3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(5):1288-1296. doi:10.7150/jca.28529 This issue Cite
Research Paper
Lower Expression of Gelsolin in Colon Cancer and Its Diagnostic Value in Colon Cancer Patients
1. Laboratory Medicine Center, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, China
2. Department of Clinical Laboratory, Beijing University of Chinese Medicine Shenzhen Hospital, Shenzhen, Guangdong, China
3. Guangdong Key Laboratory of Biochip Technology, Southern Medical University, Guangzhou, Guangdong, China
4. Huayin Medical Laboratory Center Co., Ltd., Guangzhou, Guangdong, China
# These authors contributed equally to this work.
Received 2018-7-15; Accepted 2019-1-4; Published 2019-1-30
Abstract
Colon cancer is one of the most common malignancies causing the majority of cancer-related deaths. Gelsolin (GSN) has been found to be dysregulated in various cancers. However, the secreted GSN in colon cancer remains largely unknown. In the present study, we explored the expression profile of GSN in colon cancer tissues and the diagnostic value of serum GSN in colon cancer. In addition, the effects of secreted GSN in colon cancer cells were studied. We thus found that immunoreactive GSN levels were significantly lower in colon cancer tissues than those in non-tumor colon tissues. Functional studies demonstrated that secreted GSN could restrain cell invasion and migration in vitro. Mechanistically, dose dependent recombinant GSN down-regulated the expression of MMP2 and MMP9, which might restrain the process of cell invasion and migration. Furthermore, serum levels of GSN were significantly lower in colon cancer patients than those in healthy volunteers, and ROC curves showed serum level of GSN had a better diagnostic value for colon cancer (AUC=0.932) than the traditional tumor biomarker Carcinoembryonic Antigen (CEA) or Carbohydrate Antigen 19-9 (CA199). In conclusion, our results suggest that the secreted GSN restrains the invasion and migration of colon cancer cells. Meanwhile, the serum GSN may be a new biomarker for the diagnosis of colon cancer.
Keywords: colon cancer, diagnosis, gelsolin, metastasis
Introduction
Colon cancer is one of the most common types of solid malignancies, and is a primary cause of cancer-related mortality worldwide [1]. Most cases of colon cancer achieved better prognosis if diagnosed at early stage, for the five-year overall survival rate can be up to 80-90% [2]. However, the majority of patients present with advanced disease; and the survival rate is low when metastasis occurs [3]. Due to the numerous contributing factors in the development of colon cancer, the pathogenesis remains unclear. Therefore, the investigation of novel therapeutic strategies is a key focus in colon cancer research.
Gelsolin (GSN) is an actin-binding protein that serves to cleave and cap actin filaments to regulate cytoskeletal turnover and plays an important role in cell motility [4-6]. Besides, what's mentioned above, GSN is able to regulate cell morphology, proliferation, or even apoptosis as published data indicated that GSN was downregulated in several solid tumors such as hepatic carcinoma, colon carcinoma, gastric cancer, cervical cancer, and ovarian cancer [7-11]. Two forms of GSN protein, cytosolic GSN and secreted GSN, are transcribed from a single gene [12, 13]. Previous studies have shown that plasma GSN is a biomarker of inflammation [14]. However, the role of secreted GSN in colon cancer has not been examined.
Here we examined the expression of GSN in colon cancer specimens. In vitro, we explored the impact of secreted GSN on invasion and migration of colon cancer cells, and its underlying mechanism. Additionally, we examined the serum levels of GSN in colon cancer patients to explore whether serum GSN has the diagnostic value for colon cancer.
Materials and methods
Patients and specimens
Colon cancer tissues and corresponding adjacent non-tumor tissues were obtained from 36 patients who were diagnosed with colon cancer at Nanfang Hospital, Southern Medical University (Guangzhou, China). All the samples were previously processed following routine formalin fixation and paraffin embedding protocols, and then stored at -20℃ before used. Similarly, 3 pairs of colon cancer tissues and corresponding adjacent non-tumor tissues were collected and stored at liquid nitrogen before used. For the measurement of serum GSN levels in colon cancer patients and healthy controls, we obtained 90 cases of serum samples from the colon cancer patients and 51 cases of serum samples from healthy volunteers in Nanfang Hospital from August 2016 to January 2018. All the colon cancer patients were diagnosed by histological examination, and all the healthy volunteers were recruited without any health problems during health check-ups at Nanfang hospital. All serum samples were stored at -80℃ before further analysed. Ethics approvals were granted by the Ethics Committee of Nanfang Hospital (Guangzhou, China), with all methods carried out in accordance with the approved guidelines. Written informed consents were required for all patients and healthy volunteers enrolled into the study.
