3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(16):4917-4932. doi:10.7150/jca.40673 This issue Cite
Research Paper
EXO1 Plays a Carcinogenic Role in Hepatocellular Carcinoma and is related to the regulation of FOXP3
1. Hepatic Surgery Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
2. Department of Hepatobiliary and Laparoscopic Surgery, Renmin Hospital, Wuhan University, Hubei Key Laboratory of Digestive System Disease, Wuhan, China.
Received 2019-9-26; Accepted 2020-5-29; Published 2020-6-16
Abstract
Exonuclease 1 (EXO1), a member of the RAD2 nuclease family, was first described as possessing 5' to 3' nuclease activity and 5' structure-specific endonuclease activity. Here, we show that EXO1 is significantly upregulated in HCC tumor tissues and that high EXO1 expression is significantly correlated with liver cirrhosis. We further demonstrate that EXO1 knockdown decreases proliferation and colony forming abilities of HCC cells in vitro and tumorigenicity in vivo, as well as decreases migration and invasive capabilities of HCC cells. Alternatively, EXO1 overexpression significantly increases the proliferation, colony forming ability, and migration and invasive capabilities of HCC cells in vitro. Additionally, we truncated a region upstream of the transcription start site (TSS) of EXO1 and used the region with the strongest transcriptional activity to predict that the transcription factor FOXP3 can bind to the EXO1 promoter. Bioinformatics analysis found that FOXP3 was positively correlated with EXO1 and luciferase reporter assays and RT-PCR confirmed that FOXP3 could enhance the transcriptional activity of EXO1. CCK-8 assays showed that depletion of FOXP3 further reduces cell proliferation ability after knocking down of EXO1 in vitro. Taken together, our findings indicate that EXO1 acts as an oncogene in HCC and its expression level is related to FOXP3 activity.
Keywords: EXO1, FOXP3, Hepatocellular carcinoma, Metastasis, Transcriptional regulation
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer-related deaths worldwide [1]. Most patients are initially diagnosed at an advanced stage in the disease and thus, the curative effect of hepatectomy is often unsatisfactory. The 5-year recurrence rate of HCC after hepatectomy exceeds 80%, and the 5-year survival rate is only 30%-70% [2, 3]. Therefore, in-depth study of the molecular mechanisms involved in the development of HCC is of great significance to develop effective therapeutic drugs and prevent the recurrence of HCC after hepatectomy.
Exonuclease 1 (EXO1) is a member of the Rad2/XPG family of nucleases and was originally identified in Schizosaccharomyces pombe [4]. EXO1 possess 5' to 3' exonuclease activity and structure-specific endonuclease activity and plays an extremely important role in biological processes such as DNA replication, DNA mismatch repair (MMR), DNA double-strand break repair (DSB) and telomere maintenance [5-9]. Deletion of the EXO1 gene causes genomic instability and leads to impaired DNA damage repair and meiosis defects [7, 10-13]. However, studies have shown that abnormally high expression of EXO1 is related to the occurrence, development and prognosis of various malignant tumors. Kretschmer et al. reported for the first time that EXO1 is significantly elevated in breast ductal carcinoma and invasive ductal carcinoma [14]. A recent study by de Sousa et al. has shown that EXO1 expression is significantly elevated in glioma, and high EXO1 expression is an independent risk factor for poor prognosis of patients [15]. Dai et al. reported that EXO1 is upregulated in HCC specimens and EXO1 overexpression may also be associated with poor prognosis in HCC patients [16]. In addition, several single nucleotide polymorphisms (SNPs) in the EXO1 gene are associated with tumor susceptibility in a variety of tumors including liver, gastric, ovarian, cervical, breast, and colorectal cancers as well as head and neck squamous cell carcinoma [17-25]. Among them, rs1047840 (K589E), a common SNP genotype, increases the risk for developing non-viral liver cancer, colorectal cancer and lung cancer [17, 26, 27]. However, the role of EXO1 in the invasion and metastasis of HCC and the upstream transcriptional regulation of EXO1 is still poorly understood.
Forkhead box P3 (FOXP3) is a member of the forkhead/winged-helix family of transcription factors, which was first identified in T-regulatory (Treg) cells as an essential protein for regulating immune system development and function [28]. FOXP3 is primarily known for its function in regulating the CD4+CD25+ regulatory cells and for defining their immunoregulatory phenotype [29]. Like other transcription factors, FOXP3 can bind to numerous enzymes and microRNAs to up- or downregulate a large number of genes [30]. Dysregulation of FOXP3 has been reported in the context of various tumors types indicating that it could be a poor prognostic factor in colorectal cancer and bladder cancer [31, 32], but a potential tumor suppressor gene in breast cancer [33, 34]. Nevertheless, whether FOXP3 can regulate the transcription of EXO1 is poorly understood.
In this study, we discovered that EXO1 expression is significantly higher in HCC tumor tissues. It was also found that EXO1 could promote the proliferation of HCC cells in vitro and in vivo, as well as promote the migration and invasion of HCC cells in vitro. FOXP3 could lead to the upregulation of EXO1 at the transcriptional level, where it could act as an oncogene in HCC as well. Our research revealed the relationship between the abnormal expression of EXO1 and malignant biological characteristics of HCC and might lead to the development of novel anti-cancer therapeutics for HCC treatment.
Materials and Methods
Gene chip analysis and human HCC samples
99 HCC tumor tissues and matched non-tumor tissues from archived patients at the Hepatic Surgery Centre, Tongji Hospital of Huazhong University of Science and Technology (Wuhan, China) were collected between January 2017 and December 2018. The diagnosis of HCC was confirmed by pathological examination and the differentiation status was graded according to the Edmondson grading system. After surgical resection, samples were stored at -80˚C immediately. Written informed consent was obtained from all subjects, and this study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Tongji Hospital, Huazhong University of Science and Technology.
HCC tissues and adjacent non-tumor liver tissues were placed in a cryogenic tank with dry ice and were delivered to the Shanghai Biotechnology Corporation. The Agilent version 16.0 was applied to screen the gene expression profiles of DNA repair pathways between HCC tissues and their adjacent liver tissues.
Immunohistochemical (IHC) staining assays
Formalin-fixed paraffin-embedded sections were deparaffinized in xylene and rehydrated with an ethanol gradient. The sections were placed in citric acid antigen repair buffer (Wuhan Goodbio Technology Corporation, Wuhan, China) and antigen repair was performed by microwaving on a low setting for 10 min. Endogenous peroxidases were blocked by 3% H2O2. Sections were then incubated overnight with an anti-EXO1 antibody at 1:200 (abs115859, Absin, Beijing, China) at 4℃. Secondary antibodies were incubated and peroxidase activity was detected using the EnVision kit (Dako, Glostrup, Denmark). Hematoxylin (Sigma Aldrich, St. Louis, MO, USA) was used for nuclear counterstaining. IHC scores were calculated as the positive staining intensity scores multiplied by the stained positive cells scores. Overall scores of >6 and ≤6 were defined as high expression or low expression of EXO1, respectively.
Cell culture
The HCC cell lines Hep3B, Huh7, SK-Hep1, Bel-7402, and SMMC7721 were purchased from the China Center for Type Culture Collection (Wuhan, China), and HCC cell lines MHCC-97H and HCC-LM3 were obtained from the Liver Cancer Institute, Zhongshan Hospital, Fudan University (Shanghai, China). The HCC cell line HLE was obtained from the Department of Molecular Biology, Peking University (Beijing, China). The HCC cell line PLC, human fetal liver cell line HL-7702, hepatoma cell line HepG2 and lentivirus packaging cell line 293T were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Cell lines were cultured in Dulbecco's Modified Eagle's medium (DMEM, Hyclone, Utah, USA) containing 10% fetal bovine serum (Gibco, Carlsbad, CA). All cell lines were incubated in a humidified atmosphere with 5% carbon dioxide at 37 °C.
Generation of EXO1 overexpression and knockdown constructs and FOXP3 knockdown construct
Full-length EXO1 coding sequence (CDS) was purchased from Vigene Biosciences (Shandong, China). EXO1 was subcloned into pLenti-CMV-GFP-Puro (plasmid# 17448; Addgene, Cambridge, MA, USA), according to the manufacturer's instructions.
For EXO1 knockdown assays, three shRNA (shEXO1-TRCN0000039788, shEXO1-TRCN0000039789, and shEXO1-TRCN0000010331) sequences were obtained from the Sigma website (www.sigmaaldrich.com). All these shRNAs were subcloned into the lentiviral vector pLKO.1 puro (plasmid # 8453; Addgene, Cambridge, MA, USA) according to the manufacturer's instructions. Either the shEXO1 constructs, EXO1 overexpression constructs or vector control plasmids were transfected into the 293T cell line by Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, MA, USA) using the lentivirus packaging system. The plasmids pMD2.G and psPA×2 were gifts from Didier Trono (plasmids# 12259 and# 12260; Addgene).
