3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2020; 11(15):4343-4351. doi:10.7150/jca.44410 This issue Cite
Research Paper
Association of variations in platinum resistance-related genes and prognosis in lung cancer patients
1. Department of Clinical Pharmacology and National Clinical Research Center for Geriatric Disorder, Xiangya Hospital, Central South University, Changsha 410008, China
2. Institute of Clinical Pharmacology, Central South University, Hunan Key Laboratory of Pharmacogenetics, Changsha 410078, P. R. China
3. Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008, China
4. Department of National Institution of Drug Clinical Trial, Xiangya Hospital, Central South University, Changsha 410008, China
5. Department of Lung Cancer and Gastroenterology, Hunan Cancer Hospital, Affiliated Tumor Hospital of Xiangya Medical School of Central South University, Changsha, 410013, China
6. Department of the Central Laboratory, Hunan Cancer Hospital, Affiliated Tumor Hospital of Xiangya Medical School of Central South University, Changsha, 410013, China
Received 2020-1-30; Accepted 2020-3-29; Published 2020-4-27
Abstract
Purpose: We aimed to investigate the association of single-nucleotide polymorphisms (SNPs) in HMGB1, REV3L, and NFE2L2 with prognosis in lung cancer patients with platinum-based chemotherapy.
Methods: We have recruited 348 lung cancer patients treated with platinum. Log-rank test and Cox regression analysis were used to assess overall survival (OS) and progression-free survival (PFS) among SNP genotypes.
Results: The results revealed that patients carrying TC or CC genotype in REV3L rs462779 (HR=0.67, 95% CI=0.51-0.90, P=0.007) and AG or GG genotype in HMGB1 rs1045411 (HR=0.61, 95% CI=0.38-0.99, P=0.046) had a better overall survival. Additionally, carrying TC or TT genotype in rs462779 had a lower risk (OR=0.38, 95% CI=0.17-0.89, P=0.025) of lymph node metastasis, carrying AG or AA genotype in rs1045411 was significantly related to early T stage (OR=0.47, 95% CI=0.29-0.76, P=0.002). In stratified analysis, patients with TC or CC genotype in rs462779 were significantly associated with overall survival in male patients, never-smokers, patients with younger age (≤56), no family history of cancer, adenocarcinoma, advanced stage (stage III or IV), or ECOG PS 0-1. While patients with AG or GG genotype in rs1045411 were significantly associated with overall survival in patients with advanced stage (stage III or IV) or ECOG PS 0-1.
Conclusion: Our results indicate that the TC or CC genotype in rs462779 and AG or GG genotype in rs1045411 are contributed to better overall survival. The REV3L rs462779 and HMGB1 rs1045411 may serve as prognosis markers in lung cancer patients with platinum-based chemotherapy.
Keywords: lung cancer, platinum-based chemotherapy, prognosis, polymorphisms, REV3L, HMGB1
Introduction
According to recent studies, lung cancer is still the leading cause of cancer mortality both in males and females, which accounts for 18.4% of total cancer death [1]. A majority of lung cancer patients, approximately 85%, are diagnosed with non-small cell lung cancer (NSCLC) [2]. To the best of our knowledge, tobacco smoking is the main risk factor for lung cancer, and other factors are environmental- and occupational-related. Though the incidence rate has slightly decreased in recent years, the 5-year survival of lung cancer is merely 19% [3]. Most of the patients have been diagnosed with an advanced stage, the prognosis for patients with lung cancer is still depressing. Obviously, the prognosis of lung cancer patients with different stage varies greatly, it is necessary to improve survival by finding biomarkers for prognosis prediction. In addition to traditional prognostic factors including tumor microenvironment state and clinical stage, diverse outcomes of cancer patients may also attribute to genetic alterations [4].
In spite of other treatments, like surgery, targeted therapy, radiation, and immunity therapy, platinum-based chemotherapy is still the first-line treatment for lung cancer patients [5]. Platinum-based chemotherapy drugs mainly function by forming DNA adducts intrastrand and interstrand, cells recognize the adducts as DNA damage then activate DNA repair mechanisms, like nucleotide excision repair, mismatch repair, base-excision repair, non-homologous end-joining, and homologous recombination [6, 7]. The initial response of platinum in most lung cancer patients is generally good, but intrinsic or acquired resistance would influence the sensitivity of platinum. The underlying mechanisms are described as following: increased damage, decreased accumulation, detoxification, epigenetic changes, decreased apoptosis, membrane trafficking changes, and genetic variants [8-10].
Platinum resistance-related genes affect the efficacy of platinum-based chemotherapy, which may act on the prognosis of patients. There are many genes associated with platinum resistance, according to database searching and literature references, we chose HMGB1, REV3L, and NFE2L2 to study the association between genetic variations and clinical outcome. HMGB1 (high mobility group box 1) is situated in the nucleus to maintain nuclear homeostasis, which plays an indispensable role in many diseases and cellular processes, like tumor, inflammation, cell differentiation, and migration [11]. HMGB1 directly binds to lesions of DNA then pursuits to DNA repair and tends to relate with platinum toxicity and liver damage [12, 13]. There are many reports about variants of HMGB1. The rs1045411 is related to the overall survival of gastric cancer and the risk of developing lung cancer, hepatocellular carcinoma, and lymph node metastasis of breast cancer [14-17]. The rs1412125 and rs2249825 appear to be significantly related to the risk of lung cancer and platinum response and seem to relate with the progression of breast cancer [15, 17-19]. REV3L (the zeta catalytic subunit of DNA polymerase) is the most important DNA polymerase, it plays a role in maintaining genome stability in the advent of DNA damage and cell proliferation after damage, also may inhibit tumorigenesis [20, 21]. Inhibition of REV3L sensitizes lung cancer and gliomas to cisplatin chemotherapy [22, 23]. The rs462779 and rs465646 in REV3L are associated with overall survival in platinum-treated malignant mesothelioma, rs462779 is also correlated with event-free survival and risk of colorectal cancer [24, 25]. Transcription factor NFE2L2 (nuclear factor, erythroid 2 like 2), also named NRF2, functions as an oncogene or tumor suppressor, it regulates the cellular antioxidant response and promotes cancer chemotherapy resistance, metastasis, and progression [26-28]. The rs6706649 and rs6721961 in NFE2L2 may influence breast cancer prognosis, rs6721961 and rs35652124 are associated with susceptibility to colorectal cancer [29, 30].
