3.2
Impact Factor
ISSN: 1837-9664
J Cancer 2019; 10(4):1039-1051. doi:10.7150/jca.25814 This issue Cite
Research Paper
MiR-192-5p reverses cisplatin resistance by targeting ERCC3 and ERCC4 in SGC7901/DDP cells
1. Department of Oncological Radiotherapy, First Affiliated Hospital of Anhui Medical University, Hefei, China
2. Department of Oncology, First Affiliated Hospital of Anhui Medical University, Hefei, China
3. Department of Oncology, Huaibei People's Hospital, Huaibei, China
4. Department of Food and Nutrition Hygiene, School of Public Health, Anhui Medical University, Hefei, China
*These authors contributed equally to this work.
Received 2018-2-28; Accepted 2018-11-28; Published 2019-1-29
Abstract
Cisplatin chemoresistance is a clinical obstacle in the treatment of gastric cancer (GC). Enhanced DNA repair capacity may lead to cisplatin resistance. However, the detailed molecular mechanism of GC cisplatin resistance specifically involving nucleotide excision repair (NER) is not clear. However, the mechanism through which the NER pathway contributes to cisplatin resistance in GC is still unclear. In light of the crucial role of microRNAs (miRNAs) in regulating protein expression and biological behavior, we aimed to analyze the expression and function of miR-192-5p in the NER pathway and its role in cisplatin resistance in GC. Comet assays were performed to measure the amount of DNA damage and repair in the SGC7901 and SGC7901/DDP GC cell lines by observing the tail length. MiRNA expression levels in SGC7901/DDP and SGC7901 cells were detected by microarray. Quantitative real-time PCR (qRT-PCR) was carried out to confirm the expression level of miR-192-5p. Lentiviral vector transfection modifies miR-192-5p levels in SGC7901/DDP and SGC7901 cells. The IC50 values of cisplatin-treated cells were assessed by MTT assays. The protein level was determined by Western blot and immunohistochemistry. With enhanced DNA repair, the expression levels of ERCC3 and ERCC4 in SGC 7901DDP cells increased, while miR-192-5p was significantly downregulated in SGC7901/DDP compared with SGC7901 cells. ERCC3 and ERCC4 were identified as the main targets of miR-192-5p. Forced expression of miR-192-5p in SGC7901/DDP cells significantly inhibited the expression of ERCC3 and ERCC4, making GC cells more sensitive to cisplatin in vitro and in vivo. In contrast, knockdown of miR-192-5p expression in SGC7901 cells increased the expression of ERCC3 and ERCC4, resulting in cisplatin resistance in vitro and in vivo. MiR-192-5p partially reversed GC cisplatin resistance by targeting ERCC3 and ERCC4, which participate in the NER pathway, suggesting that miR-192-5p may be a potential biomarker and therapeutic target for GC cisplatin resistance.
Keywords: gastric cancer, cisplatin resistance, miR-192, NER
Introduction
Gastric cancer (GC) remains the main contributor to human malignant tumors globally. According to Global Cancer Statistics, more than 951,000 individuals worldwide were diagnosed with GC, and 723,000 patients died of GC in 2012 [1]. Approximately two-thirds of newly diagnosed GC patients suffer from disseminated disease and need chemotherapy. Currently, platinum-based chemotherapy is the most common treatment for GC patients [2, 3]. The effectiveness of chemotherapy is limited by primary or secondary cisplatin resistance; thus, identification of new predictive markers for the molecular mechanisms involved in GC cisplatin reactions is urgently needed.
The nucleotide NER is an important mechanism for DNA repair. NER proteins eliminate Pt-DNA adducts formed when activated cisplatin reacts with the N7 positions of the nucleophilic centers of guanosine and adenosine in DNA. Therefore, increased expression of NER proteins leads to cisplatin resistance. ERCC1 overexpression leads to cisplatin resistance in ovarian cancer [4]. In urothelial cancer, mutated ERCC2 is associated with a complete response to cisplatin-based chemotherapy [5]. GC patients with high ERCC1 expression benefit less from platinum-based adjuvant chemotherapy. However, there is little research on the ERCC3 and ERCC4 proteins at present. Our previous experiments showed that compared with SGC7901 cells, increased ERCC3 and ERCC4 levels enhanced the DNA repair capacity of SGC7901/DDP cells. Therefore, the roles and regulatory mechanisms of ERCC3 and ERCC4 in cisplatin resistance are worth exploring.
MiRNAs form a class of small noncoding RNA molecules with a length of 17-25 nucleotides, and they participate in posttranscriptional regulation of gene expression by directly targeting the 3′ untranslated region (3′-UTR) of mRNA transcripts to induce mRNA degradation or to inhibit translation [6-8]. In recent years, aberrantly expressed miRNAs have been confirmed to play important roles in cisplatin resistance [9-12]. Although studies show that miRNAs regulate cisplatin resistance by targeting NER pathway proteins [4, 13-15], the pathological relevance of miRNAs in GC cisplatin resistance is still unclear.
In our study, we first compared the DNA repair ability of SGC7901/DDP and SGC7901 cells and then compared the ERCC3 and ERCC4 protein expression levels in the two cell lines. We compared the miRNA expression profiles of SGC7901/DDP and SGC7901 cells by miRNA array analysis. Based on its association with the NER pathway, we mainly focused on miR-192-5p for further study, and we comprehensively investigated its molecular mechanism in cisplatin resistance in GC cells. We demonstrate that miR-192-5p expression is frequently decreased in SGC701/DDP cells. Further analyses showed that a miR-192-5p/ERCC3 and ERCC4 axis promotes cisplatin resistance in GC cells. Furthermore, these findings indicate that this miR-192-5p/NER axis is a potential therapeutic target for platinum-resistant GC.
Results
A cisplatin-resistant cell line exhibits an increased DNA damage repair phenotype
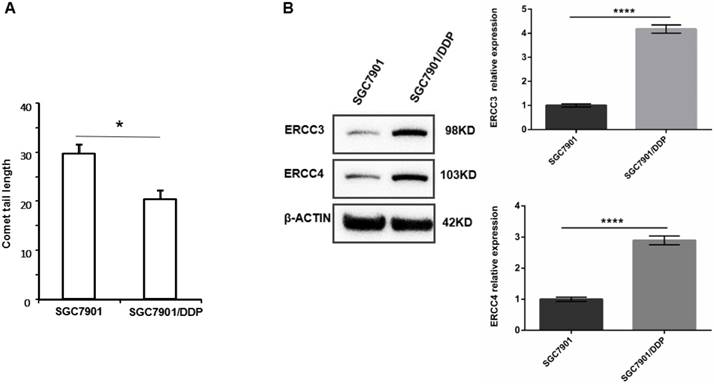
We observed the tail lengths in comet assays to determine the DNA damage repair abilities of the cell lines. As shown in Figure 1A, the DNA damage repair ability of SGC7901/DDP cells is significantly higher than that of SGC7901 cells, indicating that DNA damage repair ability is positively correlated with cisplatin resistance. Western blot analysis showed that the expression levels of ERCC3 and ERCC4 were significantly higher in SGC7901/DDP cells than in SGC7901 cells (Figure 1B). This result showed that the expression levels of the NER proteins ERCC3 and ERCC4 are positively correlated with cisplatin resistance in SGC7901/DDP cells.