The supernatant-containing lentivirus produced by transfected 293T cells was collected 72 hours later. Collected lentiviral supernatants were filtered through a 0.45 μm filter (Millipore, USA). Target cells were infected with filtered lentivirus and 8 μg/mL polybrene (Sigma-Aldrich) to generate stable cell lines; treatment with 8 μg/mL puromycin (Ann Arbor, MI, USA) for 7 days was used to screen the positive cells.
For siFOXP3 transfection assays, three siRNA (siFOXP3-1 stB0001154A, siFOXP3-2 stB0001154B and siFOXP3-3 stB0001154C) sequences were purchased from RiboBio (Guangzhou, China). About 3×105 HLF or Huh7 cells/well were seeded into a 6-well plate and infected with 5 μL of siRNAs after adherence. Lipofectamine 3000 (Invitrogen) was used for transfection.
Western blot analysis
Total protein was extracted from the cells using RIPA lysis buffer. Protein concentrations were measured with a BCA Protein Assay Kit (Thermo Fisher Scientific, MA, USA). Protein lysates were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (EMD Millipore, Billerica, MA, USA). The membrane was blocked in non-fat milk in Tris-buffered saline containing 0.1% Tween-20 for 2 h and incubated with primary antibody (EXO1, ab95012, Abcam, Cambridge, MA, USA; β-actin, Cell Signaling Technology, Danvers, MA, USA) at 4 °C overnight. Then, membranes were incubated with secondary antibodies and signals were detected by Bio-rad chemiluminescence system.
Cell proliferation assay
Cells were inoculated into 96-well plates with 1×103 cells/well, and 100 μL of 10% cell counting kit-8 (CCK-8, Dojindo, Kumamoto, Japan) solution was added into each well at 0, 1, 2, 3, 4, and 5 days after cell adherence. The optical density (OD) values were measured by an EXL-800 absorbance reader at a wavelength of 450 nm at the indicated time points (after Cell Counting Kit-8 solution was added to the wells for 2 hours).
Cell migration and matrigel invasion assays
Cell migration and invasion assays were performed using transwell chambers (Corning, USA) with or without Matrigel (BD Biosciences, CA, USA). About 1.5×104-10×104 cells in medium without FBS were seeded on transwell chambers with or without Matrigel. Below the transwell chambers, DMEM containing 10% fetal bovine serum (FBS) was added. An automated cell counter (Nexcelom Bioscience, USA) was used to quantify the number of migratory or invasive cells. Six fields at a magnification of ×200 were randomly selected for counting stained cells.
Cell colony formation
EXO1-knockdown or -overexpressing HCC cells (HLF, Huh7, Bel7402) were plated in 6-well plates (1000 cells/well) and culture media was replaced every 3 days. After 2-3 weeks when clones were visible to the naked eye, cells were fixed with 4% polyformaldehyde for 15 minutes and stained with crystal violet (Sigma-Aldrich Corporation) for 15 minutes. Photographs of the 6-well plates were taken, and the differences were counted.
Xenograft tumorigenicity assays
Animal studies were approved by the Ethics Committee of Tongji Hospital, Huazhong University of Science and Technology. All animal experiments were performed under the guidelines of the Interdisciplinary Principles and Guidelines for the Use of Animals in Research, Testing, and Education by the New York Academy of Sciences, Ad Hoc Animal Research Committee. 5-week-old male BALB/c (nu/nu) mice were obtained from the HFK BioScience Corporation (Beijing, China). All the mice were bred under specific pathogen-free (SPF) conditions at the Laboratory Animal Center of the Tongji Hospital. For subcutaneous tumorigenesis assays, HLF vector- and HLF shEXO1-788-transfected cells (2×106) were suspended in 100 μL of Dulbecco's modified Eagle's medium (DMEM) and injected subcutaneously into the left and right back of nude mice, respectively. Nude mice were monitored after the injection and sacrificed at day 30 after cell inoculation. The final tumor volumes were calculated according to the equation: V (volume, mm3) = 0.5 × L (length, mm) × W2 (width, mm2).
RNA extraction and quantitative real-time PCR
Total RNA was extracted using TRIzol Reagent (Takara, Dalian, China) and reverse transcription was performed using a reverse-transcription PCR kit (TIANGEN, Beijing, China) according to the manufacturer's instructions. Quantitative real-time PCR was carried out using SYBR Green PCR master mix (TIANGEN, Beijing, China) on a Bio-Rad CFX system (Bio-Rad, Hercules, CA, USA) with the following primers:
EXO1-forward: 5' - TGAGGAAGTATAAAGGGCAGGT -3';
EXO1-reverse: 5'- AGTTTTTCAGCACAAGCAATAGC - 3'.
FOXP3-forward: 5'- GTGGCCCGGATGTGAGAAG - 3';
FOXP3- reverse: 5'- GGAGCCCTTGTCGGATGATG - 3'.
GAPDH- forward: 5'- GGAGCGAGATCCCTCCAAAAT- 3'.
GAPDH- reverse: 5'- GGCTGTTGTCATACTTCTCATGG- 3'.
The Ct values of EXO1 were equilibrated to those of the internal control GAPDH.
Luciferase reporter assay
The region 2000 bp upstream of the EXO1 translational start site was amplified from the genomic DNA of the Huh7 cell line by PCR. EXO1 promoter regions of different lengths were subcloned into pGL4.17 (Promega, Madison, WI, USA). The open reading frame of FOXP3 and other candidate genes was amplified by PCR and then cloned into pcDNA 3.1(+) (Invitrogen, Thermo Fisher Scientific, MA, USA) to produce a series of products such as pcDNA 3.1-FOXP3 and so on. pGL4.17 and pcDNA3.1 were used as controls. pRL-TK was purchased from Promega (Madison, WI, USA). About 1 × 105 Huh7 cells/well were seeded into a 24-well plate and co-transfected with 4 ng of the pRL-TK, 200 ng of pgl4.17 and 400 ng of pcDNA3.1 after adherence. Lipofectamine 2000 (Invitrogen) was used for transfection. Cell lysates were prepared using passive lysis buffer (Promega) 48 h after transfection. A luciferase activity was performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions. The relative luciferase activity was measured using a GloMax 20/20 Luminometer (Promega).
Data acquisition and processing
Data for bioinformatics analysis were collected from The Cancer Genome Atlas (TCGA), Oncomine and Gene Expression Omnibus (GEO) databases. The cor.test () function in the R (version 3.5.2; https:// www.r-project.org/)) was used to calculate and verify the correlation coefficients between the EXO1 and FOXP3. Then R packages ggplot2 (version 3.1.1) was used for plotting.
Statistical analysis
Statistical analyses were performed with IBM SPSS Statistics 21.0 (SPSS, Chicago, IL, USA) or Prism 6.0 (GraphPad Software, La Jolla, CA) software. The results were presented as the mean ± standard deviation (SD) or mean ± standard error of mean (SEM). Quantitative data were compared by a two-tailed Student's t-test, One-way ANOVA and Mann-Whitney U test when applicable. Categorical data were analyzed by the χ2 test or Fisher's exact test. Kaplan-Meier survival analysis (log-rank test) was used to compare HCC patient survival. A two-sided value of p<0.05 was considered statistically significant.
Results
EXO1 is an upregulated DNA damage repair gene in HCC
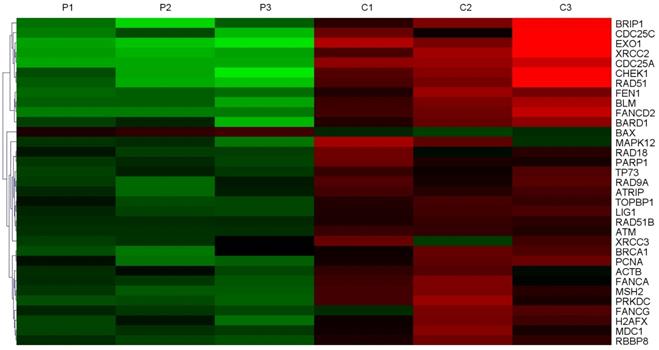
In order to screen the expression of DNA damage repair genes, three samples of HCC tissues and adjacent non-tumor tissues were collected for whole-genome sequencing. Gene chip analysis showed that 33 DNA damage repair genes were expressed at higher levels in tumor tissues compared to their corresponding non-tumor tissues (Figure 1), and in tumor tissues, 8 of them were expressed at levels greater than 10 times the normal levels. The expression of EXO1 in HCC tumor tissues was 27.2 times higher than that in peri-cancerous liver tissues (Table 1). Hence, we chose to perform follow-up studies on EXO1 and focused on its functional roles and mechanisms EXO1 in driving HCC development and metastasis.