Cell culture
The colon cancer cell lines SW480, SW620, HT29, LoVo and the normal colonic epithelial cell line FHC were purchased from the Shanghai Cell Bank of the China Academy of Sciences (Shanghai, China). Cells were maintained at 37°C in a humidified incubator containing 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM, Gibco, Gaithersburg, MD, USA) or Roswell Park Memorial Institute 1640 (RPMI-1640, Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (Biowest, Nuaillé, France).
Reagents
GSN siRNA (si-GSN) and control siRNA (si-NC) was designed from RiboBio (Ribobio, Guangzhou, China). Recombinant GSN was purchased from Cytoskeleton, Inc (Denver, CO, USA). Neutralizing antibody of GSN was purchased from R&D Systems (Minneapolis, MN, USA). Mouse anti-GSN was purchased from Sigma (Sigma-Aldrich, Burlington, USA), rabbit anti-MMP2 and rabbit anti-MMP9 were purchased from Affinity (Affinity Biosciences, OH, USA), mouse anti-GAPDH was purchased from GNI (GNI, Tokyo, Japan).
Immunohistochemistry (IHC)
Formalin-fixed and paraffin-embedded tissues were examined immunohistochemically using a monoclonal antibody directed against human GSN (Sigma-Aldrich, Burlington, USA) following the avidin-biotin complex method, as described previously [15]. Five pictures of each section were randomly captured and graded by two experienced pathologists. The intensity was assessed on a scale of 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellowish brown) and 3 (strong staining, brown). The extent of GSN staining was defined as the percentage of positive cells in total cells and was scored on a scale of 0 (0% - 1%), 1 (1% - 10%), 2 (10% - 50%) and 3 (50% - 100%). An overall protein expression score (overall score range, 0 - 9) was calculated by multiplying the intensity and positivity scores.
Cell migration and invasion assays
Uncoated costar transwells (Corning Costar Co., Corning, NY) were used for migration assays and Matrigel-coated transwells (BD Biosciences, Franklin Lakes, NJ) used for invasion assays. Cells were serum starved overnight and then seeded into the upper chamber with serum-free DMEM medium. DMEM supplemented with 10% FBS was added into the lower chamber. Cells that had migrated across the transwell membrane were stained and quantified. The ability of cells to migrate was monitored at different time points using a light microscopy and take pictures.
Measurement of serum GSN
Serum samples were prepared by centrifugation according to standard protocols, aliquoted and stored at -80°C until assayed. The levels of GSN were quantitatively determined using commercial enzyme-linked immunosorbent assays (ELISA) kits (Cusabio, Wuhan, China), according to the manufacturer's instructions. The detection limit for GSN is 62.5 pg/mL to 4000 pg/mL.
RNA isolation and qRT-PCR
RNA was extracted from cultured cells or human tissue using TRIzol Reagent (Takara, Dalian, China). For mRNAs, cDNA was generated by using the PrimeScript RT reagent kit (Takara). The RNA expression levels were measured by qRT-PCR using SYBR Green PCR Master Mix (Takara), which was performed on the Roch LightCycler® 480 Real Time PCR system (Roche, Basel, Switzerland). GAPDH were used as internal controls. All results were expressed as the means ± SD of at least three independent experiments. Comparative quantification was determined using the 2-ΔΔCt method. The primers used are presented in Supplementary Table S1.
Western blotting analysis
Total proteins were prepared from the samples by complete cell lysis (Keygen Biotech, Jiangsu, China) with protease and phosphatase inhibitors. Quantified protein lysates were separated on sodium dodecyl sulfate-polyacrylamide gel and transferred onto polyvinylidene difluoride membranes, blocked with 5% BSA for 1 h at room temperature, and incubated with primary antibodies overnight at 4°C. After incubation with the goat anti-rabbit secondary antibodies, the proteins were visualized using a chemiluminescence method (ECL Plus Western Blotting Detection System; Biostep, Burkhardtsdorf, USA). Each experiment was performed in triplicate.
Cell counting kit-8 (CCK-8) assay
The cell viability was assessed by CCK8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's protocol. All of the experiments were performed in third. The cell proliferation curves were plotted using the absorbance at each time point.
Colony formation assay
Cells were seeded in 6-well plates at a density of 100 cells/well. After incubation for 14 days, cells were washed twice with PBS, fixed with methanol and stained with crystal violet. The number of colonies containing > 50 cells was counted under a microscope.
RNA interference and transfection
HT29 cells were plated in a 6-well plate at a 30-50% confluence. After 24h, siRNA against GSN (Si-GSN) were transfected into HT29 cells using Lipofectamine 3000 reagent (Invitrogen, USA) according to the manufacturer's instructions. Cells transfected with the transfection agent, and scramble-control siRNA (NC) were used as controls. The cells were harvested 48 hours after transfection.