A. Tail lengths of the GC cell line SGC7901 and the cisplatin-resistant GC cell line SGC7901/DDP, *P < 0.05. B. Western blot showing the expression levels of ERCC3 and ERCC4 in SGC7901/DDP cells and SGC7901 cells. Values represent the means ± SEM. ****P < 0.0001.
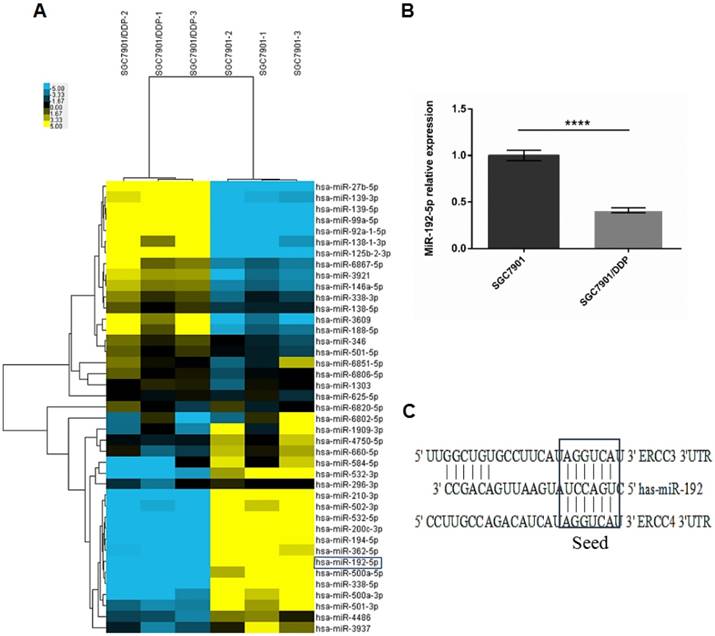
A: Two-dimensional clustering of the 41 dysregulated microRNAs; each row represents the relative levels of the individual microRNAs, and each column represents the miRNA levels in single samples. Blue: higher expression levels, Yellow: lower expression levels. (P < 0.05, FC ≥ 2). B: Real-time polymerase chain reaction (RT-PCR) analysis showing the miR-192-5p expression levels in SGC7901/DDP and SGC7901 cells. The miR-192-5p expression level was significantly downregulated in SGC7901/DDP cells compared with the parental SGC7901 cells. U6 snRNA expression was used as an internal control, ****P < 0.0001. C: Predicted miR-192-5p binding sites in the 3'-UTRs of the ERCC3 and ERCC4 mRNAs.
Identification of differentially expressed miRNAs and decreased miR-192-5p expression
MiRNA array analysis was performed to compare the miRNA expression profiles of SGC7901/DDP and SGC7901 cells. We found that 12 annotated or potential miRNAs were 2-fold upregulated and 29 miRNAs were 2-fold downregulated in SGC 7901/DDP cells compared with SGC 7901 cells (p < 0.05, Figure 2A; Table 1). These aberrant miRNAs were then bioinformatically analyzed using ten prediction programs (DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid, PICTAR4, PICTAR5, PITA, RNA22 and TargetScan) to identify miRNAs that play potential roles in NER (Table 2). The expression level of miR-192-5p was significantly decreased in SGC7901/DDP cells [fold change (FC) = 0.3, p < 0.05]. The qRT-PCR results verified the reliability of the genetic analysis results (FC = 0.41, p < 0.05, Figure 2B). According to the results of the gene analysis, we found that the 3′-UTRs of the ERCC3 and ERCC4 transcripts contain sequences matching the seed-sequence of miR-192-5p (Figure 2C). Therefore, we chose miR-192-5p for further study.
ERCC3 and ERCC4 are functional targets of miR-192-5p
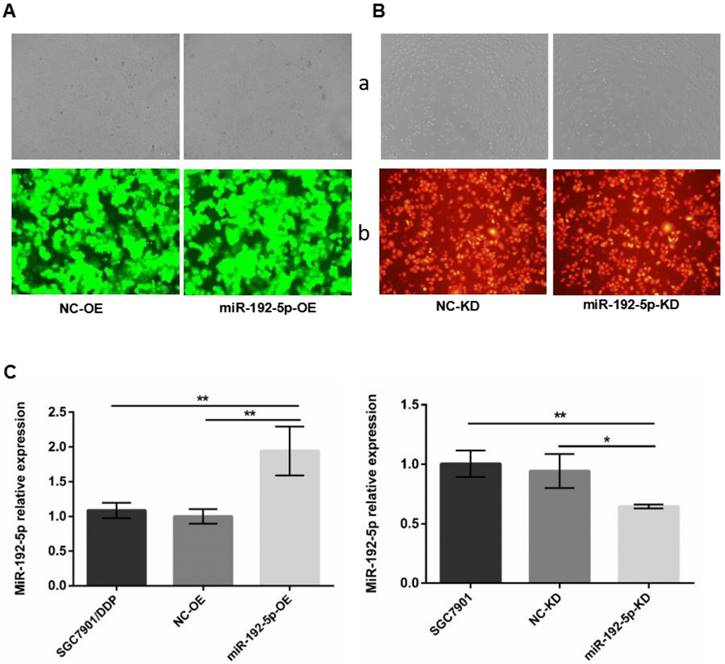
To determine whether miR-192-5p-dependent regulation of cisplatin resistance in GC cells is indeed mediated by ERCC3 and ERCC4, we first used lentiviral vectors to modify the miR-192-5p levels. Fluorescence microscopy confirmed that the lentiviral vectors were successfully transfected (Figure 3A-B). Fluorescence quantitative RT-PCR confirmed that miR-192-5p was overexpressed in miR-192-5p-OE cells compared with SGC7901/DDP cells after lentivirus transduction and that miR-192-5p was downregulated in miR-192-5p-KD cells compared with SGC7901 cells (Figure 3C).