Clinical significance of EXO1 in HCC patients
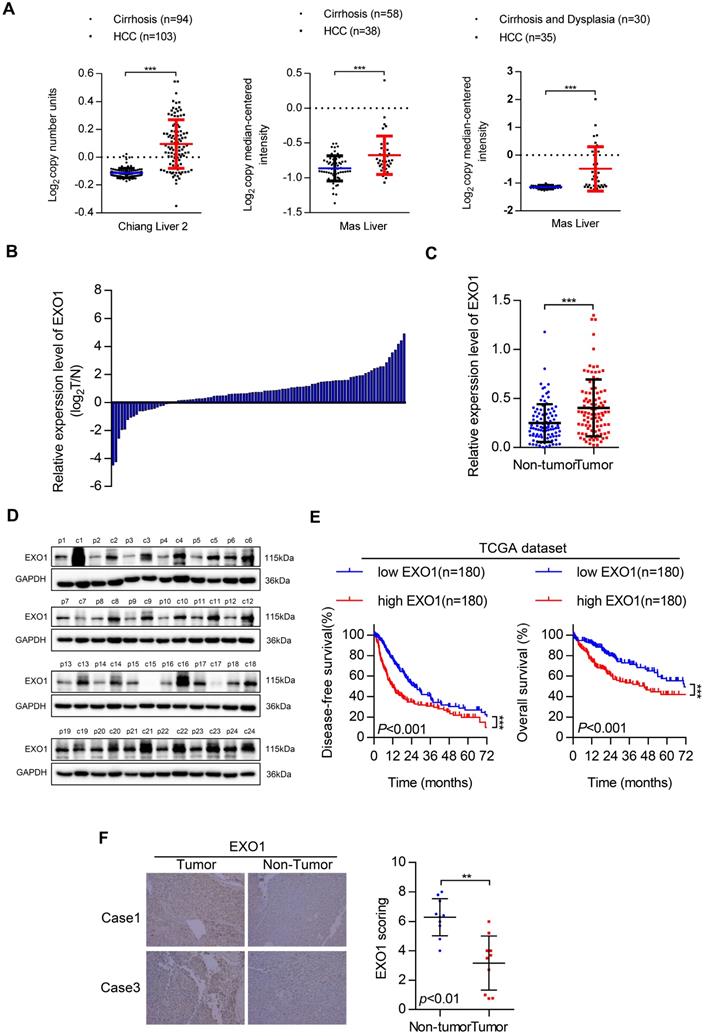
First, we investigated EXO1 expression in tumor and non-tumor liver tissues in three different clinical studies from the Oncomine database (Figure 2A). The results showed that the expression of EXO1 was significantly higher in tumor tissues than that in their adjacent non-tumor tissues. Next, the expression level of EXO1 was detected in 99 HCC samples from Tongji Hospital (Wuhan, China) by western blotting (Figure 2C, D). These results showed that the expression of EXO1 in HCC tumor tissues was higher (79%) than that in adjacent non-tumor tissues (21%) (Figure 2B). We further explored the relationship between clinicopathological features and the expression level of EXO1 in HCC patients. Ninety-nine HCC patients were divided into an EXO1 low-expression group or EXO1 high-expression group. The detailed information is summarized in Table 2. High expression of EXO1 was significantly associated with the presence of liver cirrhosis (p=0.005). However, the expression of EXO1 was not correlated with gender, age, or AFP level. We then used the TCGA database to analyze the relationship between EXO1 expression and long-term prognosis of HCC. Kaplan-Meier analysis revealed that the disease-free survival rate and overall survival rate of patients with high EXO1 expression were significantly lower than that of patients with low EXO1 expression (Figure 2E). Subsequently, the expression level of EXO1 was assessed by immunohistochemistry (IHC) in 10 pairs of tumor tissues and their adjacent non-tumor tissues. As shown in Figure 2F, EXO1 in hepatocytes was localized to the cytoplasm, and the average IHC score of EXO1 in tumor tissues was significantly higher than that in adjacent non-tumor tissues. Taken together, these results indicate that EXO1 expression is significantly upregulated in HCC tissues and high expression of EXO1 plays an important role in the prognosis of HCC patients and may contribute to the progression of HCC.
EXO1 is an up-regulated DNA damage repair gene in HCC. Heat Map of top-ranked upregulated DNA damage repair genes in HCC tissues compared with adjacent normal liver tissues detected by gene chip technology. The results showed that the expression level of 33 molecules in HCC tissues was higher than that in corresponding non-tumor tissues. (P: Pericancerous liver tissue; C: hepatocellular carcinoma tissue).
EXO1 enhances proliferation and colony formation of HCC cells in vitro, as well as tumor growth in vivo
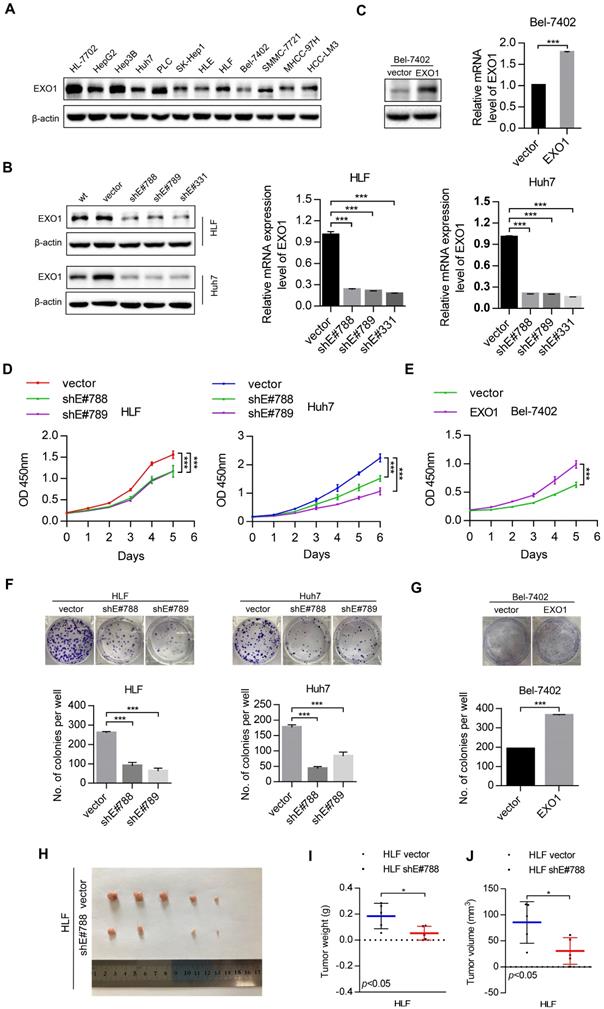
To study the tumorigenic ability of EXO1 in vitro and in vivo, western blot was used to detect the expression level of EXO1 in 10 different HCC cell lines and one immortal liver cell line, HL-7702 (Figure 3A). The results showed that EXO1 was upregulated in HLF and Huh7 cell lines, but downregulated in the Bel-7402 cell line. Hence, three short hairpin RNAs (shEXO1-788, shEXO1-789, and shEXO1-331) specifically targeting EXO1 were stably transfected into HLF and Huh7 cell lines, and an empty vector was transfected into each cell line (abbreviate as vector) and used as negative controls. The EXO1-shRNA significantly reduced EXO1 protein expression in HLF and Huh7 cells compared with their control vectors in the western blotting results and this result was also verified by RT-PCR (Figure 3B). EXO1 was also stably transduced into Bel-7402 cell lines and an empty vector was used as a control. Western blot and RT-PCR verified that EXO1 was significantly overexpressed in transfected Bel-7402 cells (Figure 3C). Using functional assays, the tumorigenicity of EXO1 was investigated. The Cell counting kit-8 (CCK-8) assay showed that EXO1 knockdown in HLF and Huh7 cells significantly inhibited cell proliferation, while EXO1 overexpression in Bel-7402 cells markedly promoted cell proliferation compared with their control vectors (p<0.01, Figure 3D, E). In the colony formation assay, the colony-forming ability of EXO1-knockdown cells was markedly lower than that of their control vectors (p<0.01, Figure 3F), whereas EXO1-overexpression in the cells showed significantly higher colony-forming ability than in the control vectors (p<0.01, Figure 3G). For in vivo tumorigenicity assays, HLF vector (control) and HLF-shEXO1-788-transfected HCC cells were injected subcutaneously into nude mice (Figure 3H). As expected, the final tumor weights and volumes of the HLF-shEXO1-788 cell-injected group were significantly reduced compared to that of the HLF vector cell-injected group (p=0.03, Figure 3I, J). These results indicate that EXO1 has a strong promotive effect on cell proliferation and colony formation in HCC cells, and even promotes HCC tumorigenicity.
