it a viable biomarker. For instance, it is highly prevalent in the disease condition, and clinicians are able to measure quantitatively the methylation status of the gene in biopsy/prostatectomy tissues and in cells isolated from serum, urine, and seminal plasma (148). If it is successfully validated, GSTP1 methylation testing of cells derived from samples of serum and urine may possess a significant clinical potential for early detection of prostate cancer and posttreatment monitoring of the disease.
Chromogranin A (CGA, GRN-A)
Chromogranin A (CGA or GRN-A), part of the granin family of proteins, is an acidic protein that has been identified in all neuroendocrine cell types studied and is produced in larger amounts than other secreted proteins by those cells. Also known as secretory protein I, it is encoded by the CHGA gene in humans (149-151). The growth of prostate cells has been found to be regulated by peptides derived from GRN-A (138). Because it is produced and secreted by prostate cells, GRN-A has been examined for its diagnostic and prognostic values as a biomarker for prostate cancer (152). However, limited evidence to this point supports its usefulness beyond traditional methods of screening (152, 153). Based on past studies, GRN-A can be used to monitor the success of cancer treatment. It can also be used to predict the outcome of the disease and prognoses that are androgen independent. These predictions would be prior to any indication of PSA progression and would show increased levels of GRN-A correlated with undesirable results and diminished overall survival (151, 154-157). Therefore, GRN-A may be very useful as a prognostic factor in patients with advanced prostate cancer (153).
Interestingly, some variations in measurements of GRN-A have been reported between two assays commercially available for discerning BPH from prostate cancer (158). Some discrepancies observed in the measurements result from the fact that the characterization of most prostate cancer by neuroendocrine cells is a temporary and reversible process; the neuroendocrine markers may frequently be undetectable, and, therefore, a subset of the neuroendocrine cells may not possess any differentiation (159).
Although an accurate distinction cannot be made between prostate cancer and BPH based on the levels of serum GRN-A, these levels do reflect the tumor stage and grade and may efficiently indicate a poor prognosis following endocrine therapy when combined with free or total PSA ratio (153, 155, 160-162).
One noted weakness of using GRN-A as a biomarker is the fact that neuroendocrine cells do not reside in all prostate tumors. Another weakness is its inability to detect the disease at a very early stage, as reported by one group (153). On the other hand, based on the expression of GRN-A in prostate cancer analyzed by serum immunoassay and tissue immunohistology procedures, it was concluded that GRN-A has clinical potential as a biomarker for early, progressive, and recurrent prostate cancer (163). Therefore, more research is needed to clearly define the clinical value of GRN-A as a serum and tumor marker for prostate cancer.
Prostate-specific Membrane Antigen (PSMA)
PSMA is a cell surface membrane that was discovered in 1987 and has been well characterized as a diagnostic and prognostic marker. It is a type II integral membrane protein that exhibits numerous enzymatic activities (40, 164). Although insufficient data exist regarding its biological role, PSMA has been seen to translocate to the plasma membrane in prostate cancer cells, whereas it is located in the cytosol in normal prostate cells (165). PSMA has been detected in prostate tissues, circulating prostate cancer cells, and serum. Its levels may correspond with poor clinical outcome; PSMA levels are higher in primary prostate cancer and metastatic disease, with more than 90% of the protein prevalent in the disease (166). The serum levels of PSMA increase with age and are considerably higher in men above 50 years of age (79). However, no concrete evidence has shown a relationship between high levels of serum PSMA and the aggressive disposition of the disease, while some have observed a decrease in advanced cases of the disease (167). In an attempt to better measure the levels of circulating PSMA, a study revealed that serum levels of PSMA in prostate cancer patients vary significantly when compared to those of healthy men and those with BPH (167). PSMA appears to be upregulated in patients with prostate cancer subsequent to hormone deprivation therapy (32), which further reveals that the levels of PSMA in men with prostate cancer is considerably higher than in those with BPH or those free of disease. Several gene therapy strategies have also used the PSMA gene promoter to transcriptionally regulate the cytotoxic genes/agents in prostate cancer cells (167). However, the prostate specificity of PSMA as a gene therapy target is limited, but it has lately been used as a target for immunotherapy (168).
One of the shortcomings in using PSMA as a serum marker is that high levels have been noticed in patients with incident case of prostate cancer and in the serum of breast cancer patients. This could make it difficult in some cases to accurately diagnose men with prostate cancer (44, 169). Another shortcoming is that the levels of serum PSMA increase with advancing age, which could result in some conflicting results if diagnosis is sought at that period in life (which is usually the case). More sufficient data is required to determine whether this biomarker can be validated clinically for use in prostate cancer detection, monitoring of treatment, or as an actual means of treatment (170).
Prostate Stem Cell Antigen (PSCA)
Prostate stem cell antigen is a membrane glycoprotein predominantly expressed in the prostate. Although the expression of PSCA has been revealed to be upregulated in the majority of prostate cancers, its biological role in prostate cancer is uncertain (159). Studies have implicated PSCA in certain stem cell functions like androgen-independent progression, metastasis, or signal transduction in many prostate cancer cells (169, 171-173). PSCA expression is associated with Gleason score, seminal vesicle invasion, and capsular invasion in prostate cancer; hence, it has potential as a therapeutic target. A correlation was detected between the increase levels of PSCA expression in most prostate cancers and higher Gleason grade and more advanced tumor stage (171). A function for PSCA in prostate cancer progression is proposed from the observation that it is jointly amplified with c-myc (an oncogene and factor in tumor progression) in locally advanced prostate cancers (159, 160). When the mRNA of other circulating prostate markers like PSA and PSMA were compared with that of PSCA, researchers observed that, although PSCA displayed inferior sensitivity and considerable inability to distinguish between malignant and benign disease, its disease specificity and independent predictive value were the highest (174).
Using human xenografts grown in SCID mice, researchers showed that anti-PSCA monoclonal antibodies inhibited tumor growth and metastasis formation (175), making PSCA potentially available for treating prostate cancer therapeutically using immunotherapeutic procedures (175-177). Despite the research revelations about PSCA and its potential, there are still no definitive conclusions regarding its being a serum marker. Factors that mitigate against PSCA as a candidate for further development include an inadequate number of published studies supporting PSCA as a valuable clinical biomarker and the lack of better measuring techniques (97). Therefore, the value of PSCA as a therapeutic target and the existing related data must await more studies to further evaluate and determine its effectiveness as a clinical prostate cancer marker.
Early Prostate Cancer Antigen (EPCA)
Early prostate cancer antigen, originally discovered in 1991 in rat prostate tissue (178), is a nuclear matrix protein linked with the nuclear transformations that occur in early prostate cancer development (179). These proteins are vital components of the nuclear matrix, a structure shown to dictate the shape and organization of the nucleus and reflect patterns of chromatin transcription. The correlation between variations in the nuclear matrix and the nuclear pleiomorphism displayed in prostate cancer was first described by Getzenberg et al. (178). Following that, EPCA was found in prostate cancer precursor lesions, specifically in prostatic intraepithelial neoplasia and proliferative inflammatory atrophy, as well as prostate cancer tissue (180). Also, the protein has been identified in men with a preliminary negative biopsy but but who later developed the cancer (180, 181). The study of Uetsuki et al. provided further evidence that EPCA can be linked with early carcinogenesis as no relationship could be found between EPCA and disease stage or Gleason score after radical prostatectomy (179). Furthermore, recent studies have verified the potential diagnostic value of serum EPCA by demonstrating the ability of EPCA antibodies to recognize prostate cancer (182). Although EPCA appears not to be present in patients devoid of prostate cancer, it has been detected in surroundings free of, but adjacent to, the cancer (180). More studies are needed to further characterize the protein as a suitable biomarker to diagnose prostate cancer.
Several studies have been conducted to evaluate and propose other tissue and circulating prostate tumor markers such as rising aneuploidy and polyploidy (associated with invasive and metastatic tumors) and biopsy ploidy, which may assist in predicting a pathologic stage (183-188). Ki-67 expression, as a marker of proliferation index, has been shown to be an independent predictor of recurrence and tumor-specific survival (93, 189). RT-PCR has shown some promise as a sensitive biomarker in identifying micrometastases in nonprostatic sites, such as PSA- and/or PSMA-positive lymph nodes, which are not detectable by conventional pathology (190). The expressions of Bcl-2 and p53 have been extensively examined as prognostic markers in prostate tissue, and these markers may aid in predicting the response of localized prostate cancer to radiotherapy. However, their utility in predicting a pathologic stage has yet to be established (191, 192). Finally, serum immunoassay for hK2 biomarker (a PSA-related protein, Table 4) combined with PSA testing provides improved discrimination among men who had total PSA levels in the 4 to 10 ng/ml range and between men with benign prostate disease and those with prostate cancer (193, 194). Some studies have indicated that serum hK2 levels aid in predicting prostate-confined disease in the staging preceding a surgical operation (194, 195).
B7-H3
B7-H3 is the first immune molecule that possibly participates in the development of prostate cancer and in predicting the recurrence and progression of cancer. B7-H3, first identified in 2001, is a member of the B7 family, a group of proteins that are important ligands interacting with known and unknown receptors to regulate the activation and function of T lymphocytes. B7 (or B7-H3) protein is believed to function as an accessory co-regulator of T cell responses subsequent to initial antigen priming (196, 197). B7 co-regulatory ligands can be abnormally expressed in human disease and may act as antigen-specific inhibitors of T-cell-mediated antitumoral immunity in cancer conditions (198). The B7-H family protein, including B7-H3 and B7-H1, can both arrest cancer growth and shield cancers from the immune system by paralyzing immune cells (199). Thereby this ligand exhibits both an immune stimulatory and inhibitory role in cancer growth.