In this study, we investigate the associations of HMGB1 (rs1045411, rs1412125, and rs2249825), REV3L (rs462779 and rs465646) and NFE2L2 (rs6706649, rs6721961, and rs35652124) with platinum-based chemotherapy prognosis in lung cancer patients.
Materials and methods
Subjects
351 lung cancer patients were recruited in the study between November 2011 and May 2013 in Xiangya Hospital of Central South University and Hunan Provincial Tumour Hospital in Changsha, Hunan Province, and clinical data were collected in the same place. No patients had received surgery, targeted therapy, radiotherapy, and/or biological therapy before first-line chemotherapy. In demographic characteristics study and outcome analysis, we excluded 3 patients who had incomplete clinical information. In tumor progression analysis, we excluded 46 patients who had incomplete TNM stage information. All subjects were Han people of Chinese and successfully genotyped after donated 5 mL blood, all patients were treated with platinum-based therapy, and each had signed informed consent. A standard follow-up was carried out to collect characteristic data. Clinical data including age, gender, smoking history, family history of cancer, histology classification, TNM stage, and Eastern Cooperative Oncology Group Performance Status. The last follow-up date was in July 2019.
SNPs selection and genotyping
HMGB1 SNPs (rs1045411, rs1412125, and rs2249825), REV3L SNPs (rs462779 and rs465646), and NFE2L2 SNPs (rs6706649, rs6721961, and rs35652124) were selected for genotyping in the Han Chinese patients. They were chosen according to the following criteria: (1) based on our previous research, HMGB1 SNPs were related with lung cancer platinum-based chemotherapy response [18], (2) association with the outcome of cancers and involvement in other cancer types in other research, (3) functional relevance of gene transcription or protein expression.
EDTA tube was used to hold 5 mL of venous blood donated from each patient. DNeasy Blood & Tissue Kit (Qiagen, Shanghai, China) or Genomic DNA Purification Kit (Promega, Madison, WI, USA) was used to extracting genomic DNA based on the instructions. Sequenom Mass Array Genotype Platform (Sequenom, San Diego, CA, USA) was used to genotype SNPs of each gene. Primers were designed using Primer-Premier 6 software (Premier Biosoft Interpairs, Palo Alto, CA). The Sequenom Mass Array Genotype Platform was used to design polymerase chain reaction (PCR), the shrimp alkaline phosphatase enzyme (SAP) reaction and associated extension reactions. The PCR system was heating for 15 minutes at 94℃, thermocycling for 45 times (94℃ for 20 seconds, 56℃ for 30 seconds, then 72℃ for 1 minute) and extension at 72℃ for 3 minutes. The SAP reaction was performed at 37℃ for 40 minutes then 85℃ for 5 minutes. The extension reaction was conducted at 94℃ for 30 seconds, 94℃ for 5 seconds, 40 cycles for 5 seconds at 52℃, five cycles for 5 seconds at 80℃, finally 72℃ for 3 minutes. The resin was used to purify the reaction product, and the Mass Array system (Sequenom) was used to resolve the data.
Statistical analysis
The SPSS version 25.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism (version 8, GraphPad Software Inc., San Diego, CA) were used for statistical analysis. Three genetic models (additive, dominant, and recessive model) were applied to evaluate the association between SNPs and prognosis of lung cancer patients. Overall survival (OS) and progression-free survival (PFS) were analyzed as endpoints and were defined as the time from diagnosis to death for any reason, the time from diagnosis to the first time when patients progressed, respectively. The association between clinical or SNPs data and OS or PFS was assessed using Cox proportional hazards regression analysis, computed as HRs with corresponding 95% CIs. Binary logistic regression was used to test the difference in disease progression between groups. The log-rank test was used to examine the difference in overall survival or progression-free survival between groups. Kaplan-Meier plot was used to visualize the results. All the P-values were two-sided, P<0.05 were supposed to be significant.
Results
Demographic characteristics of patients and prognosis analysis
A total of 348 subjects were included in this investigation, all had received platinum-based chemotherapy. As shown in Table 1, the median age was 56 years (a range of 21 to 75 years). Among them, 78 (22.4%) were females, 270 (77.6%) were males. There were 205 (58.9%) patients who had ever smoked, and the rest 143 (41.1%) patients never smoked. Most of the patients (95.4%) did not have a family history of cancer. For histology, 179 (51.4%) patients were diagnosed with adenocarcinoma, 142 (40.8%) with squamous cell carcinoma, the remainings with other types. 338 (97.1%) of the patients were in an advanced stage (stage III or IV). Likewise, most patients (94.0%) were with Eastern Cooperative Oncology Group Performance Status (ECOG PS) 0 or 1. In the univariate Cox regression analysis, histology was significantly associated with progression-free survival, the risk of progression in patients with squamous cell carcinoma (HR=0.69, 95% CI=0.54-0.89, P=0.004) was lower than that in patients with adenocarcinoma (Table 1).
Relationship between the eight SNPs and clinical outcome of platinum-based chemotherapy
Log-rank test and multivariate Cox regression analysis with adjustment for age, gender, and histology were conducted to analyze the association between the eight SNPs and OS or PFS. The rs462779 in REV3L was significantly correlated with overall survival of lung cancer patients in the additive (P for log-rank=0.018) and recessive (P for log-rank=0.005) models (Table 2). Patients carrying TC or CC genotype in rs462779 showed a markedly lower death risk (HR=0.67, 95% CI=0.51-0.90, P=0.007) when compared with TT genotype in the recessive model (Table 2). There was also a trend difference in progression risk that patients with TC or CC genotype had better progression-free survival (P for log-rank=0.034), while the P-value for Cox regression (HR=0.77, 95% CI=0.59-1.02, P=0.073) was not significant (Table 2). Additionally, patients carrying AG or GG genotype in rs1045411 were significantly related to better overall survival (HR=0.61, 95% CI=0.38-0.99, P=0.046) (Table 2). The other SNPs were not significantly associated with either OS or PFS.
Association between the eight SNPs and lung cancer progression
To investigate whether the eight single nucleotide polymorphisms were related to tumor progression, binary logistic regression was used to analyze the association. As shown in Table 3, patients carrying REV3L rs462779 TC or TT genotypes had a lower hazard (OR=0.38, 95% CI=0.17-0.89, P=0.025) of lymph node metastasis in the dominant model when compared with patients carrying CC genotype. The rs1045411 and rs2249825 in HMGB1 gene were significantly associated with T stage. Patients with AG or AA genotype in rs1045411 were significantly related to early T stage (T1 or T2) when compared with GG genotype (OR=0.47, 95% CI=0.29-0.76, P=0.002), and patients with GC or GG genotype in rs2249825 were also associated with early T stage when compared with CC genotype (OR=0.56, 95% CI=0.33-0.94, P=0.028) (Table 3).