Dysregulated miRNA in GC SGC7901/DDP cells compared with SGC7901 cells
| Transcript | Fold change | Regulation | Sequence |
|---|---|---|---|
| miR-139-5p | 3.9627 | up | UCUACAGUGCACGUGUCUCCAGU |
| miR-139-3p | 2.8456 | up | UGGAGACGCGGCCCUGUUGGAGU |
| miR-3609 | 2.6297 | up | CAAAGUGAUGAGUAAUACUGGCUG |
| miR-27b-5p | 2.6106 | up | AGAGCUUAGCUGAUUGGUGAAC |
| miR-138-1-3p | 2.3803 | up | GCUACUUCACAACACCAGGGCC |
| miR-125b-2-3p | 2.3502 | up | UCACAAGUCAGGCUCUUGGGAC |
| miR-188-5p | 2.2048 | up | CAUCCCUUGCAUGGUGGAGGG |
| miR-99a-5p | 2.0877 | up | AACCCGUAGAUCCGAUCUUGUG |
| miR-6867-5p | 2.0662 | up | UGUGUGUGUAGAGGAAGAAGGGA |
| miR-3921 | 2.0584 | up | UCUCUGAGUACCAUAUGCCUUGU |
| miR-146a-5p | 2.0484 | up | UGAGAACUGAAUUCCAUGGGUU |
| miR-92a-1-5p | 2.0356 | up | AGGUUGGGAUCGGUUGCAAUGCU |
| miR-584-5p | 0.4914 | down | UUAUGGUUUGCCUGGGACUGAG |
| miR-1909-3p | 0.4902 | down | CGCAGGGGCCGGGUGCUCACCG |
| miR-6806-5p | 0.4859 | down | UGUAGGCAUGAGGCAGGGCCCAGG |
| miR-532-5p | 0.4509 | down | CAUGCCUUGAGUGUAGGACCGU |
| miR-625-5p | 0.4481 | down | AGGGGGAAAGUUCUAUAGUCC |
| miR-6851-5p | 0.4475 | down | AGGAGGUGGUACUAGGGGCCAGC |
| miR-200c-3p | 0.4440 | down | UAAUACUGCCGGGUAAUGAUGGA |
| miR-338-3p | 0.4315 | down | UCCAGCAUCAGUGAUUUUGUUG |
| miR-532-3p | 0.4282 | down | CCUCCCACACCCAAGGCUUGCA |
| miR-6802-5p | 0.4274 | down | CUAGGUGGGGGGCUUGAAGC |
| miR-4750-5p | 0.4255 | down | CUCGGGCGGAGGUGGUUGAGUG |
| miR-1303 | 0.4198 | down | UUUAGAGACGGGGUCUUGCUCU |
| miR-6820-5p | 0.4108 | down | UGCGGCAGAGCUGGGGUCA |
| miR-210-3p | 0.3935 | down | CUGUGCGUGUGACAGCGGCUGA |
| miR-346 | 0.3795 | down | UGUCUGCCCGCAUGCCUGCCUCU |
| miR-660-5p | 0.3795 | down | UACCCAUUGCAUAUCGGAGUUG |
| miR-194-5p | 0.3763 | down | UGUAACAGCAACUCCAUGUGGA |
| miR-502-3p | 0.3732 | down | AAUGCACCUGGGCAAGGAUUCA |
| miR-362-5p | 0.3497 | down | AAUCCUUGGAACCUAGGUGUGAGU |
| hsa-miR-3937 | 0.3483 | down | ACAGGCGGCUGUAGCAAUGGGGG |
| miR-500a-3p | 0.3071 | down | AUGCACCUGGGCAAGGAUUCUG |
| miR-296-3p | 0.3033 | down | GAGGGUUGGGUGGAGGCUCUCC |
| miR-192-5p | 0.2989 | down | CUGACCUAUGAAUUGACAGCC |
| miR-500a-5p | 0.2901 | down | UAAUCCUUGCUACCUGGGUGAGA |
| miR-4486 | 0.2887 | down | GCUGGGCGAGGCUGGCA |
| miR-501-5p | 0.2836 | down | AAUCCUUUGUCCCUGGGUGAGA |
| miR-138-5p | 0.2686 | down | AGCUGGUGUUGUGAAUCAGGCCG |
| miR-501-3p | 0.2683 | down | AAUGCACCCGGGCAAGGAUUCU |
| miR-338-5p | 0.1619 | down | AACAAUAUCCUGGUGCUGAGUG |
A. Cell transfection efficiency was examined by microscope. SGC7901/DDP cells were transduced by lentivirus with GFP after puromycin selection. B. SGC7901 cells transduced by lentivirus with RFP after hygromycin selection. a: Light microscopy; b:Fluorescent microscopy with an exposure of 600 ms × 100. C: The miR-192-5p expression levels in lentivirus-transfected cells were detected by qRT-PCR. *P < 0.05, **P < 0.01.
MiRNA target prediction
| MicroRNA | gene | DIANAmT | miRanda | miRDB | miRWalk | RNAhybrid | PICTAR4 | PICTAR5 | PITA | RNA22 | TargetScan | SUM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-188-5p | ERCC3 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 5 |
| miR-192 | ERCC3 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 6 |
| miR-194 | ERCC3 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| miR-501-3p | ERCC3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| miR-501-5p | ERCC3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| miR-502-3p | ERCC3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| miR-625 | ERCC3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| miR-1303 | ERCC4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR-138 | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| miR-138-1* | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| miR-146a | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| miR-188-5p | ERCC4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR-1909 | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| miR-192 | ERCC4 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 4 |
| miR-200c | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| miR-27b* | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| miR-338-3p | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| miR-338-5p | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| miR-501-3p | ERCC4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR-502-3p | ERCC4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR-532-3p | ERCC4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR-532-5p | ERCC4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| miR-660 | ERCC4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
The expression levels of ERCC3 and ERCC4 in lentivirus-transfected cells were detected by Western blot. A: Overexpression of miR-192-5p in SGC7901/DDP cells, leading to downregulation of ERCC3 and ERCC4 expression. B: Knock-down of miR-192-5p expression in SGC7901 cells, leading to upregulation of ERCC3 and ERCC4. *P < 0.05, ** P < 0.01.
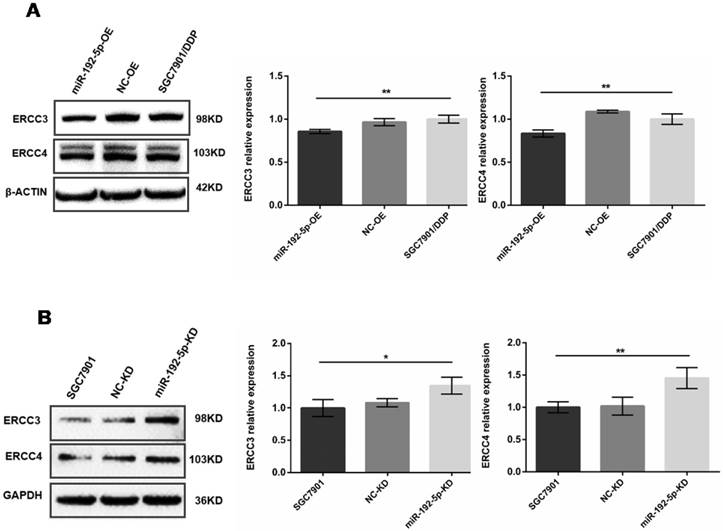
Western blot analysis showed that after miR-192-5p was overexpressed in miR-192-5p-OE cells, the ERCC3 and ERCC4 levels were lower than in SGC7901/DDP cells and NC-OE cells (Figure 4A). This negative correlation was also found between the miR-192-5p and ERCC3/4 expression levels in SGC7901 cells. Down-regulation of miR-192-5p expression in SGC7901 cells (miR-192-5p-KD cells) led to upregulation of ERCC3 and ERCC4 (Figure 4B). Collectively, these results indicate that ERCC3 and ERCC4 are targets of miR-192-5p in SGC7901/DDP cells.
