3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(17):3908-3913. doi:10.7150/jca.31615 This issue Cite
Research Paper
circGFRA1 Promotes Ovarian Cancer Progression By Sponging miR-449a
1. Department of Gynaecology and Obstetrics, The First Affiliated Hospital of University of South China, Hengyang, Hunan
2. Cancer Hospital and Cancer Research Institute, Guangzhou Medical University, Guangzhou, Guangdong
Received 2018-11-18; Accepted 2019-4-26; Published 2019-6-24
Abstract
Backgroud: Increasing studies show that circular RNAs (circRNAs) play important roles in tumor progression. However, the function of circRNAs in ovarian cancer is mostly unclear.
Methods: We detected the expression of circGFRA1 by quantitative real-time PCR (qRT-PCR) in 50 pairs of ovarian cancer tissues and adjacent normal tissues. Then, we explored the function of circGFRA1 in ovarian cancer progression, such as cell proliferation, apoptosis and invasion. Moreover, we performed luciferase reporter and RNA immunoprecipitation (RIP) assay to study the competing endogenous RNA (ceRNA) function of circGFRA1 in ovarian cancer progression.
Results: qRT-PCR showed that circGFRA1 was overexpressed in ovarian cancer tissues. Inhibition of circGFRA1 suppressed cell proliferation and invasion, but induced cell apoptosis in ovarian cancer. Luciferase reporter and RIP assay revealed that circGFRA1 could regulate the expression of GFRA1 by sponging miR-449a.
Conclusions: In summary, circGFRA1 regulated GFRA1 expression and ovarian cancer progression by sponging miR-449a. circGFRA1 could be a potential diagnostic biomarker and therapeutic target for ovarian cancer.
Keywords: circular RNAs, circGFRA1, miR-449a, GFRA1, competitive endogenous RNAs, ovarian cancer
Introduction
Ovarian cancer is a common malignant disease with poor survival in women worldwide. There are 295,414 new cases of ovarian cancer and 184,799 death in 2018 [1]. Due to the lack of effective biomarkers, many patients are diagnosed at advanced stage with poor prognosis. Exploring the molecular mechanism of ovarian cancer progression would help develop novel therapeutic targets.
Circular RNAs (circRNAs) are involved in cancer development [2]. In osteosarcoma, hsa_circ_0081001 serves as a diagnostic and prognostic biomarker [3]. In triple-negative breast cancer, circ-UBAP2 predicts poor prognosis and promotes cancer progression [4]. But the function of circRNAs in ovarian cancer is unclear.
circRNAs could act as miRNA sponges or competitive endogenous RNAs (ceRNAs) to regulate gene expression [5] and cancer progression. In gastric cancer, circRNA_001569 promotes cell proliferation by sponging miR-145 [6]. In bladder cancer, circPRMT5 promotes cancer metastasis via sponging miR-30c [7]. In triple negative breast cancer, circGFRA1 regulates the expression of GFRA1 via sponging miR-34a [8].
In this study, We found circGFRA1 was overexpressed in ovarian cancer. Downregulation of circGFRA1 inhibited cell proliferation and invasion, but induce apoptosis. Moreover, circGFRA1 could act as a sponge for miR-449a to regulate the expression of GFRA1 (GDNF family receptor alpha 1). circGFRA1 could be a potential diagnostic biomarker and therapeutic target for ovarian cancer.
Material and Methods
Cell lines and culture
Ovarian cancer cell lines OV119 and A2780 were obtained from ATCC (USA) and passaged within six months. The cells were cultured according to the manufacturer's instruction and short tandem repeat DNA profiling was performed before use.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted by TRIzol (Invitrogen, USA). Cytoplasmic and nuclear RNA were isolated by PARIS™ Kit (Invitrogen). qRT-PCR was conducted with SYBR Premix Ex Taq (Takara, Japan) and CFX96 Real-time PCR system (Bio-Rad, USA). Relative fold-change was calculated by the 2-ΔΔCt method. Primers were synthesized by Invitrogen (Supplementary Table 1).