Subgroup analysis of association between REV3L/HMGB1 polymorphisms and prognosis
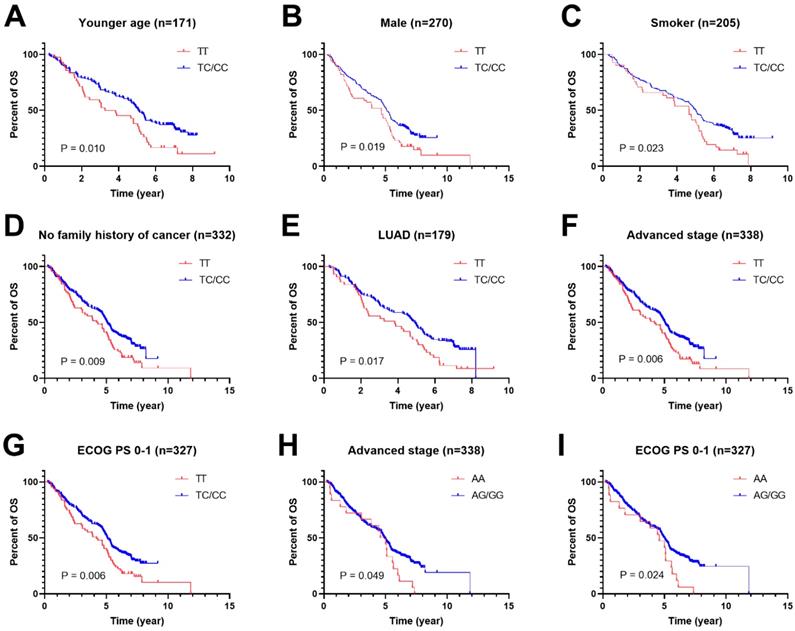
To further study the associations of rs462779 and rs1045411 with the prognosis of platinum-based chemotherapy, multivariate Cox proportional hazard analysis was performed in the recessive model stratified by clinical characteristics. In the patients with younger age (≤56 years old), we observed that the TC or CC genotype in rs462779 had significantly lower death risk (HR=0.61, 95% CI=0.42-0.91, P=0.015) when compared with TT genotype. For male patients, the TC or CC genotype also had significantly lower death risk (HR=0.69, 95% CI=0.50-0.96, P=0.027). For patients who had ever smoked, carrying TC or CC genotype had significantly lower death risk (HR=0.65, 95% CI=0.45-0.95, P=0.026). For patients without a family history of cancer, the HR for death of carrying TC or CC genotype was 0.68 (95% CI=0.51-0.92, P=0.011). In adenocarcinoma subgroup, the hazard ratio for death was 0.65 (95% CI=0.45-0.94, P=0.023). Those with advanced stage (stage III or IV) carrying TC or CC genotype had lower death risk (HR=0.68, 95% CI=0.51-0.91, P=0.009). For those who had Eastern Cooperative Oncology Group Performance Status 0 or 1 score, the HR for death of carrying TC or CC genotype was 0.67 (95% CI=0.49-0.90, P=0.008) (Supplementary Table 1, Figure 1A-1G).
For HMGB1 rs1045411, patients with advanced stage (stage III or IV) carrying AG or GG genotype had lower death risk (HR=0.62, 95% CI=0.38-1.00, P=0.050) when compared with AA genotype. For patients with ECOG PS 0 or 1 score, the hazard ratio for death of carrying AG or GG genotype was 0.57 (95% CI=0.35-0.93, P=0.025) (Supplementary Table 2, Figure 1H and 1I).
Discussion
Platinum remains the commonly used drug for lung cancer patients, it is generally used in combination with other antitumor drugs, but the appearance of drug resistance leads to unsatisfied efficacy. Stable DNA adducts formed by cisplatin lead to DNA damage then mainly pursuit to nucleotide excision repair, finally result in cell apoptosis [6, 7]. Enhanced DNA repair contributes to platinum resistance, which compromises the chemotherapy efficacy and therefore influences clinical outcome [10].
Distribution of characteristics in lung cancer patients and prognosis analysis (n=348)
| Variables | Overall survival (OS) | Progression-free survival (PFS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MST (mo) | Death/Total | P1 | HR (95% CI) | P2 | MST (mo) | Progression/Total | P1 | HR (95% CI) | P2 | |
| Age (year) | ||||||||||
| ≤56 | 59.0 | 123/171 | 0.968 | Ref. | 33.2 | 139/171 | 0.078 | Ref. | ||
| >56 | 60.1 | 131/177 | 1.00 (0.78-1.27) | 0.968 | 50.3 | 137/177 | 0.81 (0.64-1.03) | 0.079 | ||
| Gender | ||||||||||
| Female | 56.7 | 55/78 | 0.971 | Ref. | 35.5 | 61/78 | 0.890 | Ref. | ||
| Male | 60.5 | 199/270 | 1.01 (0.75-1.36) | 0.971 | 42.4 | 215/270 | 0.98 (0.74-1.30) | 0.890 | ||
| Smoking status | ||||||||||
| Never smoker | 60.1 | 101/143 | 0.401 | Ref. | 35.1 | 114/143 | 0.541 | Ref. | ||
| Ever smoker | 59.0 | 153/205 | 1.11 (0.87-1.43) | 0.401 | 46.1 | 162/205 | 0.93 (0.73-1.18) | 0.541 | ||
| Family history of cancer | ||||||||||
| No | 59.1 | 242/332 | 0.646 | Ref. | 39.1 | 263/332 | 0.808 | Ref. | ||
| Yes | 52.9 | 12/16 | 1.15 (0.64-2.05) | 0.646 | 38.6 | 13/16 | 1.07 (0.61-1.87) | 0.808 | ||
| Histology | ||||||||||
| LUAD | 56.1 | 137/179 | 0.172 | Ref. | 29.0 | 150/179 | 0.014 | Ref. | ||
| LUSC | 63.1 | 98/142 | 0.78 (0.60-1.01) | 0.062 | 56.5 | 104/142 | 0.69 (0.54-0.89) | 0.004 | ||
| Othersa | 60.5 | 19/27 | 0.92 (0.57-1.49) | 0.740 | 31.3 | 22/27 | 0.94 (0.60-1.46) | 0.767 | ||
| TNM stage | ||||||||||
| Ⅰ/Ⅱ | 66.8 | 7/10 | 0.606 | Ref. | 54.3 | 8/10 | 0.729 | Ref. | ||
| III/IV | 58.8 | 247/338 | 1.22 (0.57-2.58) | 0.607 | 36.8 | 268/338 | 1.13 (0.56-2.29) | 0.729 | ||
| Eastern Cooperative Oncology Group Performance Status | ||||||||||
| 0-1 | 59.6 | 236/327 | 0.362 | Ref. | 41.4 | 258/327 | 0.516 | Ref. | ||
| >1 | 48.6 | 18/21 | 1.25 (0.77-2.02) | 0.363 | 22.2 | 18/21 | 1.17 (0.73-1.89) | 0.517 | ||
MST, median survival time; mo, month; HR, hazard ratio; CI, confidence interval; P1, P-value for log-rank test; P2, P-value for univariate Cox hazards regression analysis; Ref., reference; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma. P < 0.05 are indicated in bold text.