Numerous normal tissues, except for dormant peripheral blood monocytes, express B7-H3 mRNA (196, 200). B7-H3 protein expression has been detected in placenta (201), and its expression can be stimulated in activated dendritic cells, monocytes, and T cells (202). B7-H3 is also expressed in numerous tumor cell lines, and the expression of B7-H3 in carcinomas of the kidney and bladder correlates with aggressive disease and significantly shortened survival time in patients. The expression of these proteins in prostate cancer has been linked to the spread of the disease and negative outcome (203-205). In contrast to PSA, B7-H3 remains bound to the surface of normal prostate cells, as well as of premalignant and cancerous prostate cells that show no apparent indication of migration (metastatic ability), thus making it an attractive therapeutic target and marker. This would be an especially promising target for antihormone therapy, which is the most frequent means of therapeutic treatment for advanced prostate cancer (203). Because B7-H3 is present in all prostate cancer tumors and marked levels predict recurrence, researchers examined diseased tissue from 338 patients who had clinically localized prostate cancer and were treated exclusively with radical prostatectomy between 1995 and 1998. They were able to predict with better accuracy the likelihood of cancer progression in spite of therapeutic intervention (203). This study revealed a link between a rising level of B7-H3 in prostate cancer and adverse clinicopathologic features of the disease. Therefore, B7-H3 may have the potential to independently predict prostate cancer progression and may be used as a diagnostic and prognostic marker to evaluate patients' disease status and their immunotherapeutic responses (203,205).
More research is necessary, however, to understand how the immune system is affected by B7-H3. For example, it would be useful to know whether anti-immune activity results from a mutation of B7-H3, which may be the mechanism by which B7-H3 promotes cancer growth. This information is critical and will help to establish the effectiveness of B7-H3 as a clinical marker of disease and target for therapy.
Sarcosine
Sarcosine, an N-methyl derivative of glycine, is a natural amino acid found in muscles and other body tissues. It is classified under the group collectively known as metabolites (a group of chemical products present throughout the body) (206). In 2009, Sreekumar et al. reported that sarcosine stimulates malignant growth of benign prostate cancer cells and can be used as an indicator of the malignancy of prostate cancer cells when detected in urine (207). Following the screening of urine, blood, and tissues, and profiling more than 1,126 metabolites related to prostate cancer, the researchers were able to differentiate between benign prostate, clinically localized prostate cancer, and metastatic disease based on the levels of sarcosine. The levels of sarcosine were high in invasive prostate cancer cell lines compared to benign prostate epithelial cells (207). Furthermore, it was observed that prostate cancer invasion was weakened when glycine-N-methyltransferase, the enzyme that catalyzes the production of sarcosine from glycine, was knocked down, whereas either knocking down the enzyme responsible for sarcosine degradation or adding exogenous sarcosine stimulated an invasive phenotype in BPH cells. These results together suggest that sarcosine may be a vital metabolic intermediary that promotes prostate cancer cells toward invasion and aggressiveness (208).
The ultimate goal of diagnosis is to detect aggressive-type prostate cancers at their premature stage. This, nevertheless, may not be possible very soon. Among the conflicting scientific points of view on whether sarcosine is a better diagnostic biomarker than PSA for detecting aggressive prostate cancer is that several researchers have been criticized for their possible investment interests in promoting sarcosine toward commercialization. Moreover, further investigations are still needed on the metabolites in many patients who are followed long-term (to see how they correlated with those who developed different forms of prostate cancer).
Caveolin-1
Caveolin-1 (Cav-1), an integral membrane protein expressed in two isoforms (caveolin-1α and caveolin-1β), is a main component of caveolae membranes in vivo (209). It has been implicated in regulating several signaling pathways and mediating intracellular processes, specifically as a negative regulator in several mitogenic pathways (210) and in oncogenesis (211). It has been proposed that Cav-1 may participate in certain steps of carcinogenesis in various types of cancer and is expressed in one-third of invasive breast cancers (212). Cav-1 seems to function as a tumor suppressor protein at early stages of cancer progression. However, Cav-1 is also found to be upregulated in several metastatic and multidrug-resistant cancer cell lines, as well as in some human tumor specimens (213).
Cav-1 is secreted by prostate cancer cells. Early and recent studies have shown that this secreted protein can promote cell survival and angiogenic activities (214-216). Cav-1 has been reported to be overexpressed in prostate cancer cells and is associated with the progression of the disease (217-218).
Studies of prostate tissue from men with only localized prostate cancer indicate a significant decrease in levels of Cav-1. It was also discovered that the protein was absent in tumor tissue from men with metastatic prostate cancer, and the reduced levels of Cav-1 were associated with a high Gleason score (220).
Research conducted on stromal Cav-1 expression in patients with BPH, primary prostate cancers, and prostate cancer metastases revealed that almost all BPH samples showed an abundant stromal Cav-1 immunostaining, while a subset of samples with primary prostate cancer had significantly decreased levels of stromal Cav-1. All metastatic tumors (either from lymph node or bone) lacked stromal Cav-1 staining (221). The concentration of preoperative serum Cav-1 showed prognostic potential in patients undergoing radical prostatectomy (220). Therefore, Cav-1 expression may be a useful prognostic marker for prostate cancer (220, 222).
Serum calcium
Prostate cancer cells express calcium-sensing, G-protein-coupled receptor, which can be activated by extracellular calcium (223). These cells also express calcium-dependent potassium channels that regulate their proliferation by controlling the entry of calcium into the cells (224). An association has been noted between high levels of total calcium in serum and the risk of fatal prostate cancer, which is to the result of a decrease in apoptosis and an increase in proliferation, which promote the growth and metastasis of prostate cancer cells (225). Researchers postulated that an increase in serum calcium or any factor that leads to it (such as high serum parathyroid hormone) would increase the possibility for terminal prostate cancer. An investigation by an independent group confirmed this association and suggested that serum calcium is a promising prospective biomarker for screening for fatal prostate cancer (225-227).
Hypermethylation of PDLIM4 gene
Hypermethylation of the PDLIM4 gene has been shown to be a sensitive molecular tool in detecting prostate tumorigenesis (228). PDLIM4 mRNA and protein expression levels were also reduced in LNCaP, LAPC4, DU145, CWR22, and PC3 prostate cancer cells and may function as a tumor suppressor by associating with actin in prostate cancer cells, thereby controlling cell proliferation (229). These findings support the potential use of hypermethylated PDLIM4 as a biomarker to predict the biochemical, local, and systemic recurrence of prostate cancer.
PCA3/DD3
The prostate cancer antigen 3 (PCA3 or DD3) gene encodes a prostate-specific mRNA that is overexpressed in cancer prostate tissue (230, 231). A measure of the quantity of PCA3 RNA copies in urine samples that are enriched with prostate cells can provide an insight into the aggressiveness of the prostate cancer (232) and predict the outcome of prostate biopsies while avoiding the need for repeated testing (233-235). Furthermore, the predictive value of PCA3 has been addressed in some clinical studies, either by urine test that is performed prior to radical prostatectomy or by extracapsular extension prior to a DRE exam (236, 237). Consequently, several published studies have supported the usefulness of PCA3 as a biomarker in the diagnosis of prostate cancer stage and grading (231-235, 239-244). Although PCA3 has a lower sensitivity than does PSA, it has a higher specificity (233-235), especially in certain cases where PSA tests fail to accurately predict disease (245, 246). Adding PCA3 to serum PSA detection contributes to the precise prediction of prostate cancer; in a clinical setting, PCA3 may be used to classify patients according to their risks for biopsy and cancer detection (238, 245-246). Similarly, combining urinary alpha-methylacyl-CoA racemase (AMACR) and PCA3 as a dual marker test improved sensitivity and accuracy (247). A recent study reported an upregulation of two new PCA3 isoforms in prostate cancer tissues; this will enhance the diagnostic ability to distinguish between prostate cancer and BPH (248).
TMPRSS2-ERG Gene Fusion Rearrangement
Transmembrane protease serine 2, also known as TMPRSS2, is an androgen-regulated, type II transmembrane-bound serine protease that is locally expressed in the prostate and overexpressed in neoplastic prostate epithelium. TMPRSS2 was thought to play a possible role in prostate tumor metastasis through the activation of protease-activated receptor-2 (PAR-2) (249). An extensive study focusing on gene fusion transcripts in prostate cancer identified the fusion between TMPRSS2 (located at 21q22.3) with the transcription factor genes ERG (21q22.2) and ETV1 (7p21.1) (250, 251). One TMPRSS2 allele loses its promoter, and one of the ERG alleles gains it, resulting in an overexpression of ETS family members) in the cancer cells (252) and consequently tumor progression (253). The gene fusion rearrangements between TMPRSS2 and ERG or ETV1 have been reported in several cancers, particularly in hematological malignancies (254). TMPRSS2-ERG is the most frequent oncogenic gene fusion rearrangement in prostate cancer (251): It has been observed in almost half of prostate cancer patients and detected in about one-quarter of patients with prostatic intraepithelial neoplasia (PIN) (255).