Statistical analysis
SPSS 19.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA) were used to analyze all data for statistical significance. Two-tailed Student's t-test was used for comparisons of two independent groups. Data were analyzed by one-way analysis of variance (ANOVA) in SPSS19.0, followed by Dunnett‑test when treated groups were compared only to the control group. Statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.001. P < 0.05 was considered statistically significant.
Results
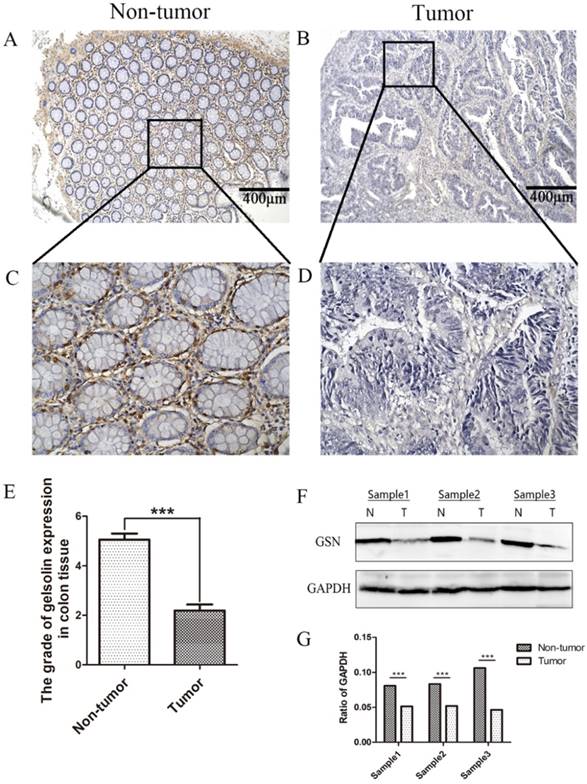
Lower expression of GSN in colon cancer tissues
The expression and subcellular localization of GSN were determined using IHC analysis. Strong GSN-positive signals were present in the non-tumor epithelial cells and extracellular interstitium. Notably, positive signals were not found in the colon cancer tissues (Fig. 1A-E). Then we examined GSN expression in 3 pairs of human colon cancer tissues and matched non-tumor colonic epithelium by Western blotting analysis. As shown in Fig. 1F-G, the colon cancer tissues (T) exhibited lower GSN expression level compared with the corresponding non-tumor controls (N).
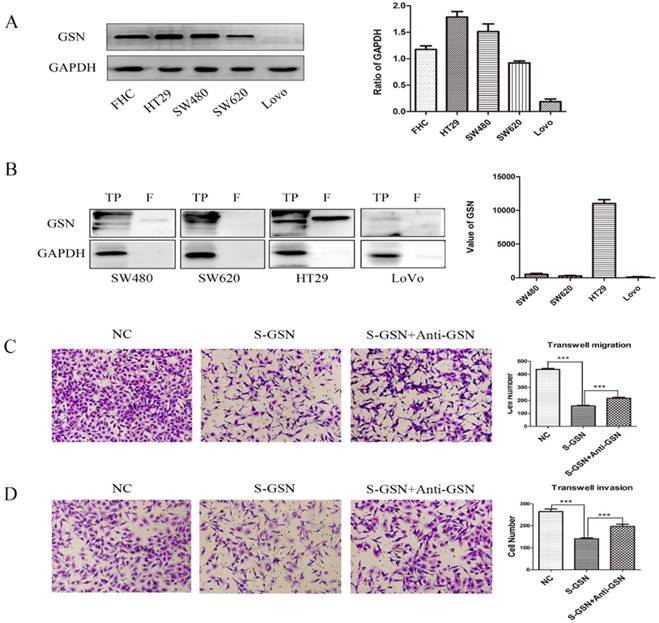
Secreted GSN decreased the invasion and migration of colon cancer cells in vitro
To further explore the role of GSN in colon cancer, we firstly examined the expression level of GSN protein in normal human colon cell line and cancer cell lines. It was confirmed that GSN protein expression was higher expressed in the normal human colon cell line (FHC) and gradually decreased in the HT29, SW480, SW620, and LoVo cancer cell lines (Fig. 2A). Next, we collected the conditioned medium (CM) of cancer cell lines and used ultrafiltration to obtain secreted GSN, and then measured the expression of GSN by Western Blotting. The results showed that secreted GSN was also highly expressed in HT29 cell line and hardly expressed in LoVo cell line (Fig. 2B).