aOther carcinomas include adenosquamous carcinoma, large cell carcinoma, bronchogenic lung cancer, and mucoepidermoid carcinoma.
Association between single nucleotide polymorphisms (SNPs) and platinum-based chemotherapy prognosis (n=348)
| Gene/SNP | Model | Genotype | Overall survival (OS) | Progression-free survival (PFS) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Death/total | MST (month) | P1 | HR (95% CI) | P2 | Progression/total | MST (month) | P1 | HR (95% CI) | P2 | |||
| HMGB1 | ||||||||||||
| rs1045411 | Additive | GG | 161/221 | 59.1 | 0.127 | Ref. | 174/221 | 41.4 | 0.698 | Ref. | ||
| AG | 75/109 | 60.0 | 0.97 (0.74-1.28) | 0.850 | 84/109 | 35.1 | 1.04 (0.80-1.35) | 0.788 | ||||
| AA | 18/18 | 57.5 | 1.62 (0.99-2.65) | 0.054 | 18/18 | 54.1 | 1.21 (0.74-1.97) | 0.444 | ||||
| Dominant | GG | 161/221 | 59.1 | 0.796 | Ref. | 174/221 | 41.4 | 0.687 | Ref. | |||
| AG/AA | 93/127 | 60.0 | 1.06 (0.82-1.37) | 0.677 | 102/127 | 35.8 | 1.06 (0.83-1.36) | 0.621 | ||||
| Recessive | AA | 18/18 | 57.5 | 0.045 | Ref. | 18/18 | 54.1 | 0.404 | Ref. | |||
| AG/GG | 236/330 | 59.1 | 0.61 (0.38-0.99) | 0.046 | 258/330 | 36.8 | 0.84 (0.52-1.35) | 0.464 | ||||
| rs1412125 | Additive | TT | 133/182 | 58.1 | 0.998 | Ref. | 145/182 | 36.6 | 0.495 | Ref. | ||
| CT | 104/143 | 59.1 | 0.99 (0.76-1.28) | 0.910 | 114/143 | 42.3 | 1.01 (0.79-1.29) | 0.947 | ||||
| CC | 17/23 | 62.1 | 1.00 (0.60-1.67) | 0.990 | 17/23 | 62.1 | 0.76 (0.46-1.26) | 0.293 | ||||
| Dominant | TT | 133/182 | 58.1 | 0.950 | Ref. | 145/182 | 36.6 | 0.812 | Ref. | |||
| CT/CC | 121/166 | 60.4 | 0.99 (0.77-1.27) | 0.922 | 131/166 | 45.6 | 0.97 (0.76-1.23) | 0.788 | ||||
| Recessive | CC | 17/23 | 62.1 | 0.991 | Ref. | 17/23 | 62.1 | 0.238 | Ref. | |||
| CT/TT | 237/325 | 58.5 | 0.99 (0.61-1.62) | 0.969 | 259/325 | 36.6 | 1.32 (0.80-2.15) | 0.275 | ||||
| rs2249825 | Additive | CC | 193/264 | 58.4 | 0.599 | Ref. | 211/264 | 36.6 | 0.476 | Ref. | ||
| GC | 53/76 | 61.1 | 0.95 (0.70-1.29) | 0.731 | 57/76 | 51.2 | 0.84 (0.62-1.13) | 0.246 | ||||
| GG | 8/8 | 64.1 | 1.38 (0.68-2.80) | 0.377 | 8/8 | 60.8 | 1.12 (0.55-2.27) | 0.762 | ||||
| Dominant | CC | 193/264 | 58.4 | 0.820 | Ref. | 211/264 | 36.6 | 0.297 | Ref. | |||
| GC/GG | 61/84 | 61.3 | 0.99 (0.74-1.33) | 0.942 | 65/84 | 54.1 | 0.87 (0.66-1.15) | 0.317 | ||||
| Recessive | GG | 8/8 | 64.1 | 0.375 | Ref. | 8/8 | 60.8 | 0.750 | Ref. | |||
| GC/CC | 246/340 | 59.0 | 0.72 (0.35-1.46) | 0.359 | 268/340 | 39.1 | 0.86 (0.43-1.75) | 0.686 | ||||
| REV3L | ||||||||||||
| rs462779 | Additive | CC | 63/90 | 61.4 | 0.018 | Ref. | 68/90 | 52.6 | 0.106 | Ref. | ||
| TC | 127/185 | 60.1 | 0.94 (0.69-1.27) | 0.671 | 141/185 | 36.6 | 1.01 (0.75-1.35) | 0.949 | ||||
| TT | 64/73 | 50.3 | 1.42 (1.00-2.02) | 0.050 | 67/73 | 27.8 | 1.30 (0.93-1.83) | 0.131 | ||||
| Dominant | CC | 63/90 | 61.4 | 0.672 | Ref. | 68/90 | 52.6 | 0.481 | Ref. | |||
| TC/TT | 191/258 | 56.0 | 1.06 (0.79-1.41) | 0.706 | 208/258 | 36.0 | 1.09 (0.83-1.43) | 0.546 | ||||
| Recessive | TT | 64/73 | 50.3 | 0.005 | Ref. | 67/73 | 27.8 | 0.034 | Ref. | |||
| TC/CC | 190/275 | 60.5 | 0.67 (0.51-0.90) | 0.007 | 209/275 | 42.3 | 0.77 (0.59-1.02) | 0.073 | ||||
| rs465646 | Additive | AA | 164/219 | 60.0 | 0.423 | Ref. | 179/219 | 33.2 | 0.252 | Ref. | ||
| GA | 84/122 | 59.0 | 0.91 (0.69-1.18) | 0.469 | 91/122 | 54.5 | 0.85 (0.66-1.10) | 0.222 | ||||
| GG | 6/7 | 60.3 | 1.37 (0.60-3.12) | 0.451 | 6/7 | 45.6 | 1.06 (0.47-2.41) | 0.888 | ||||
| Dominant | AA | 164/219 | 60.0 | 0.393 | Ref. | 179/219 | 33.2 | 0.125 | Ref. | |||
| GA/GG | 90/129 | 59.1 | 0.93 (0.72-1.20) | 0.572 | 97/129 | 54.0 | 0.86 (0.67-1.11) | 0.250 | ||||
| Recessive | GG | 6/7 | 60.3 | 0.401 | Ref. | 6/7 | 45.6 | 0.719 | Ref. | |||
| GA/AA | 248/341 | 59.1 | 0.70 (0.31-1.60) | 0.401 | 270/341 | 39.1 | 0.89 (0.40-2.02) | 0.786 | ||||
| NFE2L2 | ||||||||||||