Cell viability treated with increasing concentrations of cisplatin. A: The cisplatin concentrations were 40, 20, 2, 0.2, 0.02 and 0.002 μg/mL in the SCG7901/DDP group for 48 h; B: The cisplatin concentrations were 4, 2, 0.2, 0.02, 0.002, 0.0002 μg/mL in the SGC7901 group for 48 h. ****P < 0.0001.
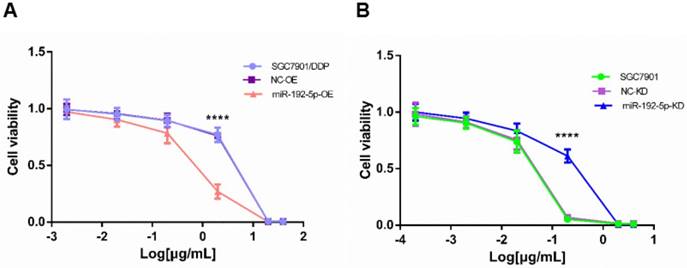
MiR-192-5p expression is inversely correlated with GC cell sensitivity to cisplatin
To investigate the biological function of miR-192-5p in the development of cisplatin resistance, changes in the cisplatin IC50 value were evaluated. Compared with SGC7901/DDP and NC-OE cells, the cisplatin IC50 was decreased in miR-192-5p-OE cells (Figure 5A). Compared with SGC7901 and NC-KD cells, the cisplatin IC50 value was increased in miR-192-5p-KD cells (Figure 5B). These results indicated that cisplatin resistance in SGC7901/DDP cells is negatively correlated with miR-192-5p expression (Table 3).
The IC50 of cisplatin in the GC cell lines (μg/mL, mean ± sd)
| Cell line | IC50 |
|---|---|
| SGC7901/DDP | 7.39 ± 0.06 |
| NC-OE | 6.86 ± 0.09 |
| miR-192-5p-OE | 1.63 ± 0.07* |
| SGC7901 | 0.21 ± 0.01 |
| NC-KD | 0.21 ± 0.007 |
| miR-192-5p-KD | 0.47 ± 0.06# |
*P < 0.05 SGC7901/DDP with NC-OE cells; #P < 0.05 comparing SGC7901 with NC-KD cells.
Therefore, these results suggest that in vitro, upregulation of miR-192-5p can reverse the resistance of SGC7901/DDP cells to cisplatin by inhibiting ERCC3 and ERCC4 expression.
MiR-192-5p reverses cisplatin resistance by targeting ERCC3 and ERCC4 in vivo
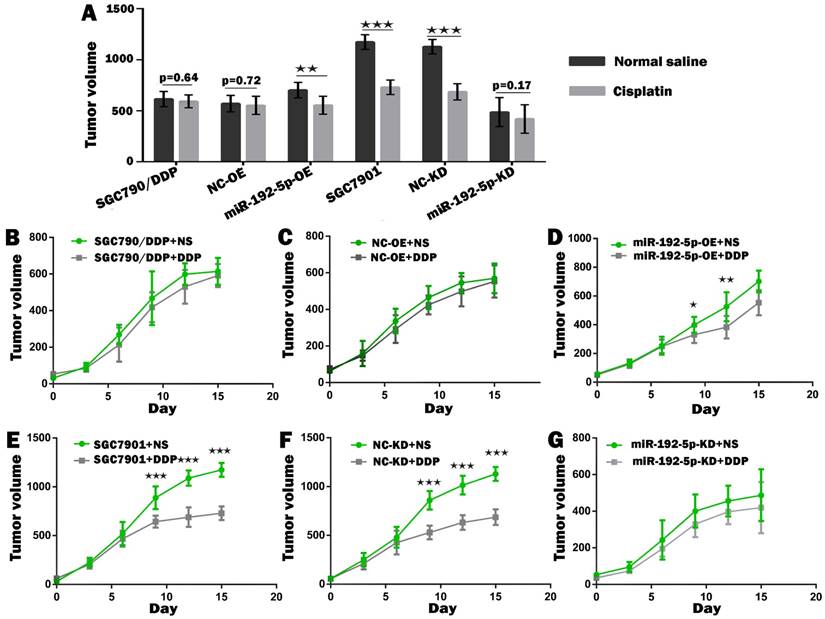
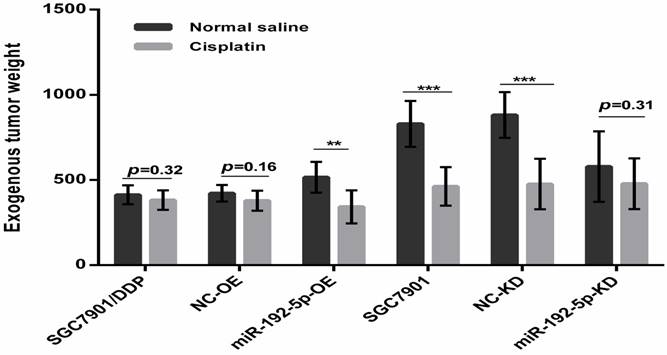
To verify our in vitro results, we tested the correlation between the miR-192-5p level and cisplatin resistance in vivo using a xenograft mice model. The mean volume and weight of the transplanted tumors in the miR-192-5p-OE+DDP group were significantly lower than those in the miR-192-5p-OE+normal saline (NS) group; this result indicated that miR-192-5p overexpression increases the effect of cisplatin treatment in vivo. However, in the miR-192-5p-KD+DDP group, the final mean tumor volume and weight were not significantly different from those of the miR-192-5p-KD+NS group, showing that the knock-down of miR-192-5p resulted in cisplatin resistance in vivo (Figures 6-8, Tables 4-5).
Tumor weight (mg, mean ± sd)
| Tumor | Cisplatin group | Normal saline group | |
|---|---|---|---|
| SGC7901/DDP | 367.36 ± 69.78 | 412.6 ± 55.59 | |
| NC-OE | 378.30 ± 58.77 | 421.69 ± 48.98 | |
| miR-192-5p-OE | 342.40 ± 96.89* | 515.74 ± 90.41 | |
| SGC7901 | 461.79 ± 113.40* | 829.01 ± 134.85# | |
| NC-KD | 475.86 ± 148.58* | 881.51 ± 134.42 | |
| miR-192-5p-KD | 477.54 ± 148.50 | 578.27 ± 206.91 | |
*P < 0.05 compared with the normal saline group; #P < 0.05 compared with the SGC7901/DDP cells.