To assess the effect of secreted GSN in colon cancer cell, the secreted GSN obtained from HT29 cell CM and GSN-neutralizing antibody (GSN NAb, 2.5ug/mL) were added into LoVo cells. Then CCK-8 assay and colony formation assay were used to verify secreted GSN on the growth characteristics of colon cancer cells in this study. Unfortunately, there was no significant difference after adding secreted GSN and GSN NAb (Supplementary Fig. 1). To gain an insight into the role of secreted GSN in cell invasion and migration, the transwell experiments were performed. We found that the addition of secreted GSN decreased the invasion and migration capacity of LoVo cells compared with control groups. Meanwhile, this phenomenon can be restrained by the GSN-neutralizing antibody (Fig. 2C-D).
Lower expression of GSN in colon cancer tissues. (A-D) Representative images of GSN expression from tumor tissue and non-cancerous tissue by immunohistochemical assays. (E) Comparison of the immunohistochemical scores of GSN expression in colon cancer tissues (n = 36) and non-tumor tissues (n = 36). (F) Proteins isolated from resected tumor and adjacent non-tumor tissue specimens were subjected to western blot analysis. T, colon cancer tissues: N, non-tumor tissues. (G) Protein expression levels were quantified by scanning densitometry and normalized to GAPDH (as an intracellular control). ***P < 0.001 compared with the control group.
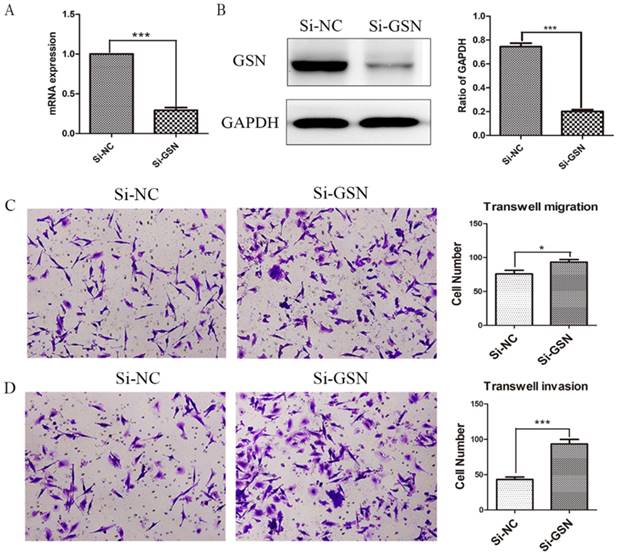
Knockdown of GSN promoted the invasion and migration of colon cancer cells
Si-GSN and Si-NC were transfected into HT29 cells and the efficiency of transfection was proved (Fig. 3A-B). Then the conditioned medium from both of them were collected and ultrafilter to obtain secreted GSN. Similarly, transwell experiments were performed. The results showed that the conditioned medium from GSN knockdown HT29 cells could promote the invasion and migration capacity of LoVo cell compared with control group (Fig. 3C-D).
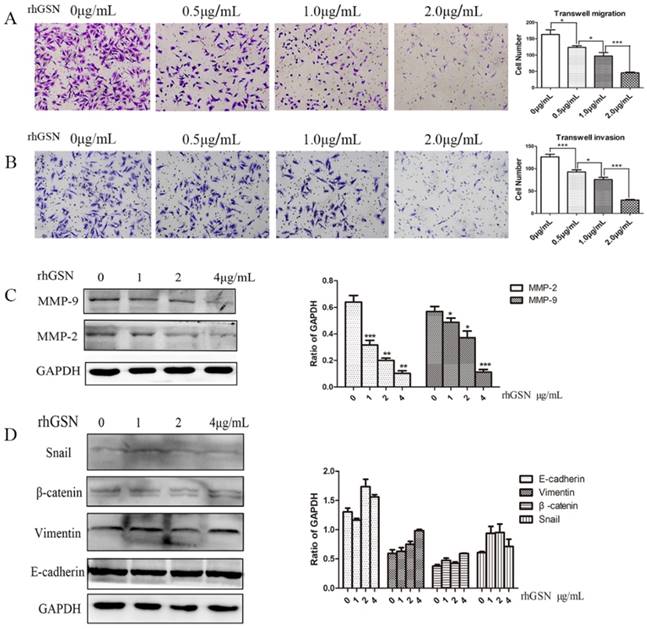
Recombinant GSN decreased the invasion and migration capacity and suppressed MMP2, MMP9 expression of colon cancer cells
LoVo cell were treated with dose dependent recombinant GSN and we also did transwell experiments. In consistent with the above results, the recombinant GSN (rhGSN) dose-dependently decreased the invasion and migration ability of colon cancer cells (Fig. 4A-B). Simultaneously, we found that the expression of matrix metalloproteinase (MMP) family members, MMP2 and MMP9, were also gradually decreased by recombinant GSN stimulation in a dose dependent manner. However, the expression of β-catenin, Vimentin, E-cadherin, and Snail, a transcription factor of E-cadherin, were not showed significant difference (Fig. 4C-D).