| rs6706649 | Additive | CC | 224/304 | 59.1 | 0.900 | Ref. | 243/304 | 41.1 | 0.873 | Ref. | ||
| CT | 29/43 | 61.1 | 0.91 (0.62-1.35) | 0.642 | 32/43 | 34.9 | 0.91 (0.63-1.33) | 0.635 | ||||
| TT | 1/1 | 64.0 | 1.01 (0.14-7.29) | 0.992 | 1/1 | 64.0 | 0.78 (0.11-5.64) | 0.808 | ||||
| Dominant | CC | 224/304 | 64.0 | 0.860 | Ref. | 243/304 | 64.0 | 0.909 | Ref. | |||
| CT/TT | 30/44 | 59.1 | 0.98 (0.14-7.04) | 0.981 | 33/44 | 39.1 | 1.26 (0.18-9.06) | 0.820 | ||||
| Recessive | TT | 1/1 | 64.0 | 0.860 | Ref. | 1/1 | 64.0 | 0.909 | Ref. | |||
| CT/CC | 253/347 | 59.1 | 0.98 (0.14-7.04) | 0.981 | 275/347 | 39.1 | 1.26 (0.18-9.06) | 0.820 | ||||
| rs6721961 | Additive | GG | 126/176 | 59.8 | 0.547 | Ref. | 138/176 | 41.4 | 0.811 | Ref. | ||
| TG | 101/140 | 58.5 | 1.08 (0.83-1.41) | 0.552 | 112/140 | 35.1 | 1.11 (0.87-1.43) | 0.399 | ||||
| TT | 27/32 | 60.1 | 1.27 (0.84-1.94) | 0.258 | 26/32 | 45.3 | 1.03 (0.68-1.57) | 0.891 | ||||
| Dominant | GG | 126/176 | 59.8 | 0.466 | Ref. | 138/176 | 41.4 | 0.567 | Ref. | |||
| TG/TT | 128/172 | 58.5 | 1.12 (0.87-1.44) | 0.378 | 138/172 | 35.8 | 1.10 (0.86-1.39) | 0.444 | ||||
| Recessive | TT | 27/32 | 60.1 | 0.311 | Ref. | 26/32 | 45.3 | 0.926 | Ref. | |||
| TG/GG | 227/316 | 59.0 | 0.81 (0.55-1.22) | 0.315 | 250/316 | 36.8 | 1.02 (0.68-1.53) | 0.927 | ||||
| rs35652124 | Additive | TT | 71/99 | 60.5 | 0.951 | Ref. | 74/99 | 39.0 | 0.675 | Ref. | ||
| CT | 124/165 | 58.5 | 1.06 (0.79-1.43) | 0.679 | 137/165 | 39.1 | 1.17 (0.88-1.55) | 0.289 | ||||
| CC | 59/84 | 58.4 | 1.02 (0.72-1.44) | 0.917 | 65/84 | 40.1 | 1.07 (0.77-1.50) | 0.693 | ||||
| Dominant | TT | 71/99 | 60.5 | 0.799 | Ref. | 74/99 | 39.0 | 0.460 | Ref. | |||
| CT/CC | 183/249 | 58.5 | 1.05 (0.80-1.38) | 0.735 | 202/249 | 39.1 | 1.13 (0.87-1.48) | 0.360 | ||||
| Recessive | CC | 59/84 | 58.4 | 0.931 | Ref. | 65/84 | 40.1 | 0.848 | Ref. | |||
| CT/TT | 195/264 | 60.3 | 1.02 (0.76-1.37) | 0.892 | 211/264 | 39.1 | 1.03 (0.78-1.36) | 0.835 | ||||
MST, median survival time; HR, hazard ratio; CI, confidence interval; P1, P-value for log-rank test; P2, P-value for multivariate Cox hazards regression with adjustment for age, gender, and histology. P ≤ 0.05 are indicated in bold text.
Additive model: comparison between minor allele subjects and major allele subjects.
Dominant model: comparison between minor allele carriers and major homozygous subjects.
Recessive model: comparison between major allele carriers and minor homozygous subjects.
Polymorphism is one of the factors that may affect prognosis, and variants of genes of drug transporters, metabolic enzymes, DNA repair system, apoptosis pathway, and folate metabolism pathway are the most studied biomarkers for predicting platinum-based chemotherapy response, while more meaningful markers still need to be discovered [10].
In the present study, we focused on the genetic alterations of HMGB1, REV3L, and NFE2L2, a total of eight SNPs, to study the prognostic effect of platinum-based chemotherapy. Among them, REV3L rs462779 and HMGB1 rs1045411 were significantly associated with overall survival; none of them was significantly associated with progression-free survival. In male patients, patients who never smoked, or patients with younger age (≤56), no family history of cancer, adenocarcinoma, advanced tumor (stage III or IV), or ECOG PS 0-1, patients carrying TC or CC genotype in rs462779 had better overall survival when compared with TT genotype. Patients carrying AG or GG genotype in rs1045411 had better overall survival in lung cancer patients with advanced stage (stage III or IV) or ECOG PS 0-1. In terms of tumor progression, carrying TC or TT genotype in REV3L rs462779 had a lower risk of lymph node metastasis when compared with CC genotype, while carrying AG or AA genotype in HMGB1 rs1045411 or carrying GC or GG genotype in HMGB1 rs2249825 were significantly related to early T stage (T1 or T2).