Expression of DNA damage repair molecules increased by more than 10-fold
| GenBank accession | Gene name | Description | Fold change |
|---|---|---|---|
| NM_130398 | EXO1 | Exonuclease 1 | 27.2* |
| NM_005431 | XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | 17.5 |
| NM_001789 | CDC25A | Cell division cycle 25 homolog A | 15.8 |
| NM_002875 | RAD51 | RAD51 homolog | 13.8 |
| NM_001274 | CHEK1 | CHK1 checkpoint homolog | 13.6 |
| NM_032043 | BRIP1 | BRCA1 interacting protein C-terminal helicase 1 | 11.4 |
| NM_012238 | SIRT1 | Sirtuin 1 | 11.4 |
| NM_001790 | CDC25C | Cell division cycle 25 homolog C | 10.8 |
EXO1 promotes tumor invasion and metastasis in vitro
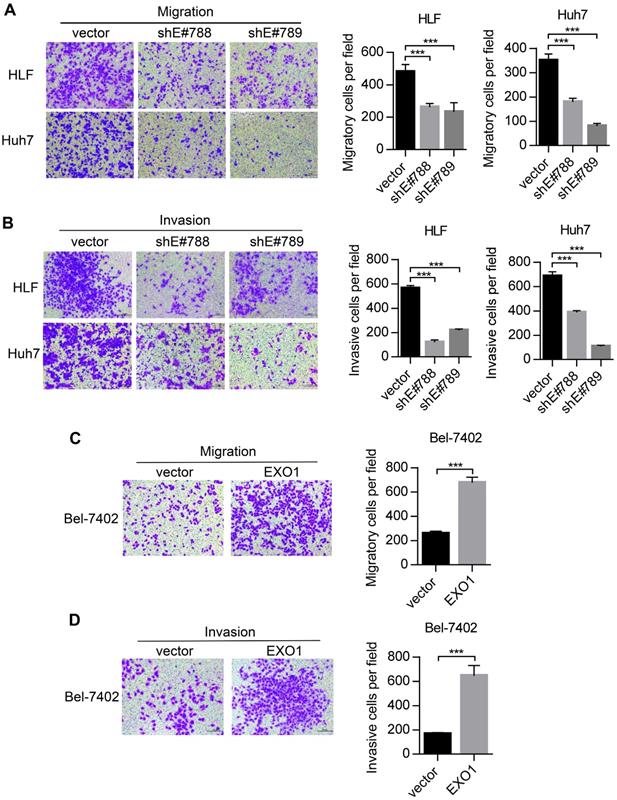
Metastasis is one of the reasons for poor outcomes in HCC. To further explore whether EXO1 can affect the invasion and metastatic abilities of HCC cells, we performed transwell assays. As shown in Figure 4A, migratory abilities were markedly decreased when EXO1 expression was downregulated by shRNA in HLF and Huh7 cells compared to their respective controls. Additionally, as shown in Figure 4B, invasive abilities of HLF-shEXO1 and Huh7-shEXO1 were observably reduced. In contrast, the migratory and invasive abilities of Bel7402-EXO1 cells overexpressing EXO1 were much higher than their corresponding controls (Figure 4C, D). Altogether, these results demonstrate that EXO1 plays an important role in the invasion and metastasis of HCC.
Prediction of upstream regulators of EXO1 via bioinformatics analysis
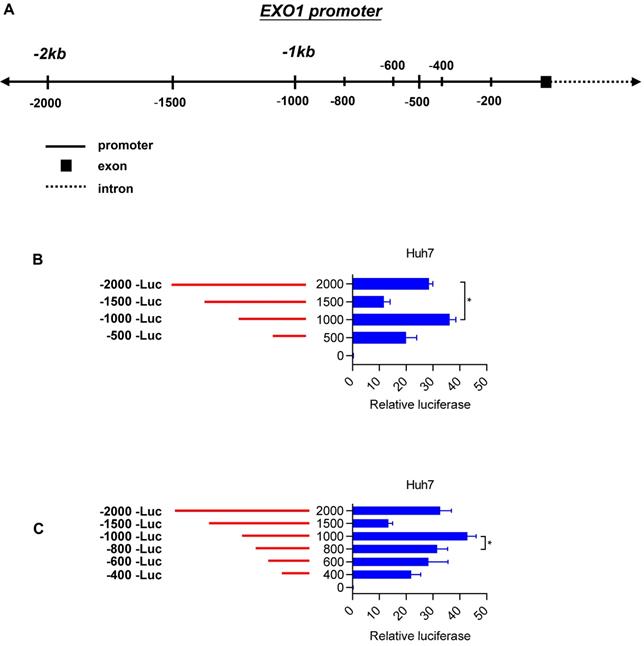
To elucidate the molecular mechanisms of EXO1 overexpression in HCC tumor tissues, we concentrated on the region 2 kb upstream of the transcription start site (TSS) of EXO1 (Figure 5A). Introduction of this 2-kb region upstream of firefly luciferase in pGL4.17-basic yielded significantly greater reporter activity compared with the promoter-less construct in Huh7 cell lines (28-fold) (Figure 5B). With serial truncations of the 2 kb sequence at 500-bp intervals, we generated a set of luciferase reporter constructs (500 bp, 1000 bp, 1500 bp, 2000 bp). The luciferase reporter assay also showed a remarkable rise in luciferase activity when the upstream sequence of TSS of EXO1 was 1000 bp long, and its luciferase activity was the strongest of the four sequences (p<0.05, Figure 5B). To further determine whether there is a region with higher transcriptional activity involved in the 1000-bp sequence, we continued to generate luciferase reporter constructs with truncations of the 1000-bp sequence at 200-bp intervals, which were 200 bp, 400 bp, 600 bp, and 800 bp upstream of the TSS of EXO1. As shown in Figure 5C, the relative reporter activity of the 1000-bp sequence was still the highest compared to other sequences. These results suggest that the 1000-bp region directly upstream of the TSS of EXO1 possesses the highest transcriptional activity, and transcription factors possibly bind to the 1000-bp segment of EXO1 promoter.
FOXP3 could influence the expression of EXO1 in HCC
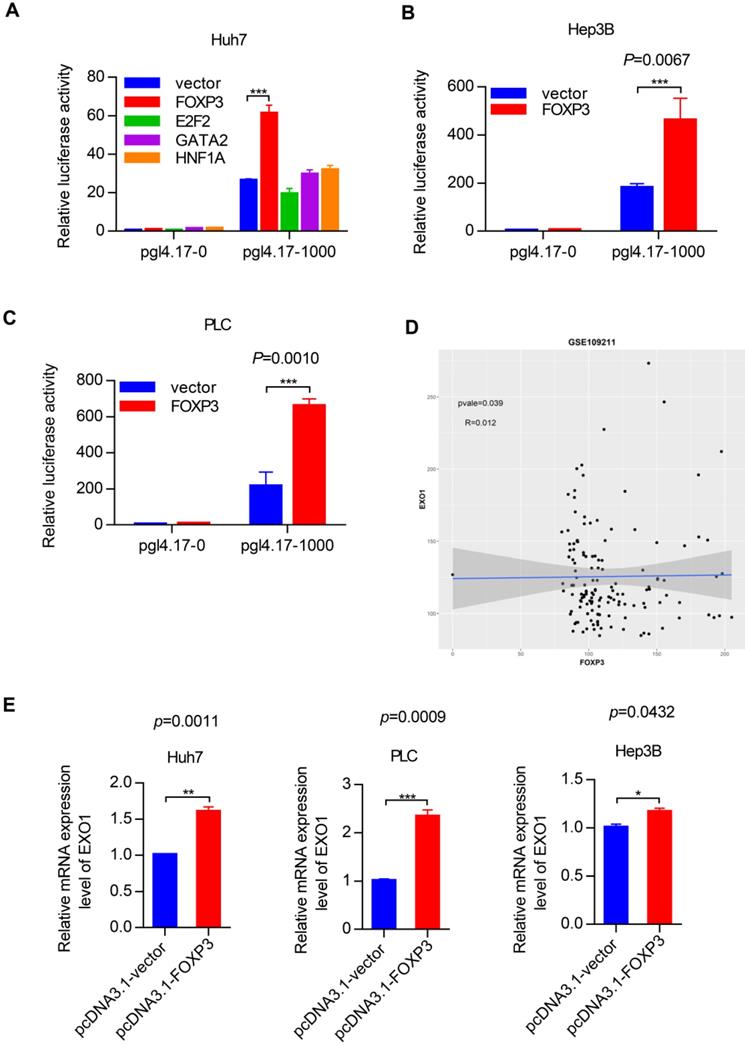
To identify the transcription factors that could bind to the promoter region of EXO1, we used the PROMO and JASPA websites to make predictions. According to the scoring system provided by the website and gene function itself, we focused on four transcription factors named FOXP3, E2F2, GATA2 and HNF1A. Plasmids were constructed with four coding sequences of these transcription factors mentioned above using pcDNA3.1 as a vector. We detected the transcriptional activity of these four transcription factors in the Huh7 cell line by luciferase reporter assay. Interestingly, as Figure 6A shows, FOXP3 had a significantly high reporter activity level compared to the other transcription factors. Based on this crucial result, we performed the same luciferase reporter assay in the Hep3B and PLC cell lines. As shown in Figure 6B and 6C, compared with co-transfection of the pcDNA 3.1 vector and pGL4.17-1000, when co-transfected with pcDNA 3.1-FOXP3 and PGL 4.17-1000, there was strong reporter activity. These results strongly suggest that FOXP3 may be involved in the transcriptional regulation of the EXO1 gene. Through bioinformatics analysis in the GEO database GSE109211, we found that the expression of FOXP3 and EXO1 had a positive correlation in HCC (Figure 6D). Hence, we transiently transfected the pcDNA3.1-FOXP3 plasmid into the three different HCC cell lines (Huh7, PLC, Hep3B) and transfected a pcDNA3.1 vector as a control. Consistently, the mRNA level of EXO1 increased with FOXP3 transfection (Figure 6E), as detected by RT-PCR. Together, these results show that FOXP3 promotes the expression of EXO1 mRNA levels and may regulate the expression of EXO1.