Secreted GSN decreased the invasion and migration of colon cancer cells. (A) GSN expression was detected by western blot with GAPDH used as the internal control. Protein expression levels quantified by scanning densitometry were shown. (B) GSN expression in cellular supernatant of cell lines. TP, total proteins. F, cellular supernatant after ultrafiltration. GAPDH was used as the internal control. GSN expression levels quantified by scanning densitometry normalized to GAPDH were showed. (C-D) Transwell migration assay and transwell invasion assay were performed in LoVo cell. NC, LoVo cell; S-GSN, LoVo cell added secreted GSN; S-GSN+Anti-GSN, LoVo cell added secreted GSN and GSN-neutralizing antibody. Data are presented as mean ± SD for at least three independent experiments. ***P < 0.001.
The diagnostic value of serum GSN in colon cancer patients
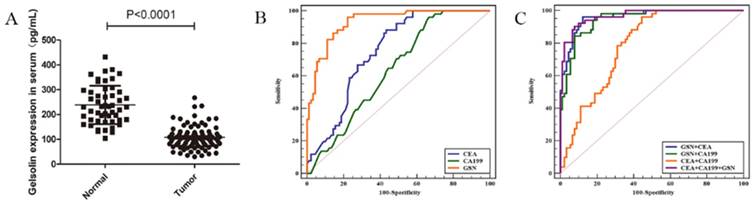
Given above results, we measured the serum expression of GSN in colon cancer patients and healthy controls by using a commercially available ELISA kit. It was found that serum levels of GSN were significantly lower in colon cancer patients (108.01 ± 44.9 pg/mL) compared with those in healthy controls (238.40 ± 76.3 pg/mL) (P < 0.001), which was in accordance with aforementioned immunohistochemical results (Fig. 5A). Next, we explored whether serum GSN has the diagnostic value for colon cancer by using ROC curve analysis. It was noteworthy that the area under the curve (AUC) of GSN was 0.932 (95% CI: 0.877 - 0.967), which was significantly higher than that of CEA (AUC = 0.751) and CA199 (AUC = 0.638). Furthermore, ROC curve analyses for combined biomarkers were compared. It was found that both the combined AUC of GSN and CA199 (AUC = 0.946) or GSN and CEA (AUC = 0.960) showed significantly higher than combined AUC of CEA and CA199 (AUC = 0.787). Moreover, the combination of GSN, CEA, and CA199 yielded the highest AUC value (AUC = 0.966) (Fig. 5B-C). Taken together, these results demonstrated that serum GSN might have the potential value in the diagnosis of colon cancer.
In addition, to determine whether the serum levels of GSN are correlated with colon cancer progression, we analyzed the relationship between GSN levels and the clinicopathological characteristics of colon cancer patients. All of the samples were classified into 2 groups with lower or higher serum levels of GSN according to the tertiles (119.32 pg/mL). Statistical analyses revealed that lower serum levels of GSN were associated with advanced Dukes' stage (P = 0.028) (Table 1), while no significant correlations were observed between serum GSN levels and other clinicopathological features.
Clinicopathologic characteristics of the colon cancer patients and their correlation with serum GSN expression
| Variables | N (%) | GSN (pg/mL) | P value | |
|---|---|---|---|---|
| ≥ 119.32 (N = 30) | < 119.32 (N = 60) | |||
| Sex | 0.635b | |||
| Male | 60 | 21 (23.3%) | 39 (43.3%) | |
| Female | 30 | 9 (10.0%) | 21 (23.3%) | |
| Age, y | 1.000b | |||
| ≥ 50 | 57 | 19 (21.1%) | 38 (42.2%) | |
| < 50 | 33 | 11 (12.2%) | 22 (24.4%) | |
| T Stage (tumor size and invasiveness) | 0.425b | |||
| T1 + T2 + T3 | 29 | 8 (8.9%) | 21 (23.3%) | |
| T4 | 61 | 22 (24.4%) | 39 (43.3%) | |
| N Stage (lymph node metastasis) | 0.353b | |||
| N0 + N1 | 57 | 21 (23.3%) | 9 (10.0%) | |
| N2 + N3 | 33 | 36 (40.0%) | 24(26.7%) | |
| M Stage (distant metastasis) | 0.652b | |||
| M0 | 51 | 18 (20.0%) | 33 (36.7%) | |
| M1 | 39 | 12 (13.3%) | 27 (30.0%) | |
| Dukes' Stage | 0.028b* | |||
| A + B | 31 | 15 (16.7%) | 16 (17.8%) | |
| C + D | 59 | 15 (16.7%) | 44 (48.9%) | |
| Degree of differentiation | 0.350a | |||
| Poor | 6 | 1 (1.1%) | 5 (5.6%) | |
| Moderate | 70 | 26 (28.9%) | 44 (48.9%) | |
| High | 14 | 3 (3.3%) | 11 (12.2%) | |
aP values were determined using Fisher's exact test for categorical variables;
bP values were determined using Chi-square test for categorical variables;
*P < 0.05 was considered statistically significant.