This TMPRSS2-ERG fusion is usually found in prostate cancer tissue from men undergoing prostatectomy, and especially among men in North America with prostate cancer on biopsy; however, it is not present in benign prostate biopsy. The TMPRSS2-ERG fusion can be detected in the urine after DRE with a 37% sensitivity and a 93% specificity (256). The addition of TMPRSS2-ERG detection also increased the sensitivity of the urine PCA3 test from 62% to 73% (256) and a greater prediction of positive tumors with a higher Gleason score (257). The results suggest that surveillance of these fusion gene transcripts improves the sensitivity of the PCA3 detection in urine samples: The combination of TMPRSS2-ERG detection with PCA3 can be very useful in accurately predicting prostate cancer development during screening (252, 258).
Exosomes
Exosomes are nanometer-sized vesicles secreted by a broad range of normal and neoplastic cell types (259). They contain both functional mRNA and microRNA, called exosomal shuttle RNA (esRNA) that are often transported from cell to cell where they can continue to be functional (260). Exosmes are constituents of urine, with a degree of variability in urine specimens. Because they often carry genetic components that come directly from tumors, such vesicles may be a useful noninvasive source of markers in renal diseases (251), including cancer of the prostate.
A recent study reported the presence of PCA3 and TMPRSS2-ERG fusion, two known prostate cancer biomarkers, in exosomes from urine samples of prostate cancer patients (261, 262). A second study reported the presence of the gene fusion TMPRSS2-ERG product in isolated exosomes in the serum from mice grafted with human PCA xenografts (263), which could shed more light on the genetics of the particular tumor and provide clinically valuable data. Although the presence of exosomes in urine samples varies in concentration, making it difficult to assess, its presence and quantification may be a potential noninvasive source of tumor markers that could be used to diagnose and monitor the status of prostate cancer.
Ki-67
Ki-67, a cell-proliferation associated marker and probably the only protein with an expression pattern under a level of cell cycle regulation (255, 264), has been described as one of the most promising biomarkers of prostate cancer. Ki-67 has been suggested as a prolific predictive biomarker for men who have low-grade, low-stage prostate cancer for their PSA relapse after radical prostatectomy (265). In a 6-year study involving 808 patients diagnosed with prostate cancer, an immunohistochemical assessment of Ki-67 expression was evaluated for its relationship to the specificity of the cancer and overall survival. Compared to information from the Gleason score and PSA, Ki-67 provided additional prognostic information (266, 267). In another study of a group of men treated with radiotherapy and androgen deprivation for prostate cancer, Ki-67 expression levels in conjunction with MDM2 were found to be correlated to distant metastasis and survivability (268). Nevertheless, further studies will be needed to validate these results and explore the possibility of combining Ki-67 with existing prognostic tools as a powerful biomarker for localized prostate cancer (269).
GOLPH2
Golgi phosphoprotein 2, or GOLPH2, is a gene that codes for type II Golgi membrane antigen GOLPH2/GP73. It is usually expressed in various epithelial cells and reported to be frequently overexpressed in cancer of the prostate, although its function is currently unknown (270). A study has observed a higher level of GOLPH2 and MYO6 (myosin VI) in the Golgi apparatus in prostate cancer cells compared to normal Golgi, thereby indicating that GOLPH2 can be used as a biomarker in distinguishing between normal cells and cancer cells (271).
A recent study, which explored the expression of some potential prostate cancer biomarkers, revealed that an increase in the levels of GOLPH2 (as with some of the other biomarkers assayed), was not only a critical indicators of prostate cancer but performed better than PSA or PCA3 alone in revealing it (272).
A comparative study of GOLPH2 protein, the basal cell marker p63, and AMACR in benign and malignant prostate lesions revealed that GOLPH2 expression was considerably higher in prostate cancer tissues compared with normal tissues, and in about 90% of the cases studied, GOLPH2 protein was upregulated. Furthermore, this upregulation was noticed in about 85% of prostate cancer cases that were negative for AMACR (273).
DAB2IP
DAB2 interacting protein (DAB2IP) is a Ras GTPase-activating protein that functions as a tumor suppressor. The human DAB2IP gene is located on chromosome 9q33.1-q33.3 (274) and is frequently observed to be downregulated in prostate cancer cell lines (275, 276). Studies have shown that loss of expression of DAB2IP may be a result of altered epigenetic regulations, for example DNA methylation and histone modification (277). The abnormal methylation in the promoter region of the DAB2IP gene has been reported to be responsible for transcriptional silencing and consequently performs a significant function in the progression of prostate cancer (278). Duggan et al. in their 2007 study reported a link between a genetic variation in DAB2IP and the risk of aggressive prostate cancer (279). This research indicates that DAB2IP protein, after further studies, can potentially be used as a very effective novel biomarker for prostate cancer diagnosis.
7. FUTURE CHALLENGES AND PERSPECTIVES
The increasing importance of biomarkers in screening for prostate cancer to reduce invasive follow-up procedures is reflected proportionally in the rapidly increasing number of research publications in this field. The application of biomarkers to prostate cancer is at the forefront of the research field because of the distinctive relationship between the genomic changes in the cancer cells and the disease progress.
The technology used in the effort to discover ideal biomarkers has advanced significantly, making it easier to study many potential biomarkers in a single trial. While several biomarkers have displayed some potential in early-phase studies, none so far appears likely to possess the appropriate level of sensitivity and specificity in terms of determining the choice and course of therapeutic treatment for prostate cancer (98). This may explain why only a small number of biomarkers are routinely validated for use in drug development or qualified for clinical applications, despite the apparent progress in this research field. Despite some advancement, several limitations still exist with the current technology that hinders the discovery and development of new biomarkers for all forms of cancer including prostate cancer. Some of these impediments may be overcome through the development of new technologies and improved strategies. For example, one strategy proposed would pair the diagnostic test with the therapeutic agent (6). Another strategy calls for more attention on studies that can generate quantified biomarkers related to cell-signaling pathways, as these biomarkers can be applied across a wide range of tumor types and diseases, as well as in different tests and drugs (10, 12).
Because of the fact that the entire process from identifying to validating a reliable biomarker is expensive and long, concerns have been raised over the profit-making incentives to develop novel biomarkers. An expensive search in the long run would result in a high cost for any reliable test developed in the future, adding even more cost to the already high cost of drug development (12). Consequently, pharmaceutical companies may gradually become reluctant to invest in diagnostic development in the earlier phases of testing because of the uncertainty of validation and approval by the FDA, without which there will be no financial return on all of their investment.
A critical point that has been reiterated is the fact that an ideal biomarker has to show a high level of specificity and sensitivity to prevent false-positive screening tests, which will create anxiety in patients and lead to more expensive and invasive testing. Thus far, studies, although inconclusive, have found that the likelihood of identifying a biomarker with such sensitivity and specificity may be slim, at least for the immediate future. Therefore, combining markers is thought to be the next best thing to improve the accuracy of diagnosing, treating, and surveillance of recurrence of prostate cancer (152). Some have suggested using multiple biomarkers with different qualities: for instance, combining a “biomarker with high sensitivity and low specificity (to detect potentially deadly cancers but would result in many false positives) with a second biomarker having less sensitivity but higher specificity” (84).
In summary, substantial discovery still awaits to be made in this field, and methodologies for the clinical evaluation of existing and novel biomarkers have yet to be explored. While much could be gained from the discovery of more novel biomarkers for early detection of prostate cancer, prediction of the malignant potential of the disease, and guidance of individualized therapy for patients, the near future of prostate cancer prognosis may eventually come to count on a few “elite club” biomarkers, which hopefully will accurately predict the incidence, stage, and progression of the disease, as well as reliably evaluate drug development. While extensive clinical validation of these novel biomarkers remains as one of the most significant and daunting challenges, overcoming this impediment will by no means eliminate all the problems hampering the identification and development of biomarkers for this disease. However, in the process of searching for novel biomarkers, we may discover valuable insights into the mechanisms of prostate cancer that could possibly lead to a cure in the long run.