EXO1 expression is upregulated in HCC and associated with poor outcomes in HCC patients. (A) Dot chart of analysis of EXO1 expression in HCC tumor tissues and non-tumor tissues in the Oncomine database. (B) The expression of EXO1 in 99 paired HCC tissues and adjacent non-tumor tissues was detected by western blotting. The bar chart shows the expression level of EXO1 in HCC tissues, which were quantified and normalized to the corresponding EXO1 levels in the adjacent non-tumor tissues. (C) Dot chart of EXO1 expression in the 99 HCC samples. (D) Representative images of Western blots of EXO1 in 99 HCC tumor tissues and their adjacent non-tumor tissues. (E) Kaplan-Meier analysis showed the disease-free survival curve and overall survival curve of two different groups: patients with high EXO1 expression and patients with low EXO1 expression. The data comes from TCGA database. (F) IHC analysis of EXO1 expression in 10 paired HCC tissues. Results of case 1 and case 3 are used as representative images in the left panels and statistical analysis of EXO1 expression in the HCC samples in the right panel. (20×, magnification) (*p<0.05; **p<0.01; *** p<0.001).
EXO1 promotes cell proliferation and colony formation in HCC cell lines and proliferation in vivo. (A) The expression levels of EXO1 in one healthy liver cell line, ten HCC cell lines and one HCC lung metastasis cell line were detected by western blotting. (B), (C) Western blotting confirmed that EXO1 expression was effectively repressed by shRNA in HLF and Huh7 cell lines and EXO1 was overexpressed in the Bel-7402 cell line. The bar chart shows that EXO1 mRNA levels were significantly decreased or increased in the corresponding cell lines compared to the vector group. The data are shown as the mean ± SEM. (D), (E) CCK-8 assays indicated that knockdown of EXO1 significantly reduced cell proliferation abilities while overexpression of EXO1 increased cell proliferation abilities in the corresponding cell lines. The data are shown as the mean ± SD (n=6) (F), (G) Representative image of colony formation assays of EXO1 knockdown and EXO1 overexpression in the corresponding cell lines. The bar chart shows that EXO1 knockdown reduced colony formation abilities but overexpression of EXO1 enhanced colony formation abilities compared to their vector groups. Data are represented as the mean ± SD (n=3). (H) Representative image of subcutaneous tumors (n=5). (I) Dot chart of final tumor weight (n=5). (J) Dot chart of final tumor volume (n=5). (*p<0.05; **p<0.01; ***p<0.001 when compared with the vector group).
EXO1 promotes migration and invasion of HCC cells in vitro. (A), (B) Representative images of migration and invasion of HLF and Huh7 cells by transwell migration and matrigel invasion assays. Bar chart shows that migration and invasion abilities were significantly reduced when EXO1 was downregulated compared to the vector groups. The data are shown as the mean ± SD (n=6). Scale bar: 200 µm. (B), (C) Representative images of migration and invasion of Bel-7402 cells by transwell migration and matrigel invasion assays. The bar chart shows the migration and invasion abilities were significantly increased when EXO1 was overexpressed compared to the vector groups. The data are shown as the mean ± SD (n=6). Scale bar: 200 µm (***p<0.001 when compared to the vector group).
Depletion of FOXP3 enhances the proliferation effects of EXO1 in vitro
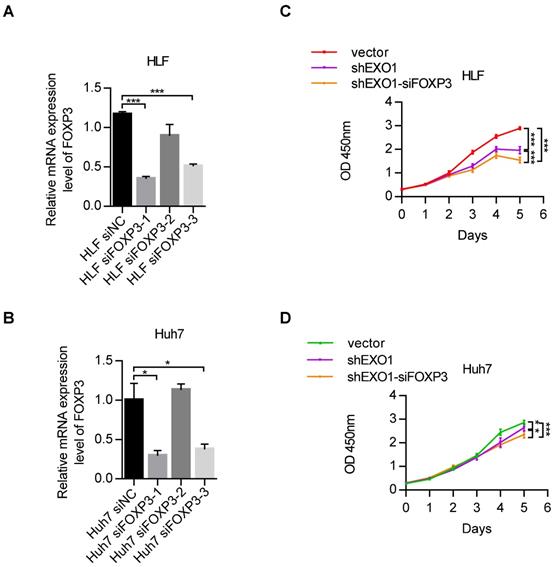
Due to the regulatory impact of FOXP3 on EXO1, we wanted to explore whether FOXP3 can affect the proliferation activity of EXO1 in HCC cells. RT-PCR showed that among the three siFOXP3 (siFOXP3-1, siFOXP3-2, and siFOXP3-3) knockdown sequences, FOXP3 was significantly downregulated by siFOXP3-1 in HLF and Huh7 cell lines (p<0.05, Figure 7A, B). Next, we transfected siFOXP3-NC into shEXO1-vector HCC cells and shEXO1-788 HCC cells, respectively, and transfected siFOXP3-1 into shEXO1-788 HCC cells. As shown in Figure 7C and D, CCK-8 assays indicated that compared with HCC cells with only EXO1 knockdown, FOXP3 and EXO1 double knockdown significantly decreased the proliferation ability of the HCC cells (p<0.05). These results suggest that depletion of FOXP3 further reduces cell proliferation ability after knocking down of EXO1 in vitro, suggesting that FOXP3 may upregulate EXO1, and depletion of FOXP3 may reduce its expression.