Knockdown of GSN promoted the invasion and migration of colon cancer cells. (A) GSN expression was detected in HT29 cells by qRT-PCR after transfected with control siRNA or GSN siRNA (si-GSN). (B) GSN expression in HT29 cells was detected by western blot after transfected with si-GSN and si-NC. GSN expression levels quantified by scanning densitometry normalized to GAPDH (as an intracellular control) were shown. (C-D) Transwell migration assay and transwell invasion assay were performed in LoVo cell. NC, HT29 cell transfected with si-NC. Si-GSN, HT29 cell transfected with si-GSN. Data are presented as mean ± SD for at least three independent experiments. *P<0.05, ***P < 0.001.
Recombinant GSN decreased the invasion and migration, and suppressed the expression of MMP2 and MMP9 of LoVo cell. (A-B) Transwell migration assay and transwell invasion assay were performed in LoVo cells with increasing concentration of recombinant GSN. (C) The expression of MMP2 and MMP9 were examined by western blot analysis in HT29 cell with increasing concentration of recombinant GSN. (D) The expression of E-cadherin, Vimentin, β-catenin and Snail1 were examined by western blot analysis in HT29 cell with concentration of recombinant GSN. Proteins expression levels were quantified by scanning densitometry normalized to GAPDH (as an intracellular control) were shown. Data were presented as mean ± SD for at least three independent experiments. *P < 0.05, ***P < 0.001.
Detection of serum GSN in patients with colon cancer. (A) Relative expression of serum GSN between healthy control group (n = 51) and colon cancer group (n = 90). (B) ROC curves for serum GSN (AUC =0.932), CEA (AUC = 0.751) and CA199 (AUC = 0.638). (C) ROC curves for combination of serum GSN, CEA and CA199 (AUC = 0.966).
Discussion
Colon cancer is one of the most common cancers worldwide with a poor prognosis. The clinical symptoms are not commonly observed in the early stage of colon cancer development in most cases. Thus, the exploration of early biomarkers and therapeutic targets for colon cancer is of great importance. Many studies have shown that gelsolin is involved in cancer progression, including colon cancer [7-11]. Two forms of GSN protein, cytosolic GSN and secreted GSN, have been detected in previous research [12, 13]. Despite the role of cytosolic GSN in tumors had been studied, as reported in previous studies [7-11], the effect of secreted GSN in colon cancer is unknown.
In the present study, it was confirmed that GSN was highly expressed in non-tumor tissues while lower in colon cancer tissues, which was consistent with the previous study [8], and suggested that GSN might play a negative role in colon cancer progression. Next, we detected the expression of GSN in colon cancer cell lines. The results showed that GSN was highly expressed in HT29 and SW480 cell lines, while gradually decreased in SW620 and LoVo cell lines. HT29 and SW480 were highly differentiated, SW620 was moderately differentiated, while LoVo was poorly differentiated [16-18], which suggested that the GSN expression had a negative correlation with cancer cell differentiation.
In Western blotting analyses, secreted GSN could be detected in the conditioned medium of cancer cell lines after ultrafiltration, and the expression trend of the secreted GSN expression in cancer cell lines were consistent with the cytosolic GSN. Malignant cells possess key hallmarks, namely, uncontrolled growth potentials and the ability to invade surrounding tissues and metastasize [19]. In vitro, we took assays to study the impact of secreted GSN in cell proliferation, but unfortunately, we found that it had no influence on the colon cancer cell proliferation. However, we found that the secreted GSN, which was obtained from CM, could restrain the invasion and migration capacity of colon cancer cells. And this result was further confirmed in the models of GSN knockdown cells and its control cells. Moreover, the regulating ability of invasion and migration was dose dependently by recombinant GSN stimulation, which was in line with the previous study [20]. These results collectively demonstrated that the secreted GSN could suppress the metastasis of colon cancer cells.