REV3L is an important DNA polymerase involved in DNA replication, repair, recombination, and has an increased mRNA expression in non-small cell lung cancer tissue, the C-terminal portion encompasses conserved DNA polymerase domain, and the N-terminal domain contains residues direct contacting to DNA [20, 31-33]. Pol zeta plays a major extender role in translesion DNA synthesis (TLS) after DNA lesions due to its capability to extend mismatched or distorted primer templates of sorts, the lesions were directly bypassed by specialized DNA polymerases, such as DNA polymerases zeta [34, 35]. Loss expression of REV3L increases the frequency of chromosome translocation and break, thus results in genomic instability, and it acts on as a tumor suppressor because of inhibition of spontaneous tumor formation [31, 35, 36]. REV3L rs462779 is a nonsynonymous SNP (p.Thr1224Ile) that may influence protein function, REV3L rs465646 alters the microRNA binding site in the 3´-UTR, thus may affect REV3L expression [24].
Genetic variations in REV3L have been reported that it was associated with tumor risk or survival in multiple kinds of tumors. Patients carrying the TC or TT genotype in rs462779 had significantly increased colorectal cancer risk, and REV3L rs462779 and RAD18 rs373572 seemed to have a strong cumulative relation with CRC risk, while carrying the TC or CC genotype in rs465646 had significantly decreased lung cancer risk [25, 37]. Carrying the TC or CC genotype in rs462779 or carrying the AG or GG genotype in rs465646 was associated with good overall survival in platinum-treated malignant mesothelioma patients (n=139), the TC or CC genotype in rs462779 had poor overall survival in osteosarcoma patients (n=66), the TC or CC genotype in rs462779 had poor event-free survival in aggressive breast cancer patients (n=738) [24, 38, 39].
Association between single nucleotide polymorphisms (SNPs) and lung cancer progression (n=305)
| Gene/SNP | Genotype | T stage | N stage | M stage | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1/T2 | T3/T4 | OR (95% CI) | P | N0 | N1/N2/N3 | OR (95% CI) | P | M0 | M1 | OR (95% CI) | P | ||
| HMGB1 | |||||||||||||
| rs1045411 | GG | 59 | 130 | Ref. | 39 | 150 | Ref. | 51 | 138 | Ref. | |||
| AG/AA | 57 | 59 | 0.47 (0.29-0.76) | 0.002 | 16 | 100 | 1.63 (0.86-3.07) | 0.134 | 37 | 79 | 0.79 (0.48-1.31) | 0.358 | |
| rs1412125 | TT | 58 | 100 | Ref. | 35 | 123 | Ref. | 45 | 113 | Ref. | |||
| CT/CC | 58 | 89 | 0.89 (0.56-1.41) | 0.622 | 20 | 127 | 1.81 (0.99-3.30) | 0.054 | 43 | 104 | 0.96 (0.59-1.58) | 0.882 | |
| CC | 6 | 11 | Ref. | 2 | 15 | Ref. | 3 | 14 | Ref. | ||||
| CT/TT | 110 | 178 | 0.88 (0.32-2.45) | 0.811 | 53 | 235 | 0.59 (0.13-2.66) | 0.494 | 85 | 203 | 0.51 (0.14-1.83) | 0.302 | |
| rs2249825 | CC | 79 | 150 | Ref. | 45 | 184 | Ref. | 65 | 164 | Ref. | |||
| GC/GG | 37 | 39 | 0.56 (0.33-0.94) | 0.028 | 10 | 66 | 1.61 (0.77-3.39) | 0.205 | 23 | 53 | 0.91 (0.52-1.61) | 0.754 | |
| REV3L | |||||||||||||
| rs462779 | CC | 28 | 48 | Ref. | 7 | 69 | Ref. | 18 | 58 | Ref. | |||
| TC/TT | 88 | 141 | 0.94 (0.55-1.60) | 0.805 | 48 | 181 | 0.38 (0.17-0.89) | 0.025 | 70 | 159 | 0.71 (0.39-1.28) | 0.252 | |
| TT | 30 | 38 | Ref. | 16 | 52 | Ref. | 20 | 48 | Ref. | ||||
| TC/CC | 86 | 151 | 1.39 (0.80-2.40) | 0.242 | 39 | 198 | 1.56 (0.81-3.01) | 0.183 | 68 | 169 | 1.04 (0.57-1.87) | 0.908 | |
| rs465646 | AA | 77 | 118 | Ref. | 34 | 161 | Ref. | 50 | 145 | Ref. | |||
| GA/GG | 39 | 71 | 1.19 (0.73-1.93) | 0.486 | 21 | 89 | 0.90 (0.49-1.64) | 0.718 | 38 | 72 | 0.65 (0.39-1.09) | 0.100 | |
| NFE2L2 | |||||||||||||
| rs6706649 | CC | 99 | 165 | Ref. | 46 | 218 | Ref. | 76 | 188 | Ref. | |||
| CT/TT | 17 | 24 | 0.85 (0.43-1.65) | 0.627 | 9 | 32 | 0.75 (0.34-1.68) | 0.484 | 12 | 29 | 0.98 (0.47-2.01) | 0.950 | |
| rs6721961 | GG | 64 | 88 | Ref. | 33 | 119 | Ref. | 44 | 108 | Ref. | |||
| TG/TT | 52 | 101 | 1.41 (0.89-2.25) | 0.145 | 22 | 131 | 1.65 (0.91-2.99) | 0.098 | 44 | 109 | 1.01 (0.62-1.66) | 0.971 | |
| TT | 13 | 18 | Ref. | 7 | 24 | Ref. | 8 | 23 | Ref. | ||||
| TG/GG | 103 | 171 | 1.20 (0.56-2.55) | 0.637 | 48 | 226 | 1.37 (0.56-3.37) | 0.489 | 80 | 194 | 0.84 (0.36-1.97) | 0.693 | |
| rs35652124 | TT | 38 | 52 | Ref. | 19 | 71 | Ref. | 24 | 66 | Ref. | |||
| CT/CC | 78 | 137 | 1.28 (0.78-2.12) | 0.330 | 36 | 179 | 1.33 (0.72-2.47) | 0.367 | 64 | 151 | 0.86 (0.49-1.49) | 0.586 | |
| CC | 26 | 41 | Ref. | 12 | 55 | Ref. | 24 | 43 | Ref. | ||||
| CT/TT | 90 | 148 | 1.04 (0.60-1.82) | 0.883 | 43 | 195 | 0.99 (0.49-2.01) | 0.976 | 64 | 174 | 1.52 (0.85-2.70) | 0.156 | |
OR, odds ratio; CI, confidence interval; P, P-value for binary logistic regression analysis; Ref., reference. P < 0.05 are indicated in bold text.