CCK8 assay
Cells were seeded 24 hours before transfection. 48 hours after transfection, CCK-8 solution (10 μL) was added and cells were incubated for 2 hours before measuring the absorbance at 490 nM with Spectra Max 250 spectrophotometer (Molecular Devices, USA).
Apoptosis assay
Cells were transfected and apoptotic rate was detected. Annexin V/propidium iodide staining and flow cytometry were performed with Annexin V-fluorescein isothiocynate Apoptosis Detection Kit (KeyGen, China).
Transwell assay
Cells were seeded on the EC matrix (Millipore, Germany) and 20% fetal bovine serum was added in the lower chamber as an attractant. Non-invading cells were gently removed while invasive cells were fixed and stained, imaged and counted 48 hours later.
Mouse xenograft and metastasis assays
4-week old BALB/c nude mice (five per group to provied a power of 90% for a significance level of 0.05 with a two-tailed t test.) were subcutaneously injected with 2 × 106 OV119 cells then intratumorally injected with 40 μL si-NC or si-circGFRA1. For metastasis assays, OV119 cells were injected into nude mice (five per group to provied a power of 90% for a significance level of 0.05 with a two-tailed t test.) through tail vein. 4 weeks later the mice were sacrificed and lung metastatic nodules were counted.
Luciferase reporter assay
Luciferase reporter vector with the 3′-UTR of GFRA1 or circGFRA1 were constructed. Cells were co-transfected with luciferase reporter vector and miR-449a mimics by Lipofectamine 2000 (Invitrogen). 48 hours after incubation, the luciferase activities were detected by dual-luciferase reporter assay (Promega, USA).
RNA immunoprecipitation (RIP) assay
Cells were transfected with MS2bs-circGFRA1, MS2bs-circGFRA1mt (the miR-449a binding site was mutant) or control MS2bs-Rluc together with MS2bp-GFP. RIP was conducted by Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) 48 hours later.
Western blot analysis
Briefly, proteins were separated by 10% SDS-PAGE and transferred to PVDF membrane (Millipore) then incubated with 5% skim milk powder. Then incubated with antibody against GFRA1 (1:1000, CST, USA) and secondary antibody (1:3000, CST). Anti-β- actin antibody (1:1000, Affinity, USA) served as a control.
Statistical analysis
Statistical analysis was performed with SPSS 19.0 software. Comparisons between groups were analyzed with t tests and χ2 tests. Unless otherwise indicated, data are presented as the mean ± s.e.m. of triplicate independent experiments. P < 0.05 was considered statistically significant.
Results
circGFRA1 is overexpressed and promotes ovarian cancer progression in vitro
To detect the expression of circGFRA1 in ovarian cancer, we performed qRT-PCR on 50 pairs of ovarian cancer tissues and adjacent normal tissues. And circGFRA1 was found overexpressed in ovarian cancer tissues (Figure 1A). Due to circGFRA1 was overexpressed, we knockdowned the expression of circGFRA1 to explore its function in ovarian cancer. qRT-PCR indicated that the knockdown was successful (Figure 1B). CCK-8 assay showed that knockdown of circGFRA1 significantly inhibited cell proliferation (Figure 1C). Apoptosis assay revealed that knockdown of circGFRA1 promoted cell apoptosis (Figure 1D). Transwell assay indicated that knockdown of circGFRA1 inhibited cell invasion (Figure 1E). All these findings indicate that knockdown of circGFRA1 suppresses ovarian cancer progression in vitro.
circGFRA1 promotes ovarian cancer progression in vivo
To further explore the function of circGFRA1 in vivo, we performed mouse xenograft experiments. Knockdown of circGFRA1 caused a significant decrease of tumor proliferation (Figure 2A). Moreover, lung metastatic nodules decreased after knockdown of circGFRA1 (Figure 2B). All these findings indicate that knockdown of circGFRA1 suppresses ovarian cancer progression in vivo.