Positive expression of ERCC3 and ERCC4 in tissues shown by immunohistochemistry (%)
| Tumor tissue | ERCC3 | ERCC4 |
|---|---|---|
| SGC7901/DDP | 63.33 ± 5.77 | 56.67 ± 5.77 |
| NC-OE | 66.67 ± 15.27 | 60.00 ± 10.00 |
| miR-192-5p-OE | 36.30 ± 5.77* | 26.70 ± 5.77* |
| SGC7901 | 23.30 ± 5.77 | 30.00 ± 5.77 |
| NC-KD | 26.70 ± 5.77 | 33.30 ± 5.77 |
| miR-192-5p-KD | 40.00 ± 5.77# | 53.30 ± 5.77# |
*P < 0.05 comparing SGC7901/DDP with NC-OE cells; #P < 0.05 comparing SGC7901 with NC-KD cells.
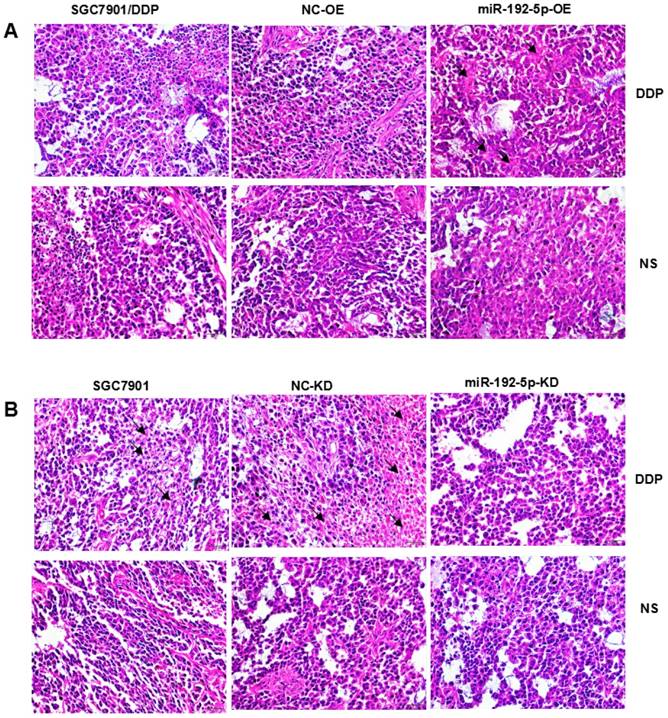
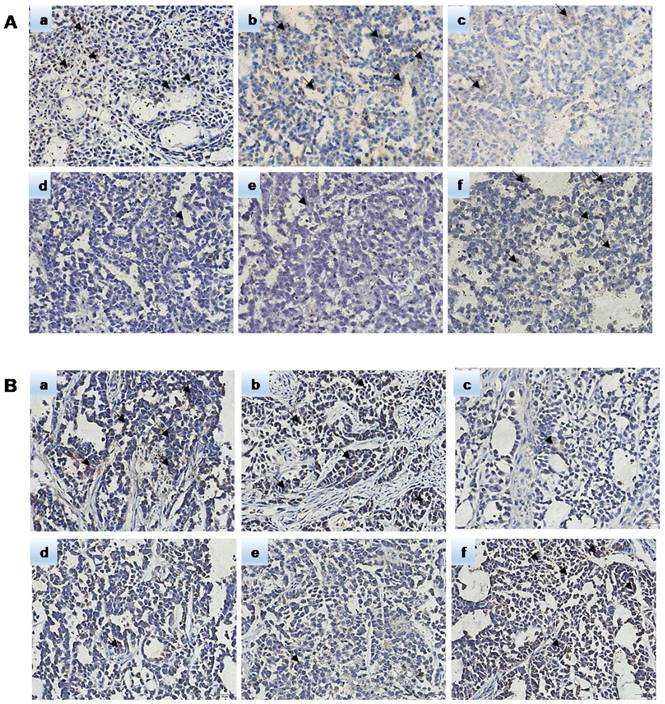
Hematoxylin-eosin (HE) staining showed subcutaneous tumor tissue (Figure 9). Pathological analysis showed that the immunohistochemically stained tissue was subcutaneously transplanted GC. Immunohistochemistry confirmed that the ERCC3 and ERCC4 levels were significantly lower in the miR-192-5p-OE group compared with those in the SGC7901/DDP and NC-OE group, while the ERCC3 and ERCC4 levels in the miR-192-5p-KD group were significantly higher than those in the SGC7901 and NC-KD groups (Figure 10). These data suggest that miR-192-5p expression is negatively correlated with ERCC3/4 expression and positively correlated with cisplatin sensitivity in vivo.
Discussion
In recent years, studies have shown that abnormally expressed miRNAs target many downstream proteins to regulate various molecular mechanisms of cisplatin resistance in many cancers. Microarrays are a high-throughput method for exploring miRNA profiles and for identifying abnormally expressed miRNAs [16-18]. Therefore, we used isolated miRNA microarrays to analyze the differential regulation of 775 miRNAs in SGC7901/DDP compared with SGC7901 cells. We found that compared with SGC7901 cells, miR-192-5p was downregulated in SGC7901/DDP cells. Further experiments showed that modulation of the miR-192-5p expression level could regulate cisplatin resistance by targeting ERCC3 and ERCC4 in vitro and in vivo. These findings suggest that miR-192-5p can be used as a new therapeutic target to reverse cisplatin resistance.