Acknowledgements
We thank Jordan Masters and Dr. David L. Armbruster for reviewing the manuscript.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225-49
2. Home page of Prostate Cancer Foundation. Prostate Cancer Foundation. http://www.prostatecancerfoundation.org
3. Stanford JL, Stephenson RA, Coyle LM. et al. Prostate Cancer Trends 1973-1995, NIH Pub No 99-4543. Bethesda, MD: SEER Program, National Cancer Institute. 1999
4. Prostate Cancer Screening: A Decision Guide for African Americans. US Department of Health and Human Services, Centers for Disease Control and Prevention (CDC). http://www.ustoo.org/PDFs/CDC_PCa_Screen_Guide_AA.pdf
5. Atkinson AJJr, Colburn WA, DeGruttola VG, DeMets DL. et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Biomarkers Definitions Working Group. Clinical Pharmacol & Therapeutics. 2001;69:89-95
6. A Framework for Biomarker and Surrogate Endpoint Use in Drug Development. Woodcock J. http://www.fda.gov/ohrms/dockets/ac/04/slides/2004-4079S2_03_Woodcock.ppt
7. Dictionary of Cancer Terms. National Cancer Institute. http://www.cancer.gov/dictionary/?searchTxt=biomarker
8. Rolan P. The contribution of clinical pharmacology surrogates and models to drug development: a critical appraisal. Br J Clin Pharmacol. 1997;44:219-25
9. Ciarmiello A, Del Vecchio S, Silvestro P. et al. Tumor clearance of technetium 99m-sestamibi as a predictor of response to neoadjuvant chemotherapy for locally advanced breast cancer. J Clin Oncol. 1998;16:1677-83
10. Committee on Developing Biomarker-Based Tools for Cancer Screening Diagnosis and Treatment. Cancer Biomarkers: The Promises and Challenges of Improving Detection and Treatment. Washington DC: National Academies Press. 2007
11. Lagakos SW, Hoth DF. Surrogate markers in AIDS: where are we? Where are we going? Ann Intern Med. 1992;116:599-601
12. Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548-52
13. Ross RK, Pike MC, Coetzee GA. et al. Androgen metabolism and prostate cancer: establishing a model of genetic susceptibility. Cancer Res. 1998;58:4497-504
14. Smith RA, Mettlin CJ, Eyre H. Cancer Medicine: Cancer Screening and Early Detection. In: (ed.) Kufe DW. et al. Cancer Medicine. Hamilton, Canada: BC Decker Inc. 2003
15. Prostate Cancer - Biology, Diagnosis, Pathology, Staging, and Natural History. Theodorescu D, Krupski T. http://www.emedicine.com
16. Bostwick DG. The pathology of early prostate cancer. CA Cancer J Clin. 1989;39:376-93
17. Stamey TA, Freiha FS, McNeal JE. et al. Localized prostate cancer treatment: relationship of tumor volume to clinical significance for treatment of prostate cancer. Cancer. 1993;71:933-8
18. Epstein JI, Walsh PC, Carmichael M. et al. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA. 1994;271:368-74
19. Ohori M, Wheeler TM, Dunn JK. et al. The pathologic features and prognosis of prostate cancers detectable with current diagnostic tests. J Urol. 1994;152:1714-20
20. Montie JM, Wood DP, Pontes JE. et al. Adenocarcinoma of the prostate in cystoprostatectomy specimens removed for bladder cancer. Cancer. 1989;63:381-5
21. Smith DS, Catalona WJ. The nature of prostate cancer detected through prostate specific antigen based screening. J Urol. 1994;152:1732-6
22. Oh WK, Hurwitz M, D'Amico AV. et al. Neoplasms of the Prostate. In: (ed.) Kufe DW. et al. Cancer Medicine. Hamilton, Canada: BC Decker Inc. 2003
23. Yano M, Imamoto T, Suzuki H. et al. The clinical potential of pretreatment serum testosterone level to improve the efficiency of prostate cancer screening. Eur Urol. 2007;51:375-80
24. Fielding LP, Genglio-Presier CM, Freedman LS. The future of prognostic factors in outcome prediction for patients with cancer. Cancer. 1992;70:2367-77
25. Jewett HJ. The present status of radical prostatectomy for stages A and B prostatic cancer. Urol Clin North Am. 1975;2:105-24
26. American Joint Committee on Cancer. Prostrate: in AJCC Cancer Staging Manual, 6th ed. New York, NY: Springer. 2002:309-16
27. Bostwick DG, Myers RP, Oesterling JE. Staging of prostate cancer. Semin Surg Oncol. 1994;10:60-72
28. American Joint Committee on Cancer. Prostate cancer. In: (ed.) Fleming ID, Cooper JS, Henson DE, Hutter RVP. AJCC cancer staging manual. 5th ed. Philadelphia: Lippincott. 1997:219-24
29. Montie JE. Staging of prostate cancer: current TNM classification and future prospects for prognostic factors. Cancer. 1995;75(7 Suppl):1814-8
30. Epstein JI, Walsh PC, Brendler CB. Radical prostatectomy for impalpable prostate cancer: The Johns Hopkins experience with tumours found on transurethral resection (stages T1A and T1B) and on needle biopsy. J Urol. 1994;152:1721-9
31. Mettlin C, Lee F, Drago J. et al. The American Cancer Society National Prostate Cancer Detection Project. Findings on the detection of early prostate cancer in 2425 men. Cancer. 1991;67:2949-58
32. Catalona WJ, Smith DS, Ratliff TL. et al. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N Engl J Med. 1991;324:1156-61
33. Basler JW, Thompson IM. Lest we abandon digital rectal examination as a screening test for prostate cancer. J NCI. 1998;90:1761-3
34. Mahon SM. Screening for prostate cancer: informing men about their options. Clinical Journal of Oncology Nursing. 2005;9:5
35. Yedema CA, Kenemans P, Voorhorst F. et al. CA 125 half-life in ovarian cancer. A multivariate survival analysis. Br J Cancer. 1993;67:1361-7
36. Jacobsen SJ, Bergstralh EJ, Katusic SK. et al. Screening digital rectal examination and prostate cancer mortality: a population-based case-control study. Urology. 1998;52:173-9
37. Friedman GD, Hiatt Ram, Quesenberry CP. et al. Case-control study of screening for prostatic cancer by digital rectal examinations. Lancet. 1991;337:1526-9
38. Richert-Boe KE, Humphrey LL, Glass AG. et al. Screening digital rectal examination and prostate cancer mortality: a case-control study. J Med Screen. 1998;5:99-103
39. Harris R, Lohr KN. Screening for prostate cancer: an update of the evidence for the U.S. Preventive Services Task Force. Ann Intern Med. 2002;137:917-29
40. Hara M, Koyanagi Y, Inoue T. et al. Some physico-chemical characteristics of “-seminoprotein”, an antigenic component specific for human seminal plasma. Forensic immunological study of body fluids and secretion. VII. Nihon Hoigaku Zasshi. 1971;25:322-4
41. Li TS, Beling CG. Isolation and characterization of two specific antigens of human seminal plasma. Fertil Steril. 1973;24:134-44
42. Sensabaugh GF, Golden VL. Phenotype dependence in the inhibition of red cell acid phosphatase (ACP) by folates. Am J Hum Genet. 1978;30:553-60
43. Wang MC, Valenzuela LA, Murphy GP. et al. Purification of a human prostate specific antigen. Invest Urol. 1979;17:159-63
44. Stamey TA, Yang N, Hay AR. et al. E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med. 1987;317:909-16
45. Partin AW, Oesterling JE. The clinical usefulness of prostate specific antigen. J Urol. 1994;152:1358-68
46. Polascik TJ, Oesterling JE, Partin AW. Prostate specific antigen. A decade of discovery—what we have learned and where we are going. J Urol. 1999;162:293-306
47. Rao AR, Motiwala HG, Karim OM. The discovery of prostate-specific antigen. BJU Int. 2008;101:5-10
48. Chu MT. Prostate-specific antigen and early detection of prostate cancer. Tumor Biol. 1997;18:123-34
49. Diamandis EP, Yu H. Nonprostatic sources of prostate-specific antigen. Urol Clin North Am. 1997;24:275-82
50. Stenman UH, Leinonen J, Zhang WM. Standardization of PSA determinations. Scand J Clin Lab Invest (Suppl). 1995;221:45-51
51. Carter HB, Pearson JD, Metter EJ. et al. Longitudinal evaluation of prostate-specific antigen levels in men with and without prostate disease. JAMA. 1992;267:2215-20
52. Moul JW, Sesterhenn IA, Connelly RR. et al. Prostate-specific antigen values at the time of prostate cancer diagnosis in African-American men. JAMA. 1995;274:1277-81
53. Presti JCJr, Hovey R, Bhargava V. et al. Prospective evaluation of prostate specific antigen and prostate specific antigen density in the detection of carcinoma of the prostate: ethnic variations. J Urol. 1997;157:907-11
54. Morgan TO, Jacobsen SJ, McCarthy WF. et al. Age-specific reference ranges for prostate-specific antigen in black men. N Engl J Med. 1996;335:304-10
55. Partin AW, Yoo J, Carter HB. et al. The use of prostate specific antigen, clinical stage and Gleason score to predict pathological stage in men with localized prostate cancer. J Urol. 1993;150:110-4
56. Partin AW, Carter HB, Chan DW. et al. Prostate specific antigen in the staging of localized prostate cancer. Influence of tumor differentiation, tumor volume and benign hyperplasia. J Urol. 1990;143:747-52
57. Stamey TA, Dietrick DD, Issa MM. Large, organ confined, impalpable transition zone prostate cancer. Association with metastatic levels of prostate specific antigen. J Urol. 1993;149:510-5
58. Catalona WJ, Smith DS, Ratliff TL. et al. Detection of organ-confined prostate cancer is increased through prostate-specific antigen-based screening. JAMA. 1993;270:948-54
59. Balducci L, Pow-Sang J, Friedland J. et al. Prostate cancer. Clin Geriatr Med. 1997;13:283-306