circGFRA1 is overexpressed and promotes ovarian cancer progression in vitro A. The expression of circGFRA1 in 50 pairs of ovarian cancer tissues (Tumor) and adjacent normal tissues (Normal). B. qRT-PCR was conducted after cell transfection. C. Cells were transfected and CCK8 assay was conducted. D. Apoptosis assay was performed 48 hours after transfection. E. Transwell assay was performed. Original magnification, ×200. **P < 0.01
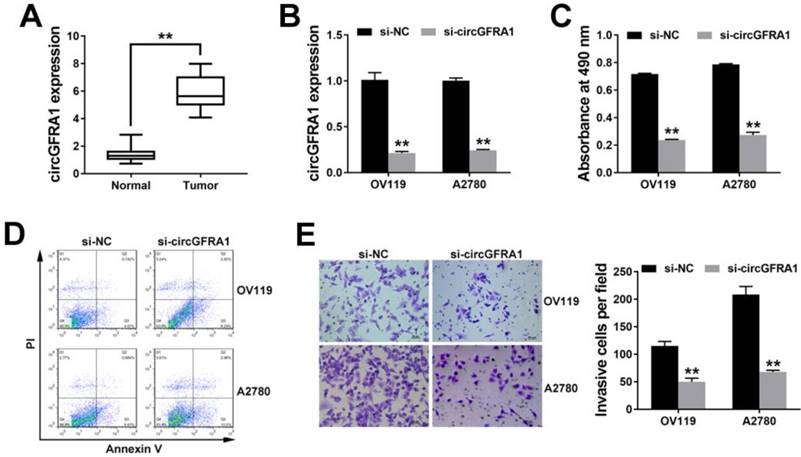
circGFRA1 promotes ovarian cancer progression in vivo A. Mouse xenograft tumors (left). The weight of tumors was summarized (right). B. HE stained lung metastatic nodules (left). The number of metastatic nodules was summarized (right). *P < 0.05, **P < 0.01
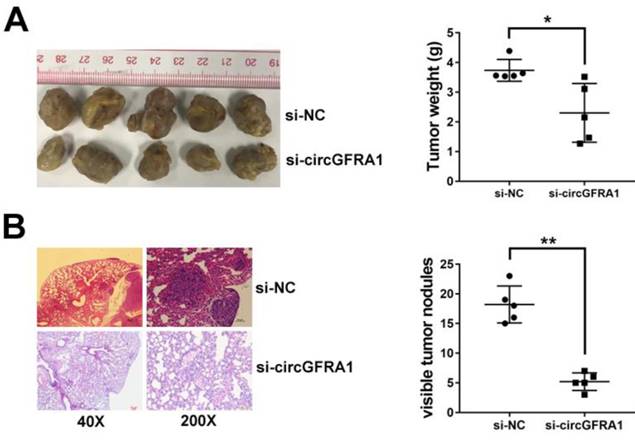
circGFRA1 acts as a decoy for miR-449a
We detected the intracellular location of circGFRA1 in ovarian cancer cell and found that circGFRA1 mainly located in cytoplasm (Figure 3A). Thus, we explored the potential circRNA/miRNA interaction by Arraystars's home-made miRNA target prediction software. Binding site of miR-449a was found within circABCC2 sequence (Figure 3B). Therefore, we detected the expression of miR-449a in ovarian cancer tissues and adjacent normal tissues and found it downregulated in ovarian cancer (Figure 3C). Next, we conducted luciferase reporter assays to explore whether miR-449Aa could bind to circGFRA1. The luciferase intensity decreased when co-transfected with luciferase reporters and miR-449a mimics (Figure 3D). To confirm the direct binding between circGFRA1 and miR-449a, MS2bp-MS2bs based RIP assay was performed. We found that miR-449a mainly enriched in MS2bs-circGFRA1 group, indicating the specific binding of circGFRA1 and miR-449a (Figure 3E). Taken together, circGFRA1 could directly interact with miR-449a and act as a decoy for miR-449a.