The correlation between EXO1 expression and clinicopathological features in HCC
| Clinical variables | No. of patients | EXO1 expression level | p value | |
|---|---|---|---|---|
| n=99 | Low (n=21) | High (n=78) | ||
| Age (years) | 0.118 | |||
| <60 | 66 | 17(81.0%) | 49(62.8%) | |
| ≥60 | 33 | 4(19.0%) | 29(37.2%) | |
| Gender | 0.123 | |||
| Male | 87 | 21(100.0%) | 66(84.6%) | |
| Female | 12 | 0(0.0%) | 12(15.4%) | |
| HBsAg | 1.000 | |||
| Positive | 88 | 19(90.5%) | 69(88.5%) | |
| Negative | 11 | 2(9.5%) | 9(11.5%) | |
| AFP(ng/ml) | 0.911 | |||
| <400 | 67 | 14(66.7%) | 53(67.9%) | |
| ≥400 | 32 | 7(21.9%) | 25(78.1%) | |
| Child-Pugh Class | 0.152 | |||
| A | 88 | 21(100.0%) | 67(85.9%) | |
| B | 11 | 0(0.0%) | 11(14.1%) | |
| Liver cirrhosis | 0.005 | |||
| No | 19 | 9(42.9%) | 10(12.8%) | |
| Yes | 80 | 12(57.1%) | 68(87.2%) | |
| Tumor size (cm) | 0.709 | |||
| ≤5 | 46 | 9(42.9%) | 37(47.4%) | |
| >5 | 53 | 12(57.1%) | 41(52.6%) | |
| Tumor number | 0.589 | |||
| Single | 86 | 17(81.0%) | 69(88.5%) | |
| Multiple | 13 | 4(19.0%) | 9(11.5%) | |
| Tumor encapsulation | 0.227 | |||
| Yes | 63 | 11(52.4%) | 52(66.7%) | |
| No | 36 | 10(47.6%) | 26(33.3%) | |
| Vascular invasion | 0.094 | |||
| Yes | 22 | 8(38.1%) | 14(17.9%) | |
| No | 77 | 13(61.9%) | 64(82.1%) | |
| Tumor differentiation | 0.903 | |||
| Well | 12 | 2(9.5%) | 10(12.8%) | |
| Middle | 52 | 11(52.4%) | 41(52.6%) | |
| Poor | 35 | 8(38.1%) | 27(34.6%) | |
| TNM stage | 0.531 | |||
| I-II | 75 | 17(81.0%) | 58(74.4%) | |
| III-IV | 24 | 4(19.0%) | 20(25.6%) | |
Discussion
It has been shown that DNA damage caused by the hepatitis B virus (HBV) infection is one of the main molecular mechanisms of hepatocellular carcinoma [35-37]. HBV-DNA can be integrated into host genomic DNA and cause insertion mutations or other genomic instabilities [38, 39]. Studies have shown that DNA double strand breaks (DSBs) induced by DNA damage are potential targets for HBV-DNA integration [40-42]. The body repairs DSBs through two major repair pathways: homologous recombination (HR) and non-homologous end-joining (NHEJ) [43]. Bill et al. previously suggested that integration of duck hepatitis B virus (DHBV) DNA into the host hepatocyte genome may be accomplished by the body's NHEJ repair of DSBs, and the integration could occur at the sites of DNA damage [44]. EXO1 plays an important role in DNA damage repair, especially in HR and NHEJ after DSBs [45-48]. As HR and NHEJ may participate in the integration of HBV-DNA, the relationship between EXO1 and HBV related HCC is worth investigating. In the present study we observed that EXO1 was frequently overexpressed in HCC samples and the same result was also seen in the Oncomine database. Analysis of EXO1 prognosis from the TCGA database showed the overall survival rate and tumor-free survival rate of patients with high EXO1 expression were significantly lower than those of patients with low EXO1 expression. Thus, we concluded that EXO1 is highly expressed in HCC and is associated with poor prognosis.
The link between EXO1 and cancer is still under continuous research and much progress has been made. As mentioned earlier, several SNPs in the EXO1 gene are related to the occurrence of certain tumors [17-25]. Some EXO1-specific mutations such as E109K and A153V are thought to be located in the region required for nuclease activity, thereby deactivating protein function and affecting cancer susceptibility [49, 50]. In the mouse model, the incidence of lymphoma in EXO1null/null increased and the survival rate decreased, but the microsatellite instability (MSI) was not affected [13]. EXO1 has also been reported to be overexpressed in several other cancers as described in the introduction [15, 16, 51-53]. In general, EXO1 is poorly expressed and increased levels of EXO1 may cause cellular damage due to its 5'-3' exonuclease activity, which may be contrary to previous studies [8, 54]. This may be due to the complex role of EXO1 in DNA damage repair and its tissue-specific expression. In humans, EXO1 is highly expressed in the testis and moderately expressed in the thymus, colon, and placenta [5, 55]. The liver has high mitochondrial activity and increased reactive oxygen species (ROS), which may lead to increased activation of the DNA damage repair pathway, resulting in a relatively high expression level of EXO1 in the liver [55]. Muthuswami et al. treated breast cancer cell line, MCF7, with different alkylating agents such as carboplatin, cyclophosphamide, etc. to induce DNA repair and found that EXO1 expression increased with increasing concentration of these alkylating agents [51]. de Sousa et al. observed that EXO1 knockdown supported a faster DNA-DSB restoration after irradiation (IR) exposure in glioblastoma cell line T98G [15]. All these, explain to some extent that EXO1 is involved in the DNA repair pathway under cancerous conditions. The overexpression of EXO1 in tumors may also increase the genetic instability and contribute to tumor initiation and progression through its involvement in recombinant events such as DSBs and telomere stabilization [56].
At present, the research on the role of EXO1 in HCC is still lacking. Previously, it was reported that SNPs K589E (rs1047840) and rs3754093 of EXO1 could increase and decrease the susceptibility of HCC, respectively [17, 18]. In 2018, Dai et al. used clonogenic assays to find that the expression of EXO1 affects the HCC cell survival under irradiation, and EXO1 overexpression has a poor prognosis for HCC patients [16]. In other types of tumors, as previously mentioned, Muthuswami et al. and de Sousa et al. investigated the possible role of EXO1 in different types of tumors through the breast cancer cell line MCF7 and the glioblastoma cell line T98G, respectively [15, 51]. In 2019, Luo et al. found the differential expression of EXO1 in five prostate cancer cell lines, and the EXO1 expression in the other four high‐invasive/metastatic prostate cancer cell lines significantly increased compared with the low‐invasive/metastatic potential cell line LNCaP [53]. These researches above led us to explore the role of EXO1 in the malignant biological characteristics of HCC by using HCC cell lines in vitro and in vivo. In this study, we observed that knockdown of EXO1 reduced cell proliferation in vitro and in vivo, while overexpression of EXO1 enhanced it. Subsequently, transwell assays showed that EXO1 could markedly increase the cell motility of HCC cells in vitro, indicating that EXO1 can affect the migration and invasion of HCC cells. Altogether, these functional assays indicate that EXO1 may play a carcinogenic role in HCC. However, previous research reported that knockdown of EXO1 in mice exhibits a mild phenotype, which may be contrary to our experimental results [57]. This may be due to complex environmental factors in vivo as well as the multiple roles the EXO1 gene itself plays in DSBs. There are still some researches suggesting that the absence of EXO1 in mice will show MMR defects, apoptotic defects, and sterility [10, 11]. Therefore, although we suggest that EXO1 may function as a tumorigenic gene in vitro, more in vivo experiments are needed to validate these conclusions.
Truncation of EXO1 promoter region. (A) Schematic diagram of the region lying 2000 bp upstream of EXO1 transcription start site and a series of truncations. (B) Schematic diagram of the serial 500- promoter truncations (red) placed upstream of the EXO1 translational start site and their corresponding luciferase reporter activities. Data are represented as the mean ± SD (n=3). (C) Schematic diagram of the serial 200- promoter truncations (red) placed upstream of the EXO1 translational start site and their corresponding luciferase reporter activity. Data are represented as the mean ± SD (n=3). (*p<0.05).
FOXP3 enhances the expression of EXO1. (A) Bar chart of luciferase activity of transcription factors possibly binding to EXO1 promoter region compared to the pcDNA3.1-vector group. Data are represented as the mean ± SD (n=3). (B), (C) pcDNA3.1 and pGL4.17 were co-transfected into Hep3B and PLC cells for 48 h. The bar chart of luciferase activity shows that when co-transfected with pcDNA3.1-FOXP3 and pGL4.17-1000, the relative reporter activity was strongest. Data are represented as mean ± SD (n=3). (D) The scatterplot diagram of positive correlation between EXO1 and FOXP3 expression level in GSE109211 (n=140). (E) pcDNA3.1-FOXP3 or pcDNA3.1-vector were transiently transfected into Huh7, PLC, and Hep3B cells for 48 h. The expression of EXO1 was examined by RT-PCR. Data are represented as mean ± SD (n=3). (*p<0.05; **p<0.01; ***p<0.001).
Depletion of FOXP3 enhances the proliferative effects of EXO1 in vitro (A), (B) RT-PCR confirmed that FOXP3 expression was effectively repressed by siFOXP3-1 in HLF and Huh7 cell lines. The data are shown as the mean ± SEM. (C), (D) CCK-8 assays indicated that knockdown of FOXP3 in EXO1 knockdown cells significantly reduced cell proliferation abilities compared to the EXO1 knockdown alone. The data are shown as the mean ± SD (n=6). (*p<0.05; **p<0.01; ***p<0.001).