60. SEER Cancer Statistics Review, 1975-2005. Ries LA, Melbert D, Krapcho M, et al. http://seer.cancer.gov/csr/1975_2005/
61. Hankey BF, Feuer EJ, Clegg LX. et al. Cancer surveillance series: Interpreting trends in prostate cancer - Part I: Evidence of the effects of screening in recent prostate cancer incidence, mortality, and survival rates. J Natl Cancer Inst. 1999;91:1017
62. Feuer EJ, Etzioni R, Cronin KA. et al. A. The use of modeling to understand the impact of screening on U.S. mortality: examples from mammography and PSA testing. Stat Methods Med Res. 2004;13:421
63. Polednak AP. Estimating the prevalence of cancer in the United States. Cancer. 1997;80:136-41
64. Kirby R, Kirby M, Fitzpatrick J. et al. Shared Care for Prostate Disease. Oxford: ISIS Medical Media. 1994
65. Brown ML, Lipscomb J, Snyder C. The burden of illness of cancer: economic cost and quality of life. Annual Review of Public Health. 2001;22:91-113
66. Hakama M, Auvinen A. Cancer screening. In: (ed.) Heggenhougen Kl, Quah SR. International Encyclopedia of Public Health. San Diego, CA: Academic Press. 2008:464-80
67. Andriole GL, Grubb RL III, Buys SS. et al. The PLCO Project Team. Mortality results from a randomized Ppostate-cancer screening trial. NEJM. 2009;360:1310-9
68. Aus G, Ahlgren G, Bergdahl S. et al. Infection after transrectal core biopsies of the prostate--risk factors and antibiotic prophylaxis. Br J Urol. 1996;77:851-5
69. Rietbergen JB, Kruger AE, Kranse R. et al. Complications of transrectal ultrasound-guided systematic sextant biopsies of the prostate: evaluation of complication rates and risk factors within a population-based screening program. Urology. 1997;49:875-80
70. Yao SL, Lu-Yao G. Population-based study of relationships between hospital volume of prostatectomies, patient outcomes, and length of hospital stay. J NCI. 1999;91:1950-6
71. Alibhai SMH, Leach M, Tomlinson G. et al. 30-Day mortality and major complications after radical prostatectomy: influence of age and comorbidity. J NCI. 2005;97:1525-32
72. Potosky AL, Davis WW, Hoffman RM. et al. Five-year outcomes after prostatectomy or radiotherapy for prostate cancer: the Prostate Cancer Outcomes Study. J NCI. 2004;96:1358-67
73. Lim AJ, Brandon AH, Fiedler J. et al. Quality of life: radical prostatectomy versus radiation therapy for prostate cancer. J Urol. 1995;154:1420-5
74. Hamilton AS, Stanford JL, Gilliland FD. et al. Health outcomes after external-beam radiation therapy for clinically localized prostate cancer: results from the Prostate Cancer Outcomes Study. J Clin Oncol. 2001;19:2517-26
75. Fowler FJ Jr, McNaughton Collins M; Walker Corkery E. et al. The impact of androgen deprivation on quality of life after radical prostatectomy for prostate carcinoma. Cancer. 2002;95:287-95
76. Tsai HK, D'Amico AV, Sadetsky N. et al. Chen, MH; Carroll, PR. Androgen deprivation therapy for localized prostate cancer and the risk of cardiovascular mortality. JNCI. 2007;99:1516-24
77. Punnen S, Nam RK. Indications and timing for prostate biopsy, diagnosis of early stage prostate cancer and its definitive treatment: A clinical conundrum in the PSA era. Surg Oncol. 2009;18:192-9
78. Cooner WH, Mosley BR, Rutherford CL Jr. et al. Prostate cancer detection in a clinical urological practice by ultrasonography, digital rectal examination and prostate specific antigen. J Urol. 1990;143:1146-52
79. Brawer MK, Chetner MP, Beatie J. et al. Screening for prostatic carcinoma with prostate carcinoma with prostate specific antigen. J Urol. 1992;147:841-5
80. Labrie F, Dupont A, Suburu R. et al. Serum prostate specific antigen as pre-screening test for prostate cancer. J Urol. 1992;147:846-51
81. Catalona WJ, Richie JP, Ahmann FR, et al.Comparison of digital rectal examination, serum prostate specific antigen in the early detection of prostate cancer. Results of a multicenter clinical trial of 6,630 men. J Urol. 1994;151:1283-90
82. Greenlee RT, Murray T, Bolden S. et al. Cancer statistics 2000. CA Cancer J Clin. 2000;50:7-33
83. Potosky AL, Miller BA, Albertsen PC. et al. The role of increasing detection in the rising incidence of prostate cancer. JAMA. 1995;273:548-52
84. Moul JW. Treatment options for prostate cancer. Part I. Staged, grade, PSA, and changes in the 1990's. Am J Managed Care. 1998;4:1031-6
85. Tarone RE, Chu KC, Brawley OW. Implications of stage-specific survival rates in assessing recent declines in prostate cancer mortality rates. Epidemiology. 2000;11:167-70
86. Smart CR. The results of prostate carcinoma screening in the U.S. as reflected in the surveillance, epidemiology, and end results program. Cancer. 1997;80:1835-44
87. Mettlin CJ, Murphy GP, Rosenthal DS. et al. The National Cancer Data Base report on prostate carcinoma after the peak in incidence rates in the U.S. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer. 1998;83:1679-84
88. Tarone RE, Chu KC, Brawley OW. Implications of stage specific survival rates in assessing recent declines in prostate cancer mortality rates. Epidemiology. 2000;11:167-70
89. Smart CR. The results of prostate carcinoma screening in the US as reflected in the Surveillance, Epidemiology, and End Results program. Cancer. 1997;80:1835-44
90. Mettlin CJ, Murphy GP, Rosenthal DS. et al. The National Cancer Data Base report on prostate carcinoma after the peak in incidence rates in the US. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer. 1998;83:1679-84
91. Bunting PS. A guide to the interpretation of serum prostate specific antigen levels. Clin Biochem. 1995;28:221-41
92. Feuer EJ, Merrill RM, Hankey BF. Cancer surveillance series: interpreting trends in prostate cancer— part II: cause of death misclassification and the recent rise and fall in prostate cancer mortality. J NCI. 1999;91:1025-32
93. Bettencourt MC, Bauer JJ, Sesterhenn IA. et al. Ki-67 expression is a prognostic marker of prostate cancer recurrence after radical prostatectomy. J Urol. 1996;156:1064-8
94. Stephenson RA. Population-based prostate cancer trends in the PSA era: data from the Surveillance, Epidemiology, and End Results Program. Monographs in Urology. 1998:3-19
95. Etzioni R, Legler JM, Feuer EJ. et al. Cancer surveillance series: interpreting trends in prostate cancer— part III: quantifying the link between population prostate specific antigen testing and recent declines in prostate cancer mortality. J NCI. 1999;91:1033-9
96. Coley CM, Barry MJ, Fleming C. et al. Early detection of prostate cancer. Part 1. Prior probability and effectiveness of tests. Ann Intern Med. 1997;126(5):394-406
97. Tricoli JV, Schoenfeldt M, Conley BA. Detection of prostate cancer and predicting progression: current and future diagnostic markers. Clin Cancer Res. 2004;10:3943-53
98. Hartwell L, Mankoff D, Paulovich A. et al. Cancer biomarkers: a systems approach. Nature Biotechnology. 2006;24:905-8
99. Cox TM. Biomarkers in lysosomal storage diseases. Fabry disease: Perspectives from 5 years of FOS. Mehta, A; Beck, M; Sunder-Plassmann, G, editors. Oxford (UK): Oxford PharmaGenesis Ltd. 2006
100. A Framework for Biomarker and Surrogate Endpoint Use in Drug Development. Woodcock J. http://www.fda.gov/ohrms/dockets/ac/04/
101. Deegan PB, Moran MT, McFarlane I. et al. Clinical evaluation of chemokine and enzymatic biomarkers of Gaucher disease. Blood Cells Mol Dis. 2005;35:259-67
102. Pepe MS, Etzioni R, Feng Z. et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054-61
103. Barker PE. Cancer biomarker validation: standards and process: roles for the National Institute of Standards and Technology (NIST). Ann NY Acad Sci. 2003;983:142-50
104. Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nature Reviews Cancer. 2005;5:845-56
105. Nadler RB, Humphrey PA, Smith DS. et al. Effect of inflammation and benign prostatic hyperplasia on elevated serum prostate specific antigen levels. J Urol. 1995;154:407-13
106. Makarov DV, Loeb S, Getzenberg RH. et al. Biomarkers for prostate cancer. Ann Rev Medicine. 2009;60:139-51
107. Taskén KA, Angelsen A, Svindland A. et al. Markers for diagnosis, prediction and prognosis of prostate cancer. Tidsskr Nor Laegeforen. 2005;125:3279-82
108. Gutman AB, Gutman EB. An “acid” phosphatase occurring in the serum of patients with metastasizing carcinoma of the prostate gland. J Clin Invest. 1938;17:473-8
109. Veeramani S, Yuan TC, Chen SJ. et al. Cellular prostatic acid phosphatase: a protein tyrosine phosphatase involved in androgen-independent proliferation of prostate cancer. Endocr Relat Cancer. 2005;12:805-22
110. Gutman AB, Gutman EB, Robinson JN. Determination of serum 'acid' phosphatase activity in differentiating skeletal metastases secondary to prostatic carcinoma from Paget's disease of bone. Am J Cancer. 1940;38:103-8
111. Doe RP, Seal US. Acid phosphatase in urology. Surg Clin North Am. 1965;45:1455-66
112. Whitesel JA, Donohue RE, Mani JH. et al. Acid phosphatase: its influence on the management of carcinoma of the prostate. J Urol. 1984;131:70-7