circGFRA1 regulates GFRA1 via miR-449a
Next, we continued to find target genes of miR-449a by TargetScan and found GFRA1 (Figure 4A). And GFRA1 was also overexpressed in ovarian cancer tissues (Figure 4B). Luciferase reporter assay showed that the luciferase activity decreased when co-transfected with miR-449 mimics and luciferase reporter (Figure 4C). And miR-449a could suppress the expression of GFRA1 (Figure 4D & 4E), indicating that GFRA1 was a downstream target of miR-449a.
Therefore, we performed RIP assay on Ago2 to explore the ceRNA function of circGFRA1. We found that circGFRA1, GFRA1 and miR-449a were all enriched to Ago2 (Figure 4F). Moreover, knockdown of circGFRA1 decreased the enrichment of Ago2 to circGFRA1, but increased that to GFRA1 (Figure 4G), indicating that circGFRA1 could function as a miRNA decoy. Thus, we investigated the expression of GFRA1 after inhibition of circGFRA1. We found that that expression of GFRA1 was suppressed, but inhibition of miR-449a could reverse the suppression (Figure 4H). All these results indicate that circGFRA1 could regulate GFRA1 expression by decoying miR-449a.
Discussion
Recently it has been reported that circRNAs play vital roles in cancer progression [9]. In prostate cancer, circRNA circ-102004 promotes cancer development and progression [10]. In clear cell renal cell carcinoma, knockdown of hsa_circ_0001451 promotes tumor growth [11]. In non-small cell lung cancer, circPTK2 inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis [12]. However, there are only a few reports exploring circRNAs in ovarian cancer. Liu N et al found that circHIPK3 was upregulated and associated with poor prognosis of epithelial ovarian cancer [13]. Ning L et al revealed that circEXOC6B and circN4BP2L2 may be used as prognostic biomarkers for epithelial ovarian cancer [14].
Here, we found that circGFRA1 was overexpressed in ovarian cancer. Subsequent experiments revealed that inhibition of circGFRA1 suppressed cell proliferation and invasion, and induced cell apoptosis, indicating that circGFRA1 could regulate ovarian cancer progression. Thus, circGFRA1 could be a potential diagnostic biomarker and therapeutic target for ovarian cancer.
circGFRA1 acts as a decoy for miR-449a A. The expression of U6, GAPDH and circGFRA1 were detected. B. The predicted binding site of miR-449a in circGFRA1. C. The expression of miR-449a in 50 pairs of ovarian cancer tissues (Tumor) and adjacent normal tissues (Normal). D. Luciferase reporter assay in cells co-transfected with miR-449a mimics and luciferase reporter. E. RIP assay was conducted after cell transfection. **P < 0.01
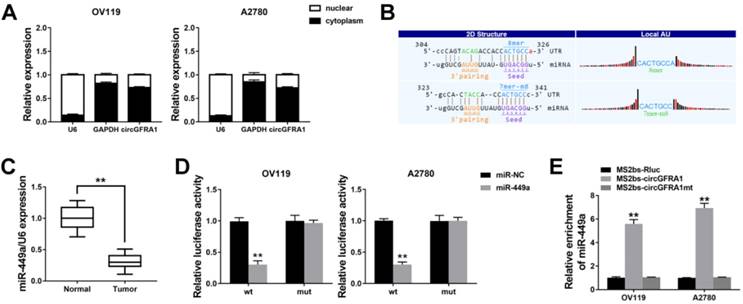
circGFRA1 regulates GFRA1 via miR-449a A. The predicted binding site of miR-449a within GFRA1 3'UTR. B. The expression of GFRA1 in 50 pairs of ovarian cancer tissues (Tumor) and adjacent normal tissues (Normal). C. Luciferase reporter assay in cells co-transfected with miR-449a mimics and luciferase reporter. D. The expression of GFRA1 was detected by qRT-PCR. E. GFRA1 expression was detected by western blot. F. RIP assay revealed the enrichment of circGFRA1, GFRA1 and miR-449a on Ago2. G. RIP assay on Ago2 was conducted. H. GFRA1 expression was detected by qRT-PCR. **P < 0.01
circRNAs have been reported to sponge miRNA to regulate cancer progression [15]. In hepatocellular carcinoma, hsa_circ_101280 promotes cell proliferation and suppressed apoptosis by sponging miR-375 [16]. In colorectal cancer, hsa_circRNA_103809 regulates cell proliferation and migration via miR-532-3p [17]. In breast cancer, hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p [18]. In ovarian cancer, circ-ITCH suppresses cancer progression by targeting miR-145/ RASA1 signaling [19]. Here, we found that circGFRA1 could bind to miR-499a and acted as a sponge for miR-499a to regulate ovarian cancer progression.