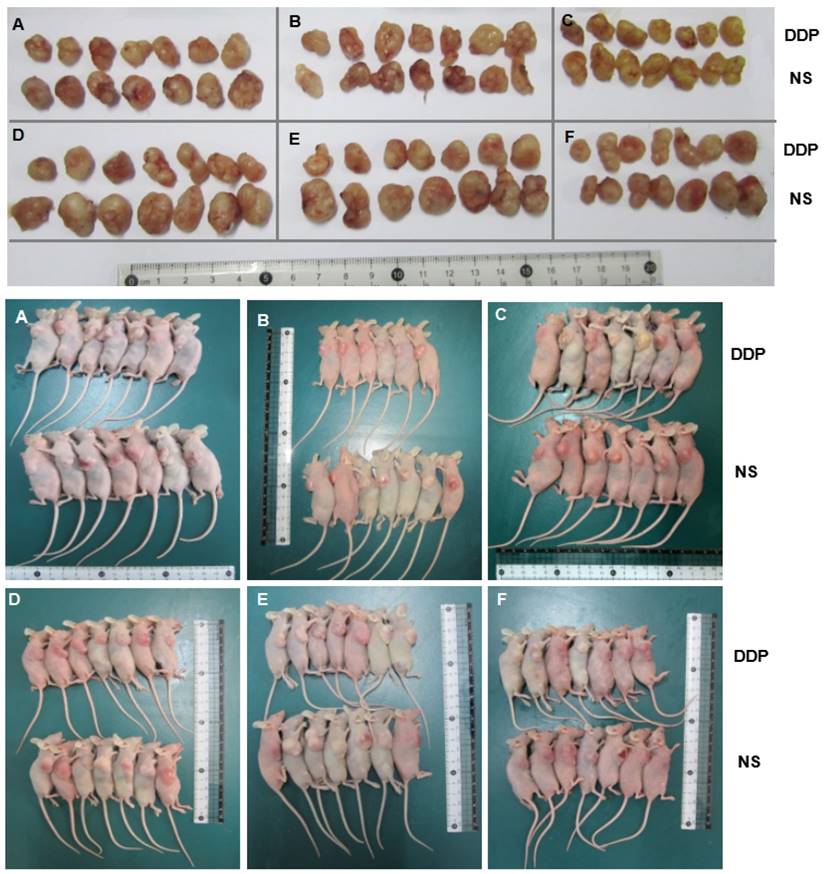
Photographs of xenografts of SGC7901 cells and SGC7901/DDP cells transduced with miR-192-5p with or without cotransduction with the vector alone into the flanks of BALB/C mice (n=7). Mice were euthanized and tumor harvested at 22 days after cell injection. DDP group mince treated with DDP by intraperitoneal injection at the dose of 4 mg/kg every three days. A: SGC7901/DDP group; B:NC-OE group; C:miR-192-5p-OE group; D:SGC7901 group; E: KD-NC group; F: miR-192-5p-KD group.
The growth curve of the gastric cancer cells subcutaneous transplantation tumors. Comparison of volume of xenografts of SGC7901 cells and SGC7901/DDP cells transduced with miR-192-5p lentivirus and NC. The data are represented as the mean±SEM (n=7) of xenografts. ★P < 0.05, ★★P < 0.01, ★★★P < 0.001, Student's t-test.
Comparison of weight of harvested xenografts of SGC7901 cells and SGC7901/DDP cells transduced with miRNA-192-5p with or without cotransduction with the vector alone. The data are represented as the mean±s.e.m. (n=7) of xenografts. ** P < 0.01, *** P < 0.001, Student's t-test.
Pathologic analysis of xenograft tumors with hematoxylin-eosin staining (×400) A:SGC7901/DDP xenograft tumors; B:SGC7901 xenograft tumors.
Cisplatin is a non-cyclical-dependent chemotherapy drug. It destroys cells by destroying DNA and is widely used to treat testicular, genital, head and neck cancers [19, 20]. However, due to a high drug resistance rate, the treatment effect can be robustly impaired. Therefore, it is urgent to analyze the mechanisms of cisplatin resistance in depth. Known mechanisms of cisplatin resistance include altered DNA repair, drug inactivation and altered cellular accumulation. The NER system is responsible for repairing cisplatin-induced DNA damage [21]. The upregulation of various NER proteins (XPA, XPB, XPC, CSB, XPD, XPF/ERCC1 and ERCC2) is accompanied by increased NER capacity, resulting in cytotoxic drug resistance, especially for cisplatin. The expression level of ERCC1 in ovarian cancer was first found to be higher than that in normal tissues, and its expression was related to cisplatin resistance [22]. ERCC1 expression could also be used to predict the response to cisplatin chemotherapy in testicular germ tumors and cervical carcinoma [23, 24]. Increased ERCC2 expression leads to cisplatin resistance in ovarian cancer [25]. MiRNAs play important roles in normal physiology, biological development, and disease by regulating various pathways and proteins. Currently, studies have shown that miRNAs regulate cisplatin resistance by regulating the expression levels of NER proteins. In ovarian cancer, miR-70-5p increases the sensitivity to cisplatin chemotherapy by regulating ERCC2 expression. Furthermore, in HepG2.2.15 cells, increased miR-192-5p levels promote cisplatin sensitivity of the tumor cells by downregulating ERCC3 and ERCC4 expression [15]. Biological prediction algorithms indicated that miR-192-5p target binding sites exist in the ERCC3 and ERCC4 mRNAs.
The expression of miR-192-5p regulated NER proteins in nude mice. Immunohistochemical method was used to detect the expression of ERCC3 and ERCC4 in xenograft tumors. A: The expression of ERCC3 B: The expression of ERCC4. a:SGC7901/DDP group b:NC-OE group c:miR-192-5p-OE group d:SGC7901 group e:NC-KD group f:miR-192-5p-KD group.
ERCC3, an ATP-dependent DNA helicase and part of the TFIIH transcription factor complex that unwinds DNA in the 3′-5′ direction, and ERCC4, which cleaves the damaged chains in the 5′ region, are the most versatile NER proteins involved in removing structure-distorting DNA damage. Our comet assay showed that the DNA repair ability of SGC7901/DDP cells increased with upregulation of ERCC3 and ERCC4 expression. Our study showed that altering the miR-192-5p level can induce abnormal expression of ERCC3 and ERC4 in vitro. Furthermore, our study showed that miR-192-5p overexpression combined with cisplatin significantly delayed tumor growth in vivo. Importantly, immunohistochemical analyses of these in vivo tumors showed that the ERCC3 and ERCC4 expression levels were decreased. We have confirmed the importance of the miR-192-5p-ERCC3/4 axis and found that miR-192-5p and ERCC3/4 are negatively correlated in tumor cells. Animal experiments also showed that increased miR-192-5p levels or decreased ERCC3/4 levels were related to cisplatin sensitivity. These data suggest that the relationship between miR-192-5p and ERCC3 and ERCC4 regulates the efficacy of cisplatin in GC.
It is worth noting that miR-192-5p overexpression can reverse the cisplatin resistance of GC cells in mice, thus identifying miR-192-5p as a potential therapeutic target. In addition, ERCC3 and ERCC4 are functional targets of miR-192-5p in GC cells and are extremely attractive therapeutic targets. Currently, lentiviruses are commonly used in gene therapy in vivo, but their safety requires further verification because random integration of viral DNA into the host genome can lead to toxic immune responses and dysfunctional gene expression. Therefore, it is necessary to further explore safer treatment strategies.
In conclusion, this study shows that miR-192-5p could contribute to the formation of cisplatin resistance in human SGC7901 cells by targeting the ERCC3 and ERCC4 NER proteins in vitro and in vivo. However, it should be noted that we only conducted our experiments only in SGC7901 cells and that we did not test all GC cell lines or models for clinical tumors. Therefore, more research is needed to elucidate the function of miR-192-5p in other cell lines and in clinical practice.