113. Johnson DE, Prout GR, Scott WW. et al. Clinical significance of serum acid phosphatase levels inadvanced prostatic carcinoma. Urology. 1976;8:123-6
114. Bishop MC, Hardy JG, Taylor MC. et al. Bone imaging and serum phosphatases in prostatic carcinoma. Br J Urol. 1985;57:317-24
115. Lilja H, Haese A, Björk T. et al. Significance and metabolism of complexed and noncomplexed prostate specific antigen forms, and human glandular kallikrein 2 in clinically localized prostate cancer before and after radical prostatectomy. J Urol. 1999;162:2029-34
116. Sokoll LJ, Partin AW, Mikolajczyk SD. et al. ProPSA in relation to free PSA gives the best performance in detecting cancer in the 2.5-4.0 ng/mL total PSA range. J Urol. 2002;167:286
117. Ercole CJ, Lange PH, Mathisen M. et al. Prostatic specific antigen and prostatic acid phosphatase in the monitoring and staging of patients with prostatic cancer. J Urol. 1987;138:1181-4
118. Pound CR, Partin AW, Eisenberger MA. et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591-7
119. Lowe FC, Trauzzi SJ. Prostatic acid phosphatase in 1993. Its limited clinical utility. Urol Clin North Am. 1993;20:589-95
120. Han M, Piantadosi S, Zahurak ML. et al. Serum acid phosphatase level and biochemical recurrence following radical prostatectomy for men with clinically localized prostate cancer. Urology. 2001;57:707-11
121. Oberaigner W, Horninger W. Klocker H. Schönitzer D; Stühlinger, W; Bartsch, G. Reduction of prostate cancer mortality in Tyrol, Austria, after introduction of prostate-specific antigen testing. Am J Epidemiol. 2006;164:376-84
122. Antenor JA, Roehl KA, Eggener SE. et al. Preoperative PSA and progression-free survival after radical prostatectomy for Stage T1c disease. Urology. 2005;66:156-60
123. Nadler RB, Loeb S, Roehl KA. et al. Use of 2.6 ng/ml prostate specific antigen prompt for biopsy in men older than 60 years. J Urol. 2005;174:2154-7
124. Thompson IM, Pauler DK, Goodman PJ. et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or = 4.0 ng per milliliter. N Engl J Med. 2004;350:2239-46
125. Han M, Partin AW, Chan DY. et al. An evaluation of the decreasing incidence of positive surgical margins in a large retropubic prostatectomy series. J Urol. 2004;171:23-36
126. Han M, Partin AW, Pound CR. et al. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy. The 15-year Johns Hopkins experience. Urol Clin North Am. 2001;28:555-65
127. Freedland SJ, Humphreys EB, Mangold LA. et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294:433-9
128. Kuefer R, Varambally S, Zhou M. et al. Alpha-methylacyl-CoA racemase: expression levels of this novel cancer biomarker depend on tumor differentiation. Am J Pathol. 2002;161:841-8
129. Zha S, Ferdinandusse S, Denis S. et al. Alpha-methylacyl-CoA racemase as an androgen-independent growth modifier in prostate cancer. Cancer Res. 2003;63:7365-76
130. Jiang Z, Wu CL, Woda BA. et al. Alpha-methylacyl-CoA racemase: a multi-institutional study of a new prostate cancer marker. Histopathology. 2004;45:218-25
131. Rubin MA, Zhou M, Dhanasekaran SM. et al. Alpha-methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662-70
132. Jiang Z, Wu CL, Woda BA. et al. P504S/alpha-methylacyl-CoA racemase: a useful marker for diagnosis of small foci of prostatic carcinoma on needle biopsy. Am J Surg Pathol. 2002;26:1169-74
133. Luo J, Zha S, Gage WR. et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220-6
134. Rogers CG, Ya G, Zha S. et al. Prostate cancer detection on urinalysis for alpha methylacyl coenzyme A racemase protein. J Urol. 2004;172:1501-3
135. Sreekumar A, Laxman B, Rhodes DR. et al. Humoral immune response to alpha-methylacyl-CoA racemase and prostate cancer. J NCI. 2004;96:834-43
136. Cardillo MR, Gentile V, Ceccariello A. et al. Can p503s, p504s and p510s gene expression in peripheral-blood be useful as a marker of prostatic cancer? BMC Cancer. 2005;5:111
137. Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445-600
138. Lee WH, Morton RA, Epstein JI. et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci USA. 1994;91:11733-7
139. Harden SV, Guo Z, Epstein JI. et al. Quantitative GSTP1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J Urol. 2003;169:1138-42
140. Jerónimo C, Varzim G, Henrique R. et al. I105V polymorphism and promoter methylation of the GSTP1 gene in prostate adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2002;11:445-50
141. Jeronimo C, Usadel H, Henrique R. et al. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J NCI. 2001;93:1747-52
142. Chu DC, Chuang CK, Fu JB. et al. The use of real-time quantitative polymerase chain reaction to detect hypermethylation of the CpG islands in the promoter region flanking the GSTP1 gene to diagnose prostate carcinoma. J Urol. 2002;167:1854-8
143. Brooks JD, Weinstein M, Lin X. et al. CG island methylation changes near the GSTP1 gene in prostatic intraepithelial neoplasia. Cancer Epidemiol Biomark Prev. 1998;7:531-6
144. Cairns P, Esteller M, Herman JG. et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7:2727-30
145. Lin X, Tascilar M, Lee WH. et al. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. Am J Pathol. 2001;159:1815-26
146. Bastian PJ, Ellinger J, Schmidt D. et al. GSTP1 hypermethylation as a molecular marker in the diagnosis of prostatic cancer: is there a correlation with clinical stage, Gleason grade, PSA value or age? Eur J Med Res. 2004;9:523-7
147. Gonzalgo ML, Pavlovich CP, Lee SM. et al. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9:2673-7
148. Gonzalgo ML, Nakayama M, Lee SM. et al. Detection of GSTP1 methylation in prostatic secretions using combinatorial MSP analysis. Urology. 2004;63:414-8
149. Simon JP, Aunis D. Biochemistry of the chromogranin A protein family. Biochem J. 1989;262:1-13
150. Helman LJ, Ahn TG, Levine MA. et al. Molecular cloning and primary structure of human chromogranin A (secretory protein I) cDNA. J Biol Chem. 1988;263:11559-63
151. Berruti A, Mosca A, Tucci M. et al. Independent prognostic role of circulating chromogranin A in prostate cancer patients with hormone-refractory disease. Endocr Relat Cancer. 2005;12:109-17
152. Deftos LJ. Granin-A, parathyroid hormone-related protein, and calcitonin gene products in neuroendocrine prostate cancer. Prostate (Suppl). 1998;8:23-31
153. Berruti A, Dogliotti L, Mosca A. et al. Circulating neuroendocrine markers in patients with prostate carcinoma. Cancer. 2000;88:2590-7
154. Zaky Ahel M, Kovacic K, Kraljic I. et al. Oral estramustine therapy in serum chromogranin A-positive stage D3 prostate cancer patients. Anticancer Res. 2001;21:1475-9
155. Ferrero-Pous M, Hersant AM, Pecking A. et al. Serum chromogranin-A in advanced prostate cancer. BJU Int. 2001;88:790-6
156. Cabrespine A, Guy L, Gachon F. et al. Circulating chromogranin A and hormone refractory prostate cancer chemotherapy. J Urol. 2006;175:1347-52
157. Sasaki T, Komiya A, Suzuki H. et al. Changes in chromogranin A serum levels during endocrine therapy in metastatic prostate cancer patients. Eur Urol. 2005;48:224-30
158. Zitella A, Berruti A, Destefanis P. et al. Gruppo Oncologico Urologico Piemontese (GOUP). Comparison between two commercially available chromogranin A assays in detecting neuroendocrine differentiation in prostate cancer and benign prostate hyperplasia. Clin Chim Acta. 2007;377:103-7
159. Ramírez ML, Nelson EC, Evans CP. Beyond prostate-specific antigen: alternate serum markers. Prostate Cancer Prostatic Diseases. 2008;11:216-29
160. Isshiki S, Akakura K, Komiya A. et al. Chromogranin a concentration as a serum marker to predict prognosis after endocrine therapy for prostate cancer. J Urol. 2002;167:512-5
161. Cheville JC, Tindall D, Boelter C. et al. Metastatic prostate carcinoma to bone: clinical and pathologic features associated with cancer-specific survival. Cancer. 2002;95:1028-36
162. Fracalanza S, Prayer-Galetti T, Pinto F. et al. Plasma chromogranin A in patients with prostate cancer improves the diagnostic efficacy of free/total prostate-specific antigen determination. Urol Int. 2005;75:57-61
163. Deftos LJ, Abrahamsson PA. Granins and prostate cancer. Urology. 1998;51:141-5
164. Riegman PH, Vlietstra RJ, Klaassen P. et al. The prostate-specific antigen gene and the human glandular kallikrein-1 gene are tandemly located on chromosome 19. FEBS Lett. 1989;247:123-6
165. Dawson NA, Vogelzang NJ. Prostate Cancer. New York: Wiley-Liss Inc. 1994
166. Myrtle JF, Klimley PG, Ivor LP. et al. Clinical utility of prostate specific antigen (PSA) in the management of prostate cancer. Advances in cancer diagnostics. San Diego, CA: Hybritech Inc. 1986:1-4
167. Xiao Z, Adam BL, Cazares LH. et al. Quantitation of serum prostate-specific membrane antigen by a novel protein biochip immunoassay discriminates benign from malignant prostate disease. Cancer Res. 2001;61:6029-33
168. Wang TY, Kawaguchi TP. Preliminary evaluation of measurement of serum prostate-specific antigen level in detection of prostate cancer. Ann Clin Lab Sci. 1986;16:461-6
169. Beckett ML, Cazares LH, Vlahou A. et al. Prostate-specific membrane antigen levels in sera from healthy men and patients with benign prostate hyperplasia or prostate cancer. Clin Cancer Res. 1999;5:4034-40