miR-499a is a known tumor suppressor that suppresses cell proliferation and promotes apoptosis [20]. It has been reported that miR-499a suppressed tumor progression in multiple cancers, such as gastric cancer [21], bladder cancer [22] and prostate cancer [23]. In hepatocellular carcinoma, miR-449a was reported to suppress epithelial-mesenchymal transition and metastasis [24]. Here, we revealed that miR-449a was downregulated in ovarian cancer.
Moreover, we found that GFRA1 was a downstream target of miR-449a. GFRA1 was overexpressed in ovarian cancer. GFRA1 has been implicated in the regulation of cancer progression. In breast cancer, high expression of GFRA1 was adversely correlated with patients outcome [25, 26]. And anti-GFRA1 antibody may provide a targeted therapeutic opportunity for breast cancer patients [27]. In osteosarcoma, Kim M et al reported that GFRA1 contributes to cisplatin induced chemoresistance by facilitating autophagy via SRC-AMPK signaling [28]. In this study, we found that circGFRA1 could regulate the expression of GFRA1 via sponging miR-449a. Knockdown of circGFRA1 suppressed the expression of GFRA1 and inhibition of miR-449a could reverse it. circGFRA1 regulates ovarian cancer progression partly through sponging miR-499a and regulating GFRA1 expression.
Conclusions
Taken together, we found that circGFRA1 is overexpressed in ovarian cancer. Knockdown of circGFRA1 suppresses cell proliferation and invasion, and induces cell apoptosis. And circGFRA1 could regulate the expression of GFRA1 through decoying miR-449a. circGFRA1 could be a potential diagnostic biomarker and therapeutic target for ovarian cancer.
Abbreviations
circRNAs: circular RNAs; ceRNA: competing endogenous RNA; qRT-PCR: quantitative real-time PCR; RIP: RNA immunoprecipitation; MS2bp: MS2- binding protein; MS2bs: MS2-binding sequences.
Supplementary Material
Supplementary tables.
Acknowledgements
This work was supported by funds from the National Natural Science Foundation of China (No. 81101526), Project of Educational Commission of Guangdong Province (No. 2016KTSCX115), Science and Technology Program of Guangzhou of Guangdong Province (No. 201707010354) and Natural Science Foundation of Guangdong Province (No. 2018B0303110015).