Materials and Methods
Cell culture
The human gastric adenocarcinoma cell lines SGC7901/DDP and SGC7901 were purchased from KeyGEN Biotechnology Company (Nanjing, Jiangsu, China). All cells were grown in RPMI-1640 medium supplemented with 10% fetal calf serum (Gibco, Grand Island, NY), 100 units/ml penicillin and 100 units/ml streptomycin in a humidified atmosphere with 5% CO2 at 37 °C. For maintenance of the cisplatin-resistance phenotype, SGC7901/DDP cells were incubated in a medium containing cisplatin (DDP, concentration of 800 ng/mL).
RNA isolation
Total RNA of the GC cells was obtained using TRIzol reagent (Invitrogen, California) according to the manufacturer's protocol. The total RNA concentrations were assessed using a NanoDrop-2000 spectrophotometer by measuring the absorbance at 260 nm. Three paired total RNA samples of SGC7901/DDP cells and SGC7901 cells were used for the microarray analysis, and the other RNA samples were used for the qRT-PCR assays.
MiRNA microarray analysis and bioinformatics prediction of miRNA target genes
MiRNA microarray expression profile analysis was performed using an Affymetrix miRNA 4.0 Array containing 775 mature human miRNA probe groups. The output data containing the normalized miRNA expression profiles were analyzed in Excel spreadsheets. The two groups of sample data were analyzed by t-tests to obtain p-values, and FDR was used to correct the p-values. FC values representing differentially expressed miRNAs were determined between SGC7901/DDP and their SGC7901 parental cells. Cluster3.0 software was used to display the differential expression modes of the miRNAs. Genes that may be targeted by the aberrantly expressed miRNAs were identified by using 10 available online tools (DIANAmT, miRanda, miRDB, miRWalk, RNAhybrid, PICTAR4, PICTAR5, PITA, RNA22 and TargetScan).
Comet assay
The comet assay under alkaline conditions was performed using our previously described methods with some modifications [26]. At least 50 images were randomly chosen from each sample and used with the comet assay analysis software to assess the DNA damage. The tail length (mean ± SD) was measured as an indicator of DNA damage [27, 28].
Lentivirus transfection
The lentiviral vector carrying miR-192-5p (pLV-miR-192-5p) and the negative control vector (pLV-miR-192-5p-NC) and the corresponding viruses (1 x 108 PFU) were constructed by GeneCopoeia (Guangzhou, Guangdong, China). Lentiviral transduction is carried out according to the manufacturer's protocol. Specifically, cells were seeded in a 12-well plate overnight at a density of 1 x 105 cells per well and then infected with lentivirus with a multiplicity of infection (MOI) of 10 pfu/cell. After 72 hours of transduction, SGC7901/DDP cells were cultured in a medium containing 2 µg/mL puromycin for 3 days. Similarly, to knock down miR-192-5p expression in SGC7901 cells, lentiviral vectors carrying RNAi sequences targeting miR-192-5p were used. After transduction, the cells were cultured in a medium containing 150 µg/mL hygromycin for 5 days. The surviving cell clones were used in the following experiments. Lentiviral vectors encoding GFP or RFP markers were used to monitor the transduction efficiency with a fluorescence microscope. In summary, we have developed miR-192-5p overexpressing (miR-192-5p-OE) cells and negative control (NC-OE) cells and produced miR-192-5p knock-down (miR-192-5p-KD) cells and negative control (NC-KD) cells from SGC7901 cells.
qRT-PCR analysis
All qRT-PCR reactions were performed using an Mx3000P system and SYBR Green qRT-PCR kit (Biomics, Nantong, China) according to the manufacturer's protocol. The qRT-PCR reaction was performed with reverse transcription for 30 minutes at 42°C, followed by 40 cycles of 20 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C, followed by incubation at 72°C to obtain the fluorescent signal. All miRNA levels were normalized to U6 expression. The relative changes were calculated using the 2-ΔΔCt [ΔΔCt=experimental (CtmiR-192-5p-Ctu6)-control (CtmiR-192-5p-Ctu6)] method. All experiments were performed in triplicate.
Western blot
Total cellular protein was extracted with RIPA lysis buffer containing protease inhibitor. Cell lysate samples containing 60 µg of total protein were electrophoresed on either a 10% or 8% sodium dodecyl sulfate-polyacrylamide gel and then transferred to PVDF membranes. The membranes were blotted with anti-ERCC3 (1:200 dilutions) and anti-ERCC4 (1:300 dilution) polyclonal antibodies (Abcam, Cambridge, MA) or with β-actin and GAPDH monoclonal antibodies (Abcam, USA) at a dilution of 1:1000, followed by incubation with goat anti-rabbit horseradish peroxidase secondary antibody (Univ-bio, Shanghai, China). The protein bands were visualized using an ECL-chemiluminescent kit (ECL-plus, Thermo Scientific). The total protein levels were normalized to GAPDH and β-actin, and the fold changes were calculated.
Assessing chemo-sensitivity to cisplatin
SGC7901/DDP, miR-192-5p-OE, NC-OE, miR-192-5p-KD, and NC-KD cells were suspended at a density of 1 × 105 cells/mL and plated into 96-well culture plates with 100 μL of RPMI-1640 medium per well. Because the human peak plasma concentration for DDP was 2.0 μg/mL, after cellular adhesion each well received freshly prepared DDP at final concentrations of 40 μg/mL, 20 μg/mL, 2 μg/mL, 0.2 μg/mL, and 0.02 μg/mL after 24 h. 48 h after DDP addition, we replaced the old medium with 180 μL of fresh medium and added 20 µl of MTT (5 mg/mL) to each well. The plate was incubated for 4 h in a humidified atmosphere before the medium was removed. Next, 150 μL of DMSO (Sigma, USA) was added to each well, and the wells were mixed for 10 minutes to dissolve the formazan crystals. The absorbance at 490 nm was measured using a spectrophotometer. The IC50 value of DDP was calculated according to the relative viability. Each experiment was performed in triplicate.
Tumor xenograft studies
The 4-week-old female BALB/c nude mice were purchased from Charles River (Beijing, China) and bred in an SPF laboratory. Logarithmic phase cells were harvested when they reached 80% confluence, and they were washed with phosphate-buffered saline (PBS) and resuspended in serum‑free medium at a concentration of 1 × 107 cells/mL. Mice were randomly divided into 6 groups. 0.2 mL of the SGC7901/DDP, miR-192-5p-OE, NC-OE, SGC7901, miR-192-5p-KD or NC-KD cell suspensions were subcutaneously (s.c.) inoculated to the flanks of the mice. The tumor volume was measured with calipers, and the tumor size was calculated using the formula a × b2/2, where a represents width and b represents length. When the tumor volume was 50-100 mm3, the xenografted tumor-bearing mice were randomly divided into experimental (DDP) and control (NS) groups. The experimental group was subjected to cisplatin (5 mg/kg) therapy and the control group was perfused with an equal volume of 0.9% NS 5 times every 3 days. The tumor volume was measured daily. All of the mice were sacrificed 25 days after cell inoculation. The exfoliated tumors were weighed and examined by pathology and immunohistochemistry. All of the mouse experimental procedures were approved by the Animal Care Committee of Hefei Institute of Physical Science, Chinese Academy of Sciences.