170. Murphy GP, Kenny GM, Ragde H. et al. Measurement of serum prostate-specific membrane antigen, a new prognostic marker for prostate cancer. Urology. 1998;51:89-97
171. Reiter RE, Gu Z, Watabe T. et al. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci USA. 1998;95:1735-40
172. Gu Z, Thomas G, Yamashiro J. et al. Prostate stem cell antigen (PSCA) expression increases with high Gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19:1288-96
173. Han KR, Seligson DB, Liu X. et al. Prostate stem cell antigen expression is associated with gleason score, seminal vesicle invasion and capsular invasion in prostate cancer. J Urol. 2004;171:1117-21
174. Hara N, Kasahara T, Kawasaki T. et al. Reverse transcription-polymerase chain reaction detection of prostate-specific antigen, prostate-specific membrane antigen, and prostate stem cell antigen in one milliliter of peripheral blood: value for the staging of prostate cancer. Clin Cancer Res. 2002;8:1794-9
175. Ross S, Spencer SD, Holcomb I. et al. Prostate stem cell antigen as therapy target: tissue expression and in vivo efficacy of an immunoconjugate. Cancer Res. 2002;62:2546-53
176. Saffran DC, Raitano AB, Hubert RS. et al. Anti-PSCA mAbs inhibit tumor growth and metastasis formation and prolong the survival of mice bearing human prostate cancer xenografts. Proc Natl Acad Sci USA. 2001;98:2658-63
177. Dannull J, Diener PA, Prikler L. et al. Prostate stem cell antigen is a promising candidate for immunotherapy of advanced prostate cancer. Cancer Res. 2000;60:5522-8
178. Getzenberg RH, Pienta KJ, Huang EY. et al. Identification of nuclear matrix proteins in the cancer and normal rat prostate. Cancer Res. 1991;51:6514-20
179. Uetsuki H, Tsunemori H, Taoka R. et al. Expression of a novel biomarker, EPCA, in adenocarcinomas and precancerous lesions in the prostate. J Urol. 2005;174:514-8
180. Dhir R, Vietmeier B, Arlotti J. et al. Early identification of individuals with prostate cancer in negative biopsies. J Urol. 2004;171:1419-23
181. Hansel DE, DeMarzo AM, Platz EA. et al. Early prostate cancer antigen expression in predicting presence of prostate cancer in men with histologically negative biopsies. J Urol. 2007;177:1736-40
182. Paul B, Dhir R, Landsittel D. et al. Detection of prostate cancer with a blood-based assay for early prostate cancer antigen. Cancer Res. 2005;65:4097-100
183. Epstein JI, Pizov G, Steinberg GD. et al. Correlation of prostate cancer nuclear deoxyribonucleic acid, size, shape and Gleason grade with pathological stage at radical prostatectomy. J Urol. 1992;148:87-91
184. Partin AW, Steinberg GD, Pitcock RV. et al. Use of nuclear morphometry, Gleason histologic scoring, clinical stage, and age to predict disease-free survival among patients with prostate cancer. Cancer. 1992;70:161-8
185. Babiarz J, Peters JM, Miles B. et al. Crissman, JD. Comparison of DNA content in primary and lymph node metastases in prostate adenocarcinoma. Anal Quant Cytol Histol. 1993;15:158-64
186. Veltri RW, Partin AW, Epstein JE. et al. Quantitative nuclear morphometry, Markovian texture descriptors, and DNA content captured on a CAS-200 Image analysis system, combined with PCNA and HER-2/neu immunohistochemistry for prediction of prostate cancer progression. J Cell Biochem (Suppl). 1994;19:249-58
187. Carmichael MJ, Veltri RW, Partin AW. et al. Deoxyribonucleic acid ploidy analysis as a predictor of recurrence following radical prostatectomy for stage T2 disease. J Urol. 1995;153:1015-19
188. Badalament RA, Miller MC, Peller PA. et al. RW. An algorithm for predicting nonorgan confined prostate cancer using the results obtained from sextant core biopsies with prostate specific antigen level. J Urol. 1996;156:1375-80
189. Bubendorf L, Tapoa C, Gasser TC. et al. Ki67 labeling index in core needle biopsies independently predicts tumor-specific survival in prostate cancer. Hum Pathol. 1998;29:949-54
190. Su SL, Boynton AL, Holmes EH. et al. Detection of extraprostatic prostate cells utilizing reverse transcription-polymerase chain reaction. Semin Surg Oncol. 2000;18:17-28
191. Scherr DS, Vaughan EDJ, Wei J. et al. BCL-2 and p53 expression in clinically localized prostate cancer predicts response to external beam radiotherapy. J Urol. 1999;162:12-6
192. Feneley MR, Partin AW. Indicators of pathololgic stage of prostate cancer and their use in clinical practice. Urol Clin North Am. 2001;28:443-58
193. Kwiatkowski MK, Recker F, Piironen T. et al. In prostatism patients the ratio of human glandular kallikrein to free PSA improves the discrimination between prostate cancer and benign hyperplasia within the diagnostic “gray zone” of total PSA 4 to 10 ng/ml. Urology. 1998;52:360-5
194. Becker C, Noldus J, Diamandis E. et al. The role of molecular forms of prostate-specific antigen (PSA or hK3) and of human glandular kallikrein 2 (hK2) in the diagnosis and monitoring of prostate cancer and in extra-prostatic disease. Crit Rev Clin Lab Sci. 2001;38:357-99
195. Haese A, Becker C, Noldus J. et al. Human glandular kallikrein 2. A potential serum marker for predicting the organ-confined versus non-organ-confined growth of prostate cancer. J Urol. 2000;163:1491-7
196. Chapoval AI, Ni J, Lau JS. et al. B7-H3: A costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol. 2001;2:269-74
197. Suh WK, Gajewska BU, Okadal H. et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat Immunol. 2003;4:899-906
198. Zang X, Allison JP. The B7 family and cancer therapy: Costimulation and coinhibition. Clin Cancer Res. 2007;13:5271-9
199. Luol L, Chapoval AI, Flies DB. et al. B7-H3 enhances tumor immunity in vivo by costimulating rapid clonal expansion of antigen-specific CD8+ cytolytic T cells. J Immunol. 2004;173:5445-50
200. Sun M, Richards S, Prasad DV. et al. Characterization of mouse and human B7-H3 genes. J Immunol. 2002;168:6294-7
201. Petroff MG, Kharatyan E, Torry DS. et al. The immunomodulatory proteins B7-DC, B7-H2, and B7-H3 are differentially expressed across gestation in the human placenta. Am J Pathol. 2005;167:465-73
202. Hofmeyer KA, Ray A, Zang X. The contrasting role of B7-H3. Proc Natl Acad Sci USA. 2008;105:10277-8
203. Roth TJ, Sheinin Y, Lohse CM. et al. B7-H3 ligand expression by prostate cancer: a novel marker of prognosis and potential target for therapy. Cancer Res. 2007;67:7893-900
204. Zang X, Thompson RH, Al-Ahmadie HA. et al. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc Natl Acad Sci USA. 2007;104:19458-63
205. Chavin G, Sheinin Y, Crispen PL. et al. RJ. Expression of immunosuppresive B7-H3 ligand by hormone-treated prostate cancer tumors and metastases. Clin Cancer Res. 2009;15:2174-80
206. Tsai G, Lane HY, Yang P. et al. Glycine transporter I inhibitor, N-methylglycine (sarcosine), added to antipsychotics for the treatment of schizophrenia. Biol Psychiatry. 2004;55:452-6
207. Sreekumar A, Poisson LM, Rajendiran TM. et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:799-800
208. Couzin J. Biomarkers. Metabolite in urine may point to high-risk prostate cancer. Science. 2009;323:865
209. Razani B, Schlegel A, Lisanti MP. Caveolin proteins in signaling, oncogenic transformation and muscular dystrophy. Cell Sci. 2000;113:2103-9
210. Okamoto T, Schlegel A, Scherer PE. et al. Caveolins, a family of scaffolding proteins for organizing "preassembled signaling complexes" at the plasma membrane. J Biol Chem. 1998;273:5419-22
211. Tirado OM, Maccarthy CM, Fatima N. et al. Caveolin-1 promotes resistance to chemotherapy-induced apoptosis in Ewing's sarcoma cells by modulating PKCalpha phosphorylation. Int J Cancer. 2010;126(2):426-36
212. Liedtke C, Kersting C, Burger H. et al. Caveolin-1 expression in benign and malignant lesions of the breast. World J Surg Oncol. 2007;5:110
213. Shatz M, Liscovitch M. Caveolin-1: a tumor-promoting role in human cancer. Int J Radiat Biol. 2008;84:177-89
214. Yang G, Addai J, Wheeler TM. et al. Correlative evidence that prostate cancer cell-derived caveolin-1 mediates angiogenesis. Hum Pathol. 2007;38:1688-95
215. Barresi V, Cerasoli S, Tuccari G. Correlative evidence that tumor cell-derived caveolin-1 mediates angiogenesis in meningiomas. Neuropathology. 2008;28:472-8
216. Tahir SA, Yang G, Goltsov AA. et al. Tumor cell-secreted caveolin-1 has proangiogenic activities in prostate cancer. Cancer Res. 2008;68:731-9
217. Tahir SA, Yang G, Ebara S. et al. Secreted caveolin-1 stimulates cell survival/clonal growth and contributes to metastasis in androgen-insensitive prostate cancer. Cancer Res. 2001;61:3882-85
218. Yang G, Truong LD, Timme TL. et al. Elevated expression of caveolin is associated with prostate and breast cancer. Clin Cancer Res. 1998;4:1873-80
219. Yang G, Troung LD, Wheeler TM. et al. Caveolin-1 expression in clinically confined human cancer: a novel prognostic marker. Cancer Res. 1999;59:5719-23
220. Thompson TC, Tahir SA, Li L. et al. The role of caveolin-1 in prostate cancer: clinical implications. Prostate Cancer Prostatic Dis. 2010;13:6-11