EXO1 has been previously reported to be regulated by a variety of molecules and to bind to certain specific proteins [58, 59]. Studies on DNA mismatch repair (MMR) have reported that EXO1 interacts with the mismatch repair factors mutL homolog 1 (MLH1), mutS homolog 2 (MSH2), mutS homolog 3 (MSH3), and proliferating cell nuclear antigen (PCNA) to excise the newly synthesized DNA sequences containing errors during the MMR process [60-63]. Checkpoint regulatory proteins 14-3-3 are widely reported as interaction partners of EXO1. Engels et al. identified in 2011 that in yeast and mammalian cells, 14-3-3 proteins interact with EXO1 in vivo to regulate the phosphorylation status of EXO1, thereby promoting EXO1-dependent fork progression after inhibiting replication [64]. In 2012, Andersen et al. demonstrated specific interactions between EXO1 and all the 7 isoforms of 14-3-3 through in vitro GST pull-down assays, and they demonstrated that the binding may involve a second unrecognized binding motif in EXO1, the amino acid S746. They also demonstrated that the 14-3-3 association may affect the nuclease activity of EXO1 [65]. In 2015, Chen et al. showed that 14-3-3 can interact with the central portion of EXO1, at least partially, by suppressing its association with PCNA, to negatively regulate EXO1 damage recruitment and thus prevent excessive DNA excision [66]. However, very little is known about the transcriptional regulation of EXO1. Muthuswami et al. discovered that transcription factors E2F and Myc may be involved in the regulation of EXO1 by using luciferase reporter assays [51]. In this study, we uncovered a new potential transcriptional regulator of EXO1, FOXP3, by truncating the promoter region of EXO1 and using bioinformatic prediction. FOXP3 has been proposed to function as a tumor suppressor in breast and prostate epithelial cells [67, 68]. However, FOXP3 could also be a prognostic factor in breast cancer and play an important role in the progression of cervical cancer [69, 70]. In a recent report from 2019, Ou et al. reported that FOXP3 silencing may be associated with the inhibition of cell proliferation and migration of HCC cells, as well as the induction of apoptosis [71]. By transiently transfecting FOXP3 into HCC cell lines, we observed that FOXP3 could promote the transcriptional activity of EXO1 by a luciferase reporter assay. We further found that EXO1 mRNA levels were significantly upregulated after transient transfection of FOXP3. Moreover, bioinformatics analysis revealed a positive correlation between the expression of EXO1 and FOXP3, and a CCK-8 assay in HCC cells proved that FOXP3 could enhance the proliferative effect of EXO1 in vitro. These results indicate that FOXP3 could upregulate the transcription of EXO1 and might further regulate the expression level of EXO1. This is consistent with the reported results of Ou et al. Further explorations are required to validate this perspective.
Nonetheless, this study has some limitations. Firstly, the size of the clinical samples in this study was relatively small, with only 99 cases. Therefore, the number of samples assessed needs to be expanded. Second, there were database limitations and the expression level of EXO1 and the prognosis of HCC patients need to be further verified by complete clinical follow-up. Finally, it is not uncommon that DNA repair genes are highly overexpressed in HCC. In addition to the possible explanations given above, X-Ray Repair Cross Complementing 2 (XRCC2) has also been reported to be up-regulated in a variety of tumors [72, 73]. It is possible that DNA damage and repair processes are more active in tumor cells. RAD51B, RAD51C, RAD51D, and XRCC2 form the BCDX2 complex, which promotes the aggregation of RAD51 protein at HR sites [74]. The mechanism of EXO1 overexpression in HCC is complex and still needs to be further explored.
In summary, our research found that EXO1 was overexpressed in HCC and played a carcinogenic role in HCC, and that FOXP3 may upregulate the expression level of EXO1. For the first time, we have uncovered a possible transcriptional regulation mechanism of EXO1. This discovery provides clues for the relationship between DNA damage repair genes and HCC and might lead to the development of novel anti-cancer therapeutics for HCC treatment.
Abbreviations
EXO1: Exonuclease 1; FOXP3: Forkhead box P3; HCC: Hepatocellular carcinoma; TNM: Tumor-node-metastasis; AFP: α-fetoprotein; ALT: alanine aminotransferase; AST: aspartate aminotransferase; SD: Standard deviation; SEM: Standard error of mean.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (81172293) and the National Science and Technology Major Project of China (2017ZX10203207-002-005).
Author contributions
Guang Yang performed the experiments and wrote the article independently. Ke-Shuai Dong conducted the bioinformatics analysis and did the statistical analysis. Zun-Yi Zhang provided technical guidance for the entire study. Er-Lei Zhang, Bin-Yong Liang collected and analyzed the clinical data. Zhi-Yong Huang directed the revision and correction of the manuscript. Xiao-Ping Chen and Zhi-Yong Huang conceived the idea and designed the study. All authors reviewed and endorsed the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
2. Jarnagin WR. Management of small hepatocellular carcinoma: a review of transplantation, resection, and ablation. Ann Surg Oncol. 2010;17:1226-33
3. Morise Z, Kawabe N, Tomishige H, Nagata H, Kawase J, Arakawa S. et al. Recent advances in liver resection for hepatocellular carcinoma. Front Surg. 2014;1:21
4. Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014-23
5. Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027-31
6. Lee BI, Wilson DM 3rd. The RAD2 domain of human exonuclease 1 exhibits 5' to 3' exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763-9
7. Tomimatsu N, Mukherjee B, Deland K, Kurimasa A, Bolderson E, Khanna KK. et al. Exo1 plays a major role in DNA end resection in humans and influences double-strand break repair and damage signaling decisions. DNA Repair (Amst). 2012;11:441-8
8. Keijzers G, Liu D, Rasmussen LJ. Exonuclease 1 and its versatile roles in DNA repair. Crit Rev Biochem Mol Biol. 2016;51:440-51
9. Keijzers G, Bakula D, Petr MA, Madsen NGK, Teklu A, Mkrtchyan G. et al. Human Exonuclease 1 (EXO1) Regulatory Functions in DNA Replication with Putative Roles in Cancer. Int J Mol Sci. 2018;20:74
10. Wei K. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes & Development. 2003;17:603-14
11. Bolderson E, Richard DJ, Edelmann W, Khanna KK. Involvement of Exo1b in DNA damage-induced apoptosis. Nucleic Acids Res. 2009;37:3452-63
12. Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK. et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38:1821-31
13. Schaetzlein S, Chahwan R, Avdievich E, Roa S, Wei K, Eoff RL. et al. Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc Natl Acad Sci U S A. 2013;110:E2470-9
14. Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15
15. de Sousa JF, Torrieri R, Serafim RB, Di Cristofaro LF, Escanfella FD, Ribeiro R. et al. Expression signatures of DNA repair genes correlate with survival prognosis of astrocytoma patients. Tumour Biol. 2017;39:1010428317694552
16. Dai Y, Tang Z, Yang Z, Zhang L, Deng Q, Zhang X. et al. EXO1 overexpression is associated with poor prognosis of hepatocellular carcinoma patients. Cell Cycle. 2018;17:2386-97
17. Bayram S, Akkiz H, Bekar A, Akgollu E, Yildirim S. The significance of Exonuclease 1 K589E polymorphism on hepatocellular carcinoma susceptibility in the Turkish population: a case-control study. Mol Biol Rep. 2012;39:5943-51
18. Tan SK, Qin RY, Zhu XN, Tan C, Song JL, Qin LY. et al. Associations between single-nucleotide polymorphisms of human exonuclease 1 and the risk of hepatocellular carcinoma. Oncotarget. 2016;7:87180-93
19. Bau DT, Wang HC, Liu CS, Chang CL, Chiang SY, Wang RF. et al. Single-nucleotide polymorphism of the Exo1 gene: association with gastric cancer susceptibility and interaction with smoking in Taiwan. Chin J Physiol. 2009;52:411-8
20. Shi T, Jiang R, Wang P, Xu Y, Yin S, Cheng X. et al. Significant association of the EXO1 rs851797 polymorphism with clinical outcome of ovarian cancer. Onco Targets Ther. 2017;10:4841-51
21. Luo XP, Hong XS, Xiong XD, Zeng LQ, Lim CED. A Single Nucleotide Polymorphism in EXO1 Gene Is Associated With Cervical Cancer Susceptibility in Chinese Patients. Int J Gynecol Cancer. 2012;22:220-5
22. Wang HC, Chiu CF, Tsai RY, Kuo YS, Chen HS, Wang RF. et al. Association of genetic polymorphisms of EXO1 gene with risk of breast cancer in Taiwan. Anticancer Res. 2009;29:3897-901
23. Yamamoto H, Hanafusa H, Ouchida M, Yano M, Suzuki H, Murakami M. et al. Single nucleotide polymorphisms in the EXO1 gene and risk of colorectal cancer in a Japanese population. Carcinogenesis. 2005;26:411-6