Metastasis, the dissemination and growth of neoplastic cells in an organ distinct from that in which they originated, is the most common cause of death in cancer patients [21, 22]. Cancer patients, both at early and late stage, dependent on life span, are likely to develop metastasis. The metastatic spread of the primary tumor accounts for over 90% of patient mortality associated with solid cancers [19]. It was reported that cytosolic GSN regulated tumor metastasis by epithelial-mesenchymal transition (EMT) [23], which was one of essential mechanism of tumor metastasis [24, 25]. Given above results, we hypothesized that secreted GSN has the same regulatory mechanism with cytosolic GSN, and then we took assays to confirm it. However, there was no obvious phenomenon after adding secreted GSN into LoVo cells (Fig. 4D). Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases which are involved in multiple biological functions. Among MMPs, MMP2 and MMP9 were recognized as the key factors of cell migration in tumors [26-28]. In this study, we found that the expression of MMP2 and MMP2 decreased on a dose dependent manner by recombinant GSN stimulation, which indicated that the secreted GSN could restrain the invasion and migration ability of colon cancer cells by decreasing the levels of MMP2 and MMP9. However, the exact involvement of secreted GSN in colon cancer still needs to be studied in the future.
Biomarkers for colon cancer have been extensively explored over the past decades. CEA and CA199 are considered as the most frequently used biomarkers for clinical colon cancer screening, yet with a low specificity or sensibility [29, 30]. In this study, we demonstrated that there was a significant difference in the serum levels of GSN between colon cancer patients and healthy controls. Thus, we predicted whether serum GSN could be a biomarker for the diagnosis of colon cancer. ROC curve analyses demonstrated that the AUC of GSN (AUC = 0.932) was significantly higher than AUC of CEA (AUC = 0.751) or CA199 (AUC = 0.638). In addition, the combined AUC of GSN, CEA and CA199 reached as high as 0.966, which was far more than the combined AUC of CEA and CA199 (AUC = 0.787). Additionally, we demonstrated that serum GSN levels were negatively associated with advanced Dukes' stage (P = 0.028). Taken together, these results suggested that serum GSN might have the potential to be a new biomarker for clinical evaluation of colon cancer.
The key finding of this study was that the secreted GSN could restrain invasion and migration of colon cancer cells. The serum levels of GSN in colon cancer patients were significantly higher than in the healthy controls, which was further confirmed to be a potential biomarker for colon cancer. However, there were still limitations in our present study. The molecular mechanisms of the role of GSN in the MMP2 and MMP9 protein expression remain unclear. In this study, our sample size for measuring serum GSN levels was small. An expanded number of serum samples were needed to be enrolled in the future.
In conclusion, we reveal that GSN is lowly expressed in colon cancer tissues and it can restrain the metastasis of colon cancers by regulating the expressions of MMP2 and MMP9. Moreover, we confirm that serum GSN is increased in colon cancer patients, which could be a candidate diagnostic biomarker and a potential target for new therapies in colon cancer.
Abbreviations
GSN, gelsolin; ROC, the area under the curve; IHC, immunohistochemistry; CM, conditioned medium; qRT-PCR, Quantitative real-time polymerase chain reaction; CCK-8, Cell Counting Kit-8; MMP, matrix metalloproteinase; CEA, Carcinoembryonic Antigen; CA199, Carbohydrate Antigen 19-9.
Supplementary Material
Supplementary figures and table.
Acknowledgements
This study was supported by the Guangdong Natural Science Foundation (2016A03-0313525); the Science and Technology Program of Guangzhou (201607010015); the Guangdong Industry-University-Research Collaboration Innovation Major Projects (201604040003); the Clinical Research Special Fund of Wu Jieping Medical Foundation (320.6750.16039).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87-108
2. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490-1502
3. Wang J, Du Y, Liu X. et al. MicroRNAs as Regulator of Signaling Networks in Metastatic Colon Cancer. BioMed Res Int. 2015;2015:823620
4. Yin HL, Stossel TP. Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature. 1979;281(5732):583-6
5. Sun HQ, Yamamoto M, Mejillano M. et al. Gelsolin, a multifunctional actin regulatory protein. J Biol Chem. 1999;274(47):33179-82
6. Li GH, Arora PD, Chen Y. et al. Multifunctional roles of gelsolin in health and diseases. Med Res Rev. 2012;32(5):999-1025