221. Di Vizio D, Morello M, Sotgia F. et al. An absence of stromal caveolin-1 is associated with advanced prostate cancer, metastatic disease and epithelial Akt activation. Cell Cycle. 2009;8:2420-24
222. Tanase CP. Caveolin-1: a marker for pancreatic cancer diagnosis. Expert Rev Mol Diagn. 2008;8:395-404
223. Lin KI, Chattopadhyay N, Bai M. et al. Elevated extracellular calcium can prevent apoptosis via the calcium-sensing receptor. Biochem Biophys Res Commun. 1998;249:325-31
224. Vanaja DK, Grossmann ME, Cheville JC. et al. PDLIM4, an actin binding protein, suppresses prostate cancer cell growth. Cancer Invest. 2009;27:264-72
225. Lallet-Daher H, Roudbaraki M, Bavencoffe A. et al. Intermediate-conductance Ca2+=activated K+ channels (IKcal) regulate human prostate cancer cell proliferation through a close control of calcium entry. Oncogene. 2009;28:1792-806
226. Liao J, Schneider A, Datta NS. et al. Extracellular calcium as a candidate mediator of prostate cancer skeletal metastasis. Cancer Res. 2006;77:9065-73
227. Schwartz GG. Is serum calcium a biomarker of fatal prostate cancer? Future Oncol. 2009;5:577-80
228. Vanaja DK, Ballman KV, Morlan BW. et al. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin Cancer Res. 2006;12:1128-36
229. Vanaja DK, Grossmann ME, Cheville JC. et al. PDLIM4, an actin binding protein, suppresses prostate cancer cell growth. Cancer Invest. 2009;27:264-72
230. Bussemakers MJ, van Bokhoven A, Verhaegh GW. et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975-9
231. de Kok JB, Verhaegh GW, Roelofs RW. et al. DD3 (PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695-98
232. Kirby RS, Fitzpatrick JM, Irani J. Prostate cancer diagnosis in the new millennium: strengths and weaknesses of prostate-specific antigen and the discovery and clinical evaluation of prostate cancer gene 3 (PCA3). BJU Int. 2009Feb;103:441-5
233. Haese A, de la Taille A, van Poppel H. et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur Urol. 2008;54:1081-88
234. Marks LS, Fradet Y, Deras IL. et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology. 2007;69:532-5
235. Chun FK, de la Taille A, van Poppel H, et al.Prostate cancer gene 3 (PCA3). development and internal validation of a novel biopsy nomogram. Eur Urol. 2009;56:659-68
236. Nakanishi H, Groskopf J, Fritsche HA. et al. PCA3 molecular urine assay correlates with prostate cancer tumor volume: implication in selecting candidates for active surveillance. J Urol. 2008;179:1804-9
237. Whitman EJ, Groskopf J, Ali A. et al. PCA3 score before radical prostatectomy predicts extracapsular extension and tumor volume. J Urol. 2008;180:1975-8
238. Wang R, Chinnaiyan AM, Dunn RL. et al. Rational approach to implementation of prostate cancer antigen 3 into clinical care. Cancer. 2009;115:3879-86
239. Phé V, Rouprêt M, Salomon L. et al. What's new in 2008 in the field of basic and clinical research in prostate cancer? Prog Urol. 2009;19:S29-42
240. Fradet Y, Saad F, Aprikian A. et al. uPM3, a new molecular urine test for the detection of prostate cancer. Urology. 2004;64:311-5
241. Hessels D, Klein Gunnewiek JM, van Oort I. et al. DD3 (PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8-15
242. Groskopf J, Aubin SM, Deras IL. et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089-95
243. Deras IL, Aubin SM, Blasé A. et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. J Urol. 2008;179:1587-92
244. Ankerst DP, Groskopf J, Day JR. et al. Predicting prostate cancer risk through incorporation of prostate cancer gene 3. J Urol. 2008;180:1303-8
245. Marks LS, Bostwick DG. Prostate cancer specificity of PCA3 gene testing: examples from clinical practice. Rev Urol. 2008;10:175-81
246. Shappell SB, Fulmer J, Arguello D. et al. PCA3 urine mRNA testing for prostate carcinoma: patterns of use by community urologists and assay performance in reference laboratory setting. Urology. 2009;73:363-8
247. Ouyang B, Bracken B, Burke B. et al. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol. 2009;181:2508-13
248. Clarke RA, Zhao Z, Guo AY. et al. New genomic structure for prostate cancer specific gene PCA3 within BMCC1: implications for prostate cancer detection and progression. PLoS One. 2009;4:e4995
249. Wilson S, Greer B, Hooper J. et al. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem J. 2005;388:967-72
250. Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497-511
251. Tomlins SA, Rhodes DR, Perner S. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644-8
252. Mwamukonda K, Chen Y, Ravindranath L. et al. Quantitative expression of TMPRSS2 transcript in prostate tumor cells reflects TMPRSS2-ERG fusion status. Prostate Cancer Prostatic Dis. 2010;13:47-5
253. Cai C, Wang H, Xu Y. et al. Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 2009;69:6027-32
254. Sardana G, Dowell B, Diamandis EP. Emerging biomarkers for the diagnosis and prognosis of prostate cancer. Clin Chem. 2008;54:1951-60
255. Gerdes J, Schwab U, Lemke H. et al. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13-20
256. Mosquera JM, Mehra R, Regan MM. et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706-11
257. Yoshimoto M, Joshua AM, Cunha IW. et al. Absence of TMPRSS2: ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451-60
258. Fradet Y. Biomarkers in prostate cancer diagnosis and prognosis: beyond prostate-specific antigen. Curr Opin Urol. 2009;19:243-6
259. Keller S, Sanderson MP, Stoeck A. et al. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102-8
260. Valadi H, Ekström K, Bossios A. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-9
261. Nilsson J, Skog J, Nordstrand A. et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603-7
262. Mitchell PJ, Welton J, Staffurth J, Court J. et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med. 2009Jan12;7:4
263. Jansen FH, Krijgsveld J, van Rijswijk A. et al. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol Cell Proteomics. 2009;8:1192-205
264. Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311-22
265. Gerdes J, Lemke H, Baisch H. et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710-5
266. Khatami A, Hugosson J, Wang W. et al. Ki-67 in screen-detected, low-grade, low-stage prostate cancer, relation to prostate-specific antigen doubling time, Gleason score and prostate-specific antigen relapse after radical prostatectomy. Scand J Urol Nephrol. 2009;43:12-8
267. Berney DM, Gopalan A, Kudahetti S. et al. Ki-67 and outcome in clinically localised prostate cancer: analysis of conservatively treated prostate cancer patients from the Trans-Atlantic Prostate Group study. Br J Cancer. 2009;100:888-93
268. Khor LY, Bae K, Paulus R. et al. MDM2 and Ki-67 predict for distant metastasis and mortality in men treated with radiotherapy and androgen deprivation for prostate cancer: RTOG 92-02. J Clin Oncol. 2009;27:3177-84
269. Jhavar S, Bartlett J, Kovacs G. et al. Biopsy tissue microarray study of Ki-67 expression in untreated, localized prostate cancer managed by active surveillance. Prostate Cancer Prostatic Dis. 2009;12:143-7
270. Wright LM, Yong S, Picken MM. et al. Decreased survival and hepato-renal pathology in mice with C-terminally truncated GP73 (GOLPH2). Int J Clin Exp Pathol. 2009;2:34-47
271. Wei S, Dunn TA, Isaacs WB. et al. GOLPH2 and MYO6: putative prostate cancer markers localized to the Golgi apparatus. Prostate. 2008;68:1387-95
272. Laxman B, Morris DS, Yu J. et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645-9
273. Kristiansen G, Fritzsche FR, Wassermann K. et al. GOLPH2 protein expression as a novel tissue biomarker for prostate cancer: implications for tissue-based diagnostics. Br J Cancer. 2008;99:939-48
274. Chen H, Pong RC, Wang Z. et al. Differential regulation of the human gene DAB2IP in normal and malignant prostatic epithelia: cloning and characterization. Genomics. 2002;79:573-81
275. Chen H, Toyooka S, Gazdar AF. et al. Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J Biol Chem. 2003;278:3121-30
276. Wang Z, Tseng CP, Pong RC. et al. The mechanism of growth-inhibitory effect of DOC-2/DAB2 in prostate cancer. Characterization of a novel GTPase-activating protein associated with N-terminal domain of DOC-2/DAB2. J Biol Chem. 2002;277:12622-31
277. Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem. 2005;280:22437-44
278. Chen H, Toyooka S, Gazdar AF. et al. Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J Biol Chem. 2003;278:3121-30
279. Duggan D, Zheng SL, Knowlton M. et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836-44
Author contact
![]() Corresponding author: Yi Lu, Ph.D., Department of Pathology, University of Tennessee Health Science Center, 19 South Manassas Street, Room 218, Memphis, TN 38163, USA, Tel: (901) 448-5436; Fax: (901) 448-5496; E-mail: yluedu
Corresponding author: Yi Lu, Ph.D., Department of Pathology, University of Tennessee Health Science Center, 19 South Manassas Street, Room 218, Memphis, TN 38163, USA, Tel: (901) 448-5436; Fax: (901) 448-5496; E-mail: yluedu

 Global reach, higher impact
Global reach, higher impact