HE staining and immunohistochemistry analysis
Xenografted tumors were fixed by Faure Marin and paraffin-embedded for the HE staining and immunohistochemistry analysis. HE staining was carried out according to the standard procedure. The ERCC3 and ERCC4 expression levels were analyzed by immunohistochemistry using the EliVisionTM super kit (Maixin, Fuzhou, Fujian, China) according to the company's protocol. The researchers evaluated ERCC3 and ERCC4 positivity without prior knowledge of the pathological data. The ERCC3 and ERCC4 expression levels were scored based on semiquantitative scoring criteria (Table 6). The total scores of the comprehensive density and intensity analysis were reported as follows: 0, negative; 1-3, ± ; 4-5, + ; 6-7, ++. +, ++ were used to determine ERCC3 and ERCC4 positivity.
The semiquantitative scoring criteria
| Score | Intensity of staining | Proportion of stained cells (%) |
|---|---|---|
| 0 | Negative (no staining) | < 5 |
| 1 | Weak (canary yellow staining) | 5-25 |
| 2 | Moderate (pale brown staining) | 25-50 |
| 3 | Strong (tan staining) | 50-75 |
| 4 | -- | > 75 |
Statistical analysis
Statistical analysis was performed with GraphPad Prism software (La Jolla, CA). The data are expressed as mean ± sd and differences were evaluated by one-way ANOVA, Student's t-test, Chi-square test and Fisher's exact test; p < 0.05 was considered statistically significant.
MiR-192-5p screening process and methods for validating it's role.
Acknowledgements
This study was supported by Projects of Foreign Science and Technology Cooperation of Anhui Province (No. 1604b0602027).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87-108
2. Wagner AD, Unverzagt S, Grothe W. et al. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010(3):CD004064
3. Ychou M, Boige V, Pignon JP. et al. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: an FNCLCC and FFCD multicenter phase III trial. J Clin Oncol. 2011;29(13):1715-21
4. Li Q, Yu JJ, Mu C. et al. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res. 2000;20(2a):645-52
5. Van Allen EM, Mouw KW, Kim P. et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4(10):1140-53
6. Arora S, Rana R, Chhabra A. et al. miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288(3-4):77-87
7. Berezikov E, Guryev V, van de Belt J. et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21-4
8. Shukla GC, Singh JBarik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3(3):83-92
9. Drayton RM, Dudziec E, Peter S. et al. Reduced expression of miRNA-27a modulates cisplatin resistance in bladder cancer by targeting the cystine/glutamate exchanger SLC7A11. Clin Cancer Res. 2014;20(7):1990-2000
10. Pouliot LM, Chen YC, Bai J. et al. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012;72(22):5945-55
11. van Jaarsveld MT, Helleman J, Boersma AW. et al. miR-141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32(36):4284-93
12. Zhu X, Shen H, Yin X. et al. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. 2016;35(3):323-32
13. Hioki M, Gotohda N, Konishi M. et al. Predictive factors improving survival after gastrectomy in gastric cancer patients with peritoneal carcinomatosis. World J Surg. 2010;34(3):555-62
14. Turchi JJ, Woods DSVanderVere-Carozza P. Testing the metal of ERCC2 in predicting the response to platinum-based therapy. Cancer Discov. 2014;4(10):1118-9
15. Xie QH, He XX, Chang Y. et al. MiR-192 inhibits nucleotide excision repair by targeting ERCC3 and ERCC4 in HepG2.2.15 cells. Biochem Biophys Res Commun. 2011;410(3):440-5
16. Freitas RCC, Bortolin RH, Lopes MB. et al. Integrated analysis of miRNA and mRNA gene expression microarrays: Influence on platelet reactivity, clopidogrel response and drug-induced toxicity. Gene. 2016;593(1):172-178
17. Pimentel F, Bonilla P, Ravishankar YG. et al. Technology in MicroRNA Profiling: Circulating MicroRNAs as Noninvasive Cancer Biomarkers in Breast Cancer. J Lab Autom. 2015;20(5):574-88
18. Wang YN, Chen ZHChen WC. Novel circulating microRNAs expression profile in colon cancer: a pilot study. Eur J Med Res. 2017;22(1):51
19. Dasari STchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364-78
20. Li J, Chen R, Ji M. et al. Cisplatin-based chronotherapy for advanced non-small cell lung cancer patients: a randomized controlled study and its pharmacokinetics analysis. Cancer Chemother Pharmacol. 2015;76(3):651-5
21. Dabholkar M, Thornton K, Vionnet J. et al. Increased mRNA levels of xeroderma pigmentosum complementation group B (XPB) and Cockayne's syndrome complementation group B (CSB) without increased mRNA levels of multidrug-resistance gene (MDR1) or metallothionein-II (MT-II) in platinum-resistant human ovarian cancer tissues. Biochem Pharmacol. 2000;60(11):1611-9
22. Dabholkar M, Bostick-Bruton F, Weber C. et al. ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J Natl Cancer Inst. 1992;84(19):1512-7
23. Mendoza J, Martinez J, Hernandez C. et al. Association between ERCC1 and XPA expression and polymorphisms and the response to cisplatin in testicular germ cell tumours. Br J Cancer. 2013;109(1):68-75
24. Sun C, Li N, Yang Z. et al. miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J Natl Cancer Inst. 2013;105(22):1750-8
25. Zhao H, Yu X, Ding Y. et al. MiR-770-5p inhibits cisplatin chemoresistance in human ovarian cancer by targeting ERCC2. Oncotarget. 2016;7:53254-68
26. Wu JC, Hseu YC, Tsai JS. et al. Fenthion and terbufos induce DNA damage, the expression of tumor-related genes, and apoptosis in HEPG2 cells. Environ Mol Mutagen. 2011;52(7):529-37
27. Huan LC, Wu JC, Chiou BH. et al. MicroRNA regulation of DNA repair gene expression in 4-aminobiphenyl-treated HepG2 cells. Toxicology. 2014;322:69-77
28. Wu Q, Beland FA, Chang CW. et al. XPC is essential for nucleotide excision repair of zidovudine-induced DNA damage in human hepatoma cells. Toxicol Appl Pharmacol. 2011;251(2):155-62
Author contact
![]() Corresponding author: Kangsheng Gu, PhD, Professor of Medicine, Department of Oncology, the First Affiliated Hospital of Anhui Medical University, 218 Jixi Road, Hefei, Anhui 230032, China. Tel/fax: +8613805692145; E-mail: 13805692145com
Corresponding author: Kangsheng Gu, PhD, Professor of Medicine, Department of Oncology, the First Affiliated Hospital of Anhui Medical University, 218 Jixi Road, Hefei, Anhui 230032, China. Tel/fax: +8613805692145; E-mail: 13805692145com

 Global reach, higher impact
Global reach, higher impact