Kaplan-Meier plots of stratified analysis of lung cancer patients with rs462779 (A-G) or rs1045411 (H-I) in recessive model. A, Overall survival of patients with age ≤56 years old. B, Overall survival of male lung cancer patients. C, Overall survival of lung cancer patients who had never smoked. D, Overall survival of lung cancer patients without a family history of cancer. E, Overall survival of lung cancer patients with adenocarcinoma. F, Overall survival of lung cancer patients with an advanced tumor (stage III or IV). G, Overall survival of lung cancer patients with Eastern Cooperative Oncology Group Performance Status 0 or 1. H, Overall survival of lung cancer patients with an advanced tumor (stage III or IV). I, Overall survival of lung cancer patients with Eastern Cooperative Oncology Group Performance Status 0 or 1.
Furthermore, REV3L rs462779 showed a slight association with severe toxicity (thrombocytopenia) in NSCLC patients treated with platinum-based chemotherapy [40]. Nonetheless, there is no report about the association between OS or PFS and polymorphisms in the platinum resistance-related REV3L gene. In our research, carrying TC or CC genotype in rs462779 had good overall survival in lung cancer patients (n=348), but no SNP was significantly associated with progression-free survival. It perhaps due to the differences in sample size and tumor type compared with previous studies. Besides, patients carrying TC or TT genotype had a lower risk of lymph node metastasis. However, the mechanisms of how rs462779 influences overall survival and lymph node metastasis still need further investigation.
HMGB1 plays nuclear factor role or extracellular signaling molecule role during cell migration and tumor metastasis. Overexpression of HMGB1 induced by chemotherapy or radiotherapy was associated with all hallmarks of cancer, which leads to tumor microenvironment disorder [12, 13, 41]. Recent studies also show that HMGB1 is involved in positive regulation and maintenance of ferroptosis in acute myeloid leukemia (AML) cells and autophagy in thyroid cancer cells, also prevents necroptosis in AML cells [42-44]. On the aspect of cancer risk, rs1045411 was significantly associated with susceptibility to hepatocellular carcinoma, oral squamous cell carcinoma, lung cancer, breast cancer, and urothelial cell carcinoma [15-17, 45, 46]. In terms of association of genetic variants of HMGB1 and prognosis, previous reports showed that carrying AG or AA genotype of rs1045411 in HMGB1 gene was significantly associated with good overall survival in gastric cancer patients [14]. HMGB1 rs1045411 is located in the 3'UTR, and was reported to decrease HMGB1 expression through hsa-miR-505-5p [45, 47]. HMGB1 rs1045411 C/T heterozygous polymorphism was associated with a significantly lower ratio of developing EGFR mutation in the smoking population; it may be a protective factor in lung adenocarcinoma [47]. In our study, carrying AG or GG genotype in rs1045411 was significantly associated with good overall survival. Moreover, rs1045411 and rs2249825 were significantly related to the early T stage.
There are also some limitations in our study. The recruitment hospitals are located in the same region, and the multi-central clinical studies may be needed to overcome results bias. It is a retrospective study with the sample size not large enough, therefore prospective or other analogous studies are warranted to validate our results. Thus we are enrolling other patients and independent validation will be done in our next studies. The underlying mechanism of rs462779 relating with cancer prognosis still needs further investigation.
In conclusion, our study suggested that REV3L rs462779 was significantly associated with overall survival and lymph node metastasis in lung cancer patients with platinum-based chemotherapy, and HMGB1 rs1045411 was related to overall survival and T stage. Genotypes of REV3L rs462779 and HMGB1 rs1045411 may be biomarkers for predicting platinum-based chemotherapy prognosis in lung cancer patients.
Supplementary Material
Supplementary tables.
Acknowledgements
Funding
This work was supported by the National Key Research and Development Program of China (2016YFC1306900), National Natural Science Foundation of China (81573508, 81874327, 81803640), The Strategy-Oriented Special Project of Central South University in China (ZLXD2017003), and Youth Science Foundation of Xiangya Hospital (2017Q02).
Human rights statement and informed consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424
2. Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel). 2018 10(8)
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7-34
4. Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer. 2016;16(8):525-37
5. Hirsch FR, Scagliotti GV, Mulshine JL. et al. Lung cancer: current therapies and new targeted treatments. The Lancet. 2017;389(10066):299-311
6. Bonanno L, Favaretto A, Rosell RJAr. Platinum drugs and DNA repair mechanisms in lung cancer. Anticancer research. 2014;34(1):493-501
7. Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364-78
8. Shen DW, Pouliot LM, Hall MD. et al. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev. 2012;64(3):706-21
9. Xiong Y, Huang BY, Yin JY. Pharmacogenomics of platinum-based chemotherapy in non-small cell lung cancer: focusing on DNA repair systems. Med Oncol. 2017;34(4):48
10. Yin J-Y, Li X, Zhou H-H, Liu Z-Q. Pharmacogenomics of platinum-based chemotherapy sensitivity in NSCLC: toward precision medicine. Pharmacogenomics. 2016;17(12):1365-78
11. Sims GP, Rowe DC, Rietdijk ST. et al. HMGB1 and RAGE in inflammation and cancer. Annual review of immunology. 2009;28:367-88
12. Tang D, Kang R, Zeh HJ 3rd. et al. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799(1-2):131-40
13. Kang R, Zhang Q, Zeh HJ 3rd. et al. HMGB1 in cancer: good, bad, or both? Clin Cancer Res. 2013;19(15):4046-57
14. Bao G, Qu F, He L. et al. Prognostic Significance of Tag SNP rs1045411 in HMGB1 of the Aggressive Gastric Cancer in a Chinese Population. PLoS One. 2016;11(4):e0154378