Ethics Committee Approval and Patient consent
This study was approved by Ethics Committees of Cancer Hospital and Cancer Research Institute, Guangzhou Medical University, and performed according to the Declaration of Helsinki. All patients' informed consents were obtained. Animal studies were approved by Institutional Animal Care and Use Committee (IACUC) of Cancer Hospital and Cancer Research Institute, Guangzhou Medical University and carried out according to the IACUC protocol.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018
2. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018;187:31-44
3. Kun-Peng Z, Chun-Lin Z, Jian-Ping H, Lei Z. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14:1513-20
4. Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y. et al. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun. 2018
5. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205-11
6. Shen F, Liu P, Li N, Yi Z, Tie X, Zhang Y. et al. CircRNA_001569 promotes cell proliferation through absorbing miR-145 in gastric cancer. J Biochem. 2018
7. Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L. et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clinical cancer research: an official journal of the American Association for Cancer Research. 2018
8. He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W. et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145
9. Yang Z, Xie L, Han L, Qu X, Yang Y, Zhang Y. et al. Circular RNAs: Regulators of Cancer-Related Signaling Pathways and Potential Diagnostic Biomarkers for Human Cancers. Theranostics. 2017;7:3106-17
10. Si-Tu J, Cai Y, Feng T, Yang D, Yuan S, Yang X. et al. Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int J Biol Macromol. 2018
11. Wang G, Xue W, Jian W, Liu P, Wang Z, Wang C. et al. The effect of Hsa_circ_0001451 in clear cell renal cell carcinoma cells and its relationship with clinicopathological features. J Cancer. 2018;9:3269-77
12. Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T. et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17:140
13. Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:3713-8
14. Ning L, Long B, Zhang W, Yu M, Wang S, Cao D. et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018
15. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Molecular cell. 2018;71:428-42
16. Cao S, Wang G, Wang J, Li C, Zhang L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol Cell Biol. 2018
17. Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun S. et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p / FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346-52
18. Zhang HD, Jiang LH, Hou JC, Zhong SL, Zhou SY, Zhu LP. et al. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother. 2018;107:1342-53
19. Hu J, Wang L, Chen J, Gao H, Zhao W, Huang Y. et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018
20. Lize M, Pilarski S, Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010;17:452-8
21. Li X, Li H, Zhang R, Liu J, Liu J. MicroRNA-449a inhibits proliferation and induces apoptosis by directly repressing E2F3 in gastric cancer. Cell Physiol Biochem. 2015;35:2033-42
22. Chen H, Lin YW, Mao YQ, Wu J, Liu YF, Zheng XY. et al. MicroRNA-449a acts as a tumor suppressor in human bladder cancer through the regulation of pocket proteins. Cancer Lett. 2012;320:40-7
23. Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H. et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714-24
24. Chen SP, Liu BX, Xu J, Pei XF, Liao YJ, Yuan F. et al. MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer. 2015;15:706
25. Fan TC, Yeo HL, Hsu HM, Yu JC, Ho MY, Lin WD. et al. Reciprocal feedback regulation of ST3GAL1 and GFRA1 signaling in breast cancer cells. Cancer Lett. 2018;434:184-95
26. Wu ZS, Pandey V, Wu WY, Ye S, Zhu T, Lobie PE. Prognostic significance of the expression of GFRalpha1, GFRalpha3 and syndecan-3, proteins binding ARTEMIN, in mammary carcinoma. BMC Cancer. 2013;13:34
27. Bhakta S, Crocker LM, Chen Y, Hazen M, Schutten MM, Li D. et al. An Anti-GDNF Family Receptor Alpha 1 (GFRA1) Antibody-Drug Conjugate for the Treatment of Hormone Receptor-Positive Breast Cancer. Mol Cancer Ther. 2018;17:638-49
28. Kim M, Jung JY, Choi S, Lee H, Morales LD, Koh JT. et al. GFRA1 promotes cisplatin-induced chemoresistance in osteosarcoma by inducing autophagy. Autophagy. 2017;13:149-68
Author contact
![]() Corresponding authors: Jing Xie, The First Affiliated Hospital of University of South China, Hengyang, Hunan, 421001, China, Email: Xj17781778com or Min Deng, Cancer Hospital and Cancer Research Institute, Guangzhou Medical University, Guangzhou, Guangdong, E-mail: dengmin2006edu.cn
Corresponding authors: Jing Xie, The First Affiliated Hospital of University of South China, Hengyang, Hunan, 421001, China, Email: Xj17781778com or Min Deng, Cancer Hospital and Cancer Research Institute, Guangzhou Medical University, Guangzhou, Guangdong, E-mail: dengmin2006edu.cn

 Global reach, higher impact
Global reach, higher impact