24. Kabzinski J, Przybylowska K, Mik M, Sygut A, Dziki L, Dziki A. et al. Association of polymorphism of Lys589Glu Exo1 gene with the risk of colorectal cancer in the Polish population. Pol Przegl Chir. 2014;86:370-3
25. Nogueira GAS, Lourenco GJ, Oliveira CBM, Marson FAL, Lopes-Aguiar L, Costa EFD. et al. Association between genetic polymorphisms in DNA mismatch repair-related genes with risk and prognosis of head and neck squamous cell carcinoma. Int J Cancer. 2015;137:810-8
26. Kabzinski J, Mucha B, Cuchra M, Markiewicz L, Przybylowska K, Dziki A. et al. Efficiency of Base Excision Repair of Oxidative DNA Damage and Its Impact on the Risk of Colorectal Cancer in the Polish Population. Oxid Med Cell Longev. 2016;2016:3125989
27. Tang J, Tang S, Liu J, Wu Q, Wan L, Xu Q. Genetic risk of lung cancer associated with a single nucleotide polymorphism from EXO1: a meta analysis. Int J Clin Exp Med. 2015;8:11132-8
28. Fontenot JD, Rudensky AY. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol. 2005;6:331-7
29. Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936-40
30. Sadlon TJ, Wilkinson BG, Pederson S, Brown CY, Bresatz S, Gargett T. et al. Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J Immunol. 2010;185:1071-81
31. Kim M, Grimmig T, Grimm M, Lazariotou M, Meier E, Rosenwald A. et al. Expression of Foxp3 in colorectal cancer but not in Treg cells correlates with disease progression in patients with colorectal cancer. PLoS One. 2013;8:e53630
32. Winerdal ME, Marits P, Winerdal M, Hasan M, Rosenblatt R, Tolf A. et al. FOXP3 and survival in urinary bladder cancer. BJU Int. 2011;108:1672-8
33. Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L. et al. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007;117:3765-73
34. Zuo T, Wang L, Morrison C, Chang X, Zhang H, Li W. et al. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129:1275-86
35. Bertoletti A, Kennedy PTF, Durantel D. HBV infection and HCC: the 'dangerous liaisons'. Gut. 2018;67:787-8
36. Buendia MA, Neuveut C. Hepatocellular carcinoma. Cold Spring Harb Perspect Med. 2015;5:a021444
37. Xu C, Zhou W, Wang Y, Qiao L. Hepatitis B virus-induced hepatocellular carcinoma. Cancer Lett. 2014;345:216-22
38. Wang Y, Wu MC, Sham JST, Tai LS, Fang Y, Wu WQ. et al. Different expression of hepatitis B surface antigen between hepatocellular carcinoma and its surrounding liver tissue, studied using a tissue microarray. J Pathol. 2002;197:610-6
39. Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. J Clin Virol. 2005;34(Suppl 1):S75-8
40. Hu X, Lin J, Xie Q, Ren J, Chang Y, Wu W. et al. DNA double-strand breaks, potential targets for HBV integration. J Huazhong Univ Sci Technolog Med Sci. 2010;30:265-70
41. Kostyushev D, Kostyusheva A, Brezgin S, Zarifyan D, Utkina A, Goptar I. et al. Suppressing the NHEJ pathway by DNA-PKcs inhibitor NU7026 prevents degradation of HBV cccDNA cleaved by CRISPR/Cas9. Sci Rep. 2019;9:1847
42. Kostyusheva AP, Kostyushev DS, Brezgin SA, Zarifyan DN, Volchkova EV, Chulanov VP. Small Molecular Inhibitors of DNA Double Strand Break Repair Pathways Increase the ANTI-HBV Activity of CRISPR/Cas9. Molecular Biology. 2019;53:274-85
43. Roos WP, Krumm A. The multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair. Nucleic Acids Res. 2016;44:10017-30
44. Bill CA, Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A. 2004;101:11135-40
45. Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981-94
46. Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241-4
47. Keijzers G, Maynard S, Shamanna RA, Rasmussen LJ, Croteau DL, Bohr VA. The role of RecQ helicases in non-homologous end-joining. Crit Rev Biochem Mol Biol. 2014;49:463-72
48. Shamanna RA, Lu H, de Freitas JK, Tian J, Croteau DL, Bohr VA. WRN regulates pathway choice between classical and alternative non-homologous end joining. Nat Commun. 2016;7:13785
49. Sun X, Zheng L, Shen B. Functional alterations of human exonuclease 1 mutants identified in atypical hereditary nonpolyposis colorectal cancer syndrome. Cancer Res. 2002;62:6026-30
50. Hansen MF, Johansen J, Bjornevoll I, Sylvander AE, Steinsbekk KS, Saetrom P. et al. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam Cancer. 2015;14:437-48
51. Muthuswami M, Ramesh V, Banerjee S, Viveka Thangaraj S, Periasamy J, Bhaskar Rao D. et al. Breast tumors with elevated expression of 1q candidate genes confer poor clinical outcome and sensitivity to Ras/PI3K inhibition. PLoS One. 2013;8:e77553
52. Chen J, Wang Z, Shen X, Cui X, Guo Y. Identification of novel biomarkers and small molecule drugs in human colorectal cancer by microarray and bioinformatics analysis. Mol Genet Genomic Med. 2019;7:e00713
53. Luo F, Wang YZ, Lin D, Li J, Yang K. Exonuclease 1 expression is associated with clinical progression, metastasis, and survival prognosis of prostate cancer. Journal of Cellular Biochemistry. 2019;120:11383-9
54. El-Shemerly M, Janscak P, Hess D, Jiricny J, Ferrari S. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Research. 2005;65:3604-9
55. Rasmussen LJ, Rasmussen M, Lee B, Rasmussen AK, Wilson DM 3rd, Nielsen FC. et al. Identification of factors interacting with hMSH2 in the fetal liver utilizing the yeast two-hybrid system. In vivo interaction through the C-terminal domains of hEXO1 and hMSH2 and comparative expression analysis. Mutat Res. 2000;460:41-52
56. Liberti SE, Rasmussen LJ. Is hEXO1 a cancer predisposing gene? Mol Cancer Res. 2004;2:427-32
57. Rein K, Yanez DA, Terre B, Palenzuela L, Aivio S, Wei KC. et al. EXO1 is critical for embryogenesis and the DNA damage response in mice with a hypomorphic Nbs1 allele. Nucleic Acids Res. 2015;43:7371-87
58. Chen X, Paudyal SC, Chin RI, You Z. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013;41:9325-38
59. Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358-69
60. Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58:4537-42
61. Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011-8
62. Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457-68
63. Goellner EM, Putnam CD, Graham WJt, Rahal CM, Li BZ, Kolodner RD. Identification of Exo1-Msh2 interaction motifs in DNA mismatch repair and new Msh2-binding partners. Nat Struct Mol Biol. 2018;25:650-9
64. Engels K, Giannattasio M, Muzi-Falconi M, Lopes M, Ferrari S. 14-3-3 Proteins regulate exonuclease 1-dependent processing of stalled replication forks. PLoS Genet. 2011;7:e1001367
65. Andersen SD, Keijzers G, Rampakakis E, Engels K, Luhn P, El-Shemerly M. et al. 14-3-3 checkpoint regulatory proteins interact specifically with DNA repair protein human exonuclease 1 (hEXO1) via a semi-conserved motif. DNA Repair (Amst). 2012;11:267-77
66. Chen X, Kim I-K, Honaker Y, Paudyal SC, Koh WK, Sparks M. et al. 14-3-3 Proteins Restrain the Exo1 Nuclease to Prevent Overresection. Journal of Biological Chemistry. 2015;290:12300-12
67. McInnes N, Sadlon TJ, Brown CY, Pederson S, Beyer M, Schultze JL. et al. FOXP3 and FOXP3-regulated microRNAs suppress SATB1 in breast cancer cells. Oncogene. 2011;31:1045-54
68. Wang L, Liu R, Li W, Chen C, Katoh H, Chen GY. et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16:336-46
69. Merlo A, Casalini P, Carcangiu ML, Malventano C, Triulzi T, Menard S. et al. FOXP3 expression and overall survival in breast cancer. J Clin Oncol. 2009;27:1746-52
70. Zeng C, Yao Y, Jie W, Zhang M, Hu X, Zhao Y. et al. Up-regulation of Foxp3 participates in progression of cervical cancer. Cancer Immunol Immunother. 2013;62:481-7
71. Ou X, Zhang GT, Tian PK, Chen JS, Lin ZW, Xie Y. et al. Forkhead box P3 gene silencing inhibits the expression of chemokines and chemokine receptors associated with cell growth, migration, and apoptosis in hepatocellular carcinoma cells. Exp Ther Med. 2019;18:1091-8
72. Zheng Z, Ng WL, Zhang X, Olson JJ, Hao C, Curran WJ. et al. RNAi-mediated targeting of noncoding and coding sequences in DNA repair gene messages efficiently radiosensitizes human tumor cells. Cancer Res. 2012;72:1221-8
73. Sullivan I, Salazar J, Majem M, Pallares C, Del Rio E, Paez D. et al. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Lett. 2014;353:160-6
74. Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ. et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296-307
Author contact
![]() Corresponding author: Professor Zhiyong Huang, Hepatic Surgical Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1095 Jie Fang Da Dao, Wuhan, 430030, China. Fax: +86 027 83663432; E-mail address: zyhuang126com.
Corresponding author: Professor Zhiyong Huang, Hepatic Surgical Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, 1095 Jie Fang Da Dao, Wuhan, 430030, China. Fax: +86 027 83663432; E-mail address: zyhuang126com.

 Global reach, higher impact
Global reach, higher impact