7. An JH, Kim JW, Jang SM. et al. Gelsolin negatively regulates the activity of tumor suppressor p53 through their physical interaction in hepatocarcinoma HepG2 cells. Biochem Biophys Res Commun. 2011;412(1):44-9
8. Li WX, Yang MX, Hong XQ. et al. Overexpression of gelsolin reduces the proliferation and invasion of colon carcinoma cells. Mol Med Rep. 2016;14(4):3059-65
9. Wang HC, Chen CW, Yang CL. et al. Tumor-Associated Macrophages Promote Epigenetic Silencing of Gelsolin through DNA Methyltransferase 1 in Gastric Cancer Cells. Cancer Immunol Res. 2017;5(10):885-97
10. Lokamani I, Looi ML, Md Ali SA. et al. Gelsolin and ceruloplasmin as potential predictive biomarkers for cervical cancer by 2D-DIGE proteomics analysis. Pathol Oncol Res. 2014;20(1):119-29
11. Noske A, Denkert C, Schober H. et al. Loss of Gelsolin expression in human ovarian carcinomas. Eur J Cancer. 2005;41(3):461-9
12. Page LJ, Huff ME, Kelly JW. et al. Ca2+ binding protects against gelsolin amyloidosis. Biochem Biophys Res Commun. 2004;322(4):1105-10
13. Bucki R, Levental I, Kulakowska A. et al. Plasma gelsolin: function, prognostic value, and potential therapeutic use. Curr Protein Pept Sci. 2008;9(6):541-51
14. DiNubile MJ. Plasma gelsolin as a biomarker of inflammation. Arthritis Res Ther. 2008;10(6):124
15. Qu L, Deng B, Zeng Y. et al. Decreased expression of the Nkx2.8 gene correlates with tumor progression and a poor prognosis in HCC cancer. Cancer Cell Int. 2014;14:28
16. Li LN, Zhang HD, Yuan SJ. et al. Differential sensitivity of colorectal cancer cell lines to artesunate is associated with expression of beta-catenin and E-cadherin. Eur J Pharmacol. 2008;588(1):1-8
17. Gatzidou E, Mantzourani M, Giaginis C. et al. Augmenter of liver regeneration gene expression in human colon cancer cell lines and clinical tissue samples. J buon. 2015;20(1):84-91
18. Hao L, Zhao Y, Wang Z. et al. Expression and clinical significance of SALL4 and beta-catenin in colorectal cancer. J Mol Histol. 2016;47(2):117-28
19. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-74
20. Yuan X, Wang W, Li J. et al. Gelsolin suppresses gastric cancer metastasis through inhibition of PKR-p38 signaling. Oncotarget. 2016;7(33):53459-70
21. Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274-84
22. Fidler IJ. Critical determinants of metastasis. Semin Cancer biol. 2012;12(2):89-96
23. Chen ZY, Wang PW, Shieh DB. et al. Involvement of gelsolin in TGF-beta 1 induced epithelial to mesenchymal transition in breast cancer cells. J Biomed Sci. 2015;22:90
24. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178-96
25. Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344):re8
26. Cui N, Hu M, Khalil RA. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog Mol Biol Transl Sci. 2017;147:1-73
27. Amar S, Smith L, Fields GB. Matrix metalloproteinase collagenolysis in health and disease. Biochim Biophys Acta. 2017;1864:1940-51
28. Zhu Y, Guo L, Wang S. et al. Association of Smoking and XPG, CYP1A1, OGG1, ERCC5, ERCC1, MMP2, and MMP9 Gene Polymorphisms with the early detection and occurrence of Laryngeal Squamous Carcinoma. J Cancer. 2018;9(6):968-77
29. Perkins GL, Slater ED, Sanders GK. et al. Serum tumor markers. Am Fam Physician. 2003;68(6):1075-82
30. McKeown E, Nelson DW, Johnson EK. et al. Current approaches and challenges for monitoring treatment response in colon and rectal cancer. J Cancer. 2014;5(1):31-43
Author contact
![]() Corresponding authors: Professor Yurong Qiu, 1Laboratory Medicine Center, Nanfang Hospital, Southern Medical University, Guangzhou Road North, Baiyun district, Guangzhou, 510515, P.R. China. 3Guangdong Key Laboratory of Biochip Technology, Southern Medical University, Guangzhou, 510515, P.R. China. 4Huayin Medical Laboratory Center Co., Ltd., Guangzhou, 510515, P.R. China. Tel/Fax: 0086-020-61642143. E-mail: qyredu.cn. Haifang Wang, PhD, Laboratory Medicine Center, Nanfang Hospital, Southern Medical University, Guangzhou Road North, Baiyun district, Guangzhou 510515, P.R. China. Tel/Fax: 0086-020-62786960. E-mail: haifang_wcom
Corresponding authors: Professor Yurong Qiu, 1Laboratory Medicine Center, Nanfang Hospital, Southern Medical University, Guangzhou Road North, Baiyun district, Guangzhou, 510515, P.R. China. 3Guangdong Key Laboratory of Biochip Technology, Southern Medical University, Guangzhou, 510515, P.R. China. 4Huayin Medical Laboratory Center Co., Ltd., Guangzhou, 510515, P.R. China. Tel/Fax: 0086-020-61642143. E-mail: qyredu.cn. Haifang Wang, PhD, Laboratory Medicine Center, Nanfang Hospital, Southern Medical University, Guangzhou Road North, Baiyun district, Guangzhou 510515, P.R. China. Tel/Fax: 0086-020-62786960. E-mail: haifang_wcom

 Global reach, higher impact
Global reach, higher impact