15. Hu W, Liu PY, Yang YC. et al. Association of HMGB1 Gene Polymorphisms with Lung Cancer Susceptibility and Clinical Aspects. Int J Med Sci. 2017;14(12):1197-202
16. Wang B, Yeh CB, Lein MY. et al. Effects of HMGB1 Polymorphisms on the Susceptibility and Progression of Hepatocellular Carcinoma. Int J Med Sci. 2016;13(4):304-9
17. Huang BF, Tzeng HE, Chen PC. et al. HMGB1 genetic polymorphisms are biomarkers for the development and progression of breast cancer. Int J Med Sci. 2018;15(6):580-6
18. Wang Y, Li XP, Yin JY. et al. Association of HMGB1 and HMGB2 genetic polymorphisms with lung cancer chemotherapy response. Clin Exp Pharmacol Physiol. 2014;41(6):408-15
19. Jiang M, Li X, Quan X. et al. Single Nucleotide Polymorphisms in HMGB1 Correlate with Lung Cancer Risk in the Northeast Chinese Han Population. Molecules. 2018 23(4)
20. Wittschieben JP, Patil V, Glushets V. et al. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010;70(7):2770-8
21. Lange SS, Bedford E, Reh S. et al. Dual role for mammalian DNA polymerase zeta in maintaining genome stability and proliferative responses. Proc Natl Acad Sci U S A. 2013;110(8):E687-96
22. Wang H, Zhang SY, Wang S. et al. REV3L confers chemoresistance to cisplatin in human gliomas: the potential of its RNAi for synergistic therapy. Neuro Oncol. 2009;11(6):790-802
23. Doles J, Oliver TG, Cameron ER. et al. Suppression of Rev3, the catalytic subunit of Pol{zeta}, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A. 2010;107(48):20786-91
24. Goričar K, Kovač V, Dolžan VJP. Polymorphisms in translesion polymerase genes influence treatment outcome in malignant mesothelioma. Pharmacogenomics. 2014;15(7):941-50
25. Pan J, Chi P, Lu X. et al. Genetic polymorphisms in translesion synthesis genes are associated with colorectal cancer risk and metastasis in Han Chinese. Gene. 2012;504(2):151-5
26. Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer. 2012;12(8):564-71
27. Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the Hallmarks of Cancer. Cancer Cell. 2018;34(1):21-43
28. Bi W, He CN, Li XX. et al. Ginnalin A from Kujin tea (Acer tataricum subsp. ginnala) exhibits a colorectal cancer chemoprevention effect via activation of the Nrf2/HO-1 signaling pathway. Food Funct. 2018;9(5):2809-19
29. Almeida M, Soares M, Ramalhinho AC. et al. Prognosis of hormone-dependent breast cancer seems to be influenced by KEAP1, NRF2 and GSTM1 genetic polymorphisms. Mol Biol Rep. 2019;46(3):3213-24
30. Gao L, Yuan F, Che G. et al. Epigenetic modifications but not genetic polymorphisms regulate KEAP1 expression in colorectal cancer. J Cell Biochem. 2019;120(8):12311-20
31. Martin SK, Wood RD. DNA polymerase zeta in DNA replication and repair. Nucleic Acids Res. 2019;47(16):8348-61
32. Makarova AV, Burgers PM. Eukaryotic DNA polymerase zeta. DNA Repair (Amst). 2015;29:47-55
33. Chen X, Zhu H, Ye W. et al. MicroRNA29a enhances cisplatin sensitivity in non-small cell lung cancer through the regulation of REV3L. Mol Med Rep. 2019;19(2):831-40
34. Burgers PMJC. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107(4):218-27
35. Sharma S, Canman CE. REV1 and DNA polymerase zeta in DNA interstrand crosslink repair. Environ Mol Mutagen. 2012;53(9):725-40
36. Raeder SB, Nepal A, Bjoras KO. et al. APIM-Mediated REV3L(-)PCNA Interaction Important for Error Free TLS Over UV-Induced DNA Lesions in Human Cells. Int J Mol Sci. 2018 20(1)
37. Zhang S, Chen H, Zhao X. et al. REV3L 3'UTR 460 T>C polymorphism in microRNA target sites contributes to lung cancer susceptibility. Oncogene. 2013;32(2):242-50
38. Varadi V, Bevier M, Grzybowska E. et al. Genetic variation in genes encoding for polymerase zeta subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Res Treat. 2011;129(1):235-45
39. Goricar K, Kovac V, Jazbec J. et al. Translesion polymerase genes polymorphisms and haplotypes influence survival of osteosarcoma patients. OMICS. 2015;19(3):180-5
40. Zheng Y, Deng Z, Yin J. et al. The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy. International journal of cancer. 2017;141(11):2336-47
41. Fu Y, Sun L-Q, Huang Y. et al. miR-142-3p Inhibits the Metastasis of Hepatocellular Carcinoma Cells by Regulating HMGB1 Gene Expression. Current molecular medicine. 2018;18(3):135-41
42. Ye F, Chai W, Xie M. et al. HMGB1 regulates erastin-induced ferroptosis via RAS-JNK/p38 signaling in HL-60/NRASQ61L cells. American journal of cancer research. 2019;9(4):730
43. Chai W, Ye F, Zeng L. et al. HMGB1-mediated autophagy regulates sodium/iodide symporter protein degradation in thyroid cancer cells. J Exp Clin Cancer Res. 2019;38(1):325
44. Liu Y, Chen P, Xu L. et al. Extracellular HMGB1 prevents necroptosis in acute myeloid leukemia cells. Biomedicine & Pharmacotherapy. 2019;112:108714
45. Lin C-W, Chou Y-E, Yeh C-M. et al. A functional variant at the miRNA binding site in HMGB1 gene is associated with risk of oral squamous cell carcinoma. Oncotarget. 2017;8(21):34630
46. Hung SC, Wang SS, Li JR. et al. Effect of HMGB1 Polymorphisms on Urothelial Cell Carcinoma Susceptibility and Clinicopathological Characteristics. Int J Med Sci. 2018;15(14):1731-6
47. Wu YL, Chien MH, Chou YE. et al. Association of EGFR mutations and HMGB1 genetic polymorphisms in lung adenocarcinoma patients. J Cancer. 2019;10(13):2907-14
Author contact
![]() Corresponding authors: Zhao-Qian Liu, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008; China; Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, Changsha 410078; China. Tel: +86 731 89753845, Fax: +86 731 82354476, E-mail: zqliuedu.cn or Juan Chen, Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008; China. E-mail: cj1028edu.cn
Corresponding authors: Zhao-Qian Liu, Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008; China; Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, Changsha 410078; China. Tel: +86 731 89753845, Fax: +86 731 82354476, E-mail: zqliuedu.cn or Juan Chen, Department of Pharmacy, Xiangya Hospital, Central South University, Changsha 410008; China. E-mail: cj1028edu.cn

 Global reach, higher impact
Global reach